Abstract
p53 is regulated at the multiple levels. We report here that p53 in multiple lines of human cancer cells is down-regulated by cardiac glycoside drugs, digoxin or ouabain, the potent inhibitors of Na+/K+-ATPase. These drugs reduced the basal levels of p53 protein at nanomolar concentrations in a dose-, time- and cancer cell line-dependent manner, but independent of p53 status of wild type (wt) or mutant. The drugs also reduced the levels of p53 induced by its activators as well as p53 transfected into human cancer cells, regardless of its status. Interestingly, the drugs had no effect on endogenous p53 in two immortalized human cell lines. Mechanistically, p53 reduction did not occur at the mRNA levels, but at the protein levels, as a result of reduced protein synthesis rather than enhanced degradation. The cellular sensitivity to drug-induced p53 reduction was not associated with the levels of α subunits of Na+/K+-ATPase in different cell lines. While lowering extracellular K+ did not reduce p53 as did ouabain and digoxin, it did potentiate both digoxin and ouabain-induced p53 reduction in sensitive lines. Finally, p53 reduction appears to be triggered by activation of Src/MAPK signaling pathways upon drug binding to the Na+/K+-ATPase and can be completely blocked by the inhibitors of Src or MEK. This is the first report that cardiac glycoside drugs, by initiating the Src/MAPK signaling pathways, reduce the p53 levels via inhibition of p53 protein synthesis. The drugs may be useful in the treatment of human cancers with a gain-of-function p53 mutation.
Keywords: p53, Src/MAPK signaling pathways, cardiac glycosides, Na+/K+ ATPase
Introduction
p53 prevents tumor formation through transcriptional dependent and independent mechanisms. Transcriptional dependent mechanism is mainly mediated by p53 upregulation of its downstream targets. Upon activation by a variety of stimuli, p53 induces the expression of pro-arrest genes, such as p21, Gadd45 or 14-3-3σ to induce growth arrest or of pro-apoptotic genes, such as PUMA, PIG-3 or DR5 to induce apoptosis (1). Through a direct binding to mitochondria and modulating BH3 family pro-apoptotic proteins, such as Bax, p53 can also regulate apoptosis in a transcriptional independent manner (2, 3). Thus, p53 acts as a guardian of the genome by inducing growth arrest to allow cells to repair the damage or apoptosis, if the damage is too severe and irreparable.
Since p53 plays a pivotal role in controlling abnormal cell growth and is inactivated by point mutations in more than 50% human cancers, p53 has been a central target for mechanism-driven cancer drug discovery (4, 5). Significant progress has been made in past decade, leading to identification and characterization of several unique classes of small molecules that modulate p53 (6, 7). They can be categorized as follows: 1) The molecules that restore wild type p53 from a mutant conformation, which include CP-31398 (8), PRIMA-1 (9) and ellipticine (10). 2) The molecules that target Mdm2 to reactivate p53, including Mdm2 E3 ubiquitin ligase inhibitor, HLI98 (11) and inhibitors that disrupt Mdm2-p53 binding, such as Nutlin (12), RITA (13), and MI-219 and its analogues (14–16). 3) The molecules that inhibits wild type p53, including pifithrin-α (17) and pifithrin-mu (18). And 4) the molecules that selectively degrade mutant p53, including Hsp90-active agents such as geldanamycin (19) and histone deacetylase inhibitors, such as trichostatin (20).
During the screening for small molecules that selectively kill mutant-p53 containing cancer cells via a synthetic lethal mechanism (21), we serendipitously found that cardiac glycoside drugs, digoxin and ouabain, reduced the p53 levels in a time and dose dependent manner in sensitive cancer cell lines. The drug sensitivity to p53 reduction is cancer cell line dependent, but independent of p53 status of a wild type or mutants. Importantly, the drugs are completely inactive in reducing wt p53 in normal “immortalized” cells. Mechanistically, the drug-induced p53 decrease occurred not at the mRNA levels, but at the protein levels, as a result of reduced synthesis, rather than enhanced degradation. The drug induced p53 reduction can be rescued by the inhibitors of Src and MEK, suggesting an involvement of Src/MAPK signaling pathways, initiated upon the drug binding to Na+/K+-ATPase. Our study revealed a novel mechanism by which activation of Src/MAPK kinase pathway could eventually lead to p53 elimination by inhibiting p53 protein synthesis.
Materials and Methods
Cell culture and drug treatment
All cell lines used in this study, except those mentioned below, were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum. H1355, HCT116 and MRC5 were maintained in RPMI 1640 Medium and McCoy’s 5A medium, respectively, containing 10% serum. NL20 cells were cultured in Ham's F12 medium as described (22). For drug treatment, subconfluent cells were treated with DG or OU alone or in combination of PP2, PD098059, or LY294002 (Sigma).
For culturing cells in low K+ conditions, A549 or H1355 cells were cultured in 10% DMEM until the cell densities reached 80%. The medium was replaced by K+-free DMEM supplemented with normal K+ concentration (5 mM) or low K+ concentrations (1 mM or 0.3 mM). Na+ was added to the low K+ medium to maintain the equal ion concentrations (23).
Western blotting analysis
The assay was performed as described previously (16). The antibodies used are p53 (Calbiochem, CA, 1:1000), p21 (BD, 1:1000), Mdm2 (Calbiochem, CA, 1:500), FAK, Src or tubulin (Santa Cruz Biotechnology, 1:1000), Src-pY418 (Invitrogen, CA, 1:1000); pERK1/2 (Cell Signaling) and β-actin (Sigma, St. Louis, MO 1:5000).
Transfection and infection
H1299 cells were plated into six-well plate at 2 ×105 cells per well and transfected the following day with 1 µg of plasmid expressing either p53-wt or p53-mutants for 24 h prior to drug treatment. For the infection of sh-RNA targeting FAK viruses (gift from Junlin-Guan), a total of 109 pfu viruses were infected into 2 ×106 A549 or H1355 cells per 100 mm dish for 72h. Cells were split, followed by exposure to drugs next day for 24 hrs. To silence endogenous wt p53, A549 or H460 cells were infected with a lenti-virus based siRNA construct, LT-p53-siRNA, as described (16).
ATPlite growth assay and IC50 determination
Cells were seeded in 96 well white plates and treated with the drugs with a range of indicated concentrations for 24 hrs. The viability of the cells was then measured using a one-step ATPlite kit (Perkin Elmer). The results were calculated and plotted in Prism 4.0 (Graphpad) to generate IC50 curves (22).
Quantitative RT-PCR
Total RNA was isolated from cells post drug treatment, using a Trizol kit (Invitrogen, Carlsbad, CA), and subjected to quantitative RT-PCR analysis, using QuantiTect SYBR green RT-PCR kit (Qiagen). Briefly, 50 µl reaction mixture was used for each reaction, which contained 2x QuantiTect SYBR Green RT-PCR Master Mix, 0.5 µl QuantiTect RT mix, 0.5 µM primer mix and 0.5 µg RNA. Cycling program was set as the following: 50 °C 30 min for RT, 95 °C 15 min for the PCR initial activation and 40 cycles of denaturation at 94 °C for 15 sec, annealing at 55 °C for 30 sec and extension at 72 °C for 30 sec. The sequences of p53 and GAPDH are as follows: hu-p53 F1□TCTGTGACTTGCACGTAC; hu-p53 R1: ATTTCCTTCCACTCGGAT. GAPDH-F1: GTTGCCATCAATGACCCCTT; GAPDH -R1: AGAGGCAGGGATGATGTTCT.
35S-Met metabolic labeling
Subconfluent cells were treated with the drugs for various time points with last hr in methionine and cysteine-free DMEM, containing 5% dialyzed FCS and 50 µM MG132. Cells were then labeled with 100 µCi/ml of [35S]-methionine (MP Biochemicals) for 5 min, followed by immunoprecipitation with anti-p53 antibody (Santa Cruz). Immunoprecipitates, along with whole cell extract, were then subjected to SDS-PAGE and autoradiography. The steady-state levels of p53 were measured by immunoprecipitated, followed by Western blotting, along with the detection of total cellular proteins by Coomassie staining.
Results
Cardiac glycosides reduced the basal levels of p53 in lung cancer cell lines
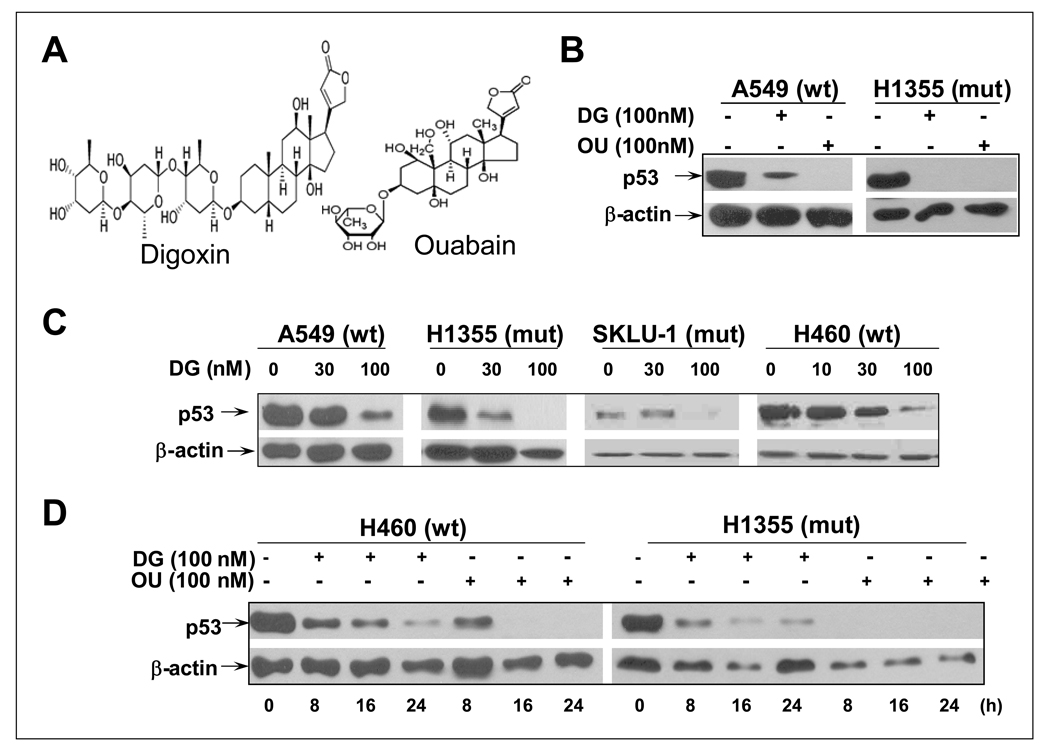
During our confirmation of candidates identified from a chemical library screen for selective killing of cancer cells with mutant p53 via synthetic lethal mechanism (21), we serendipitously found that cardiac glycosides, including digoxin (DG) and ouabain (OU) (Figure 1A for structure) are able to reduce the p53 levels. As shown in Figure 1B, DG or OU at the nanomolar concentrations reduced or eliminated the basal levels of p53 in two lung cancer lines, A549 with a wild type (wt) p53 and H1355 with a mutant p53, with OU being more potent. DG induced p53 reduction or elimination was the dose-dependent, but the p53 status independent in four lung cancer cell lines with either wt or mutant p53 status (Figure 1C). Furthermore, p53 reduction is treatment-time dependent, starting to occur at 8 hrs with a complete elimination seen by OU at 16-hrs post drug exposure (Figure 1D). Finally, cell line dependent, but p53 status-independent p53 reduction or elimination by DG or OU can be extended to multiple human cancer cell lines. These include colon cancer lines, DLD1, but not HCT116, nor HT29; breast cancer lines, MCF7, but not MDA-MB231; all three head and neck squamous carcinoma lines, but not three glioblastoma lines tested (Supplemental materials, Figure S1). Digoxigenin, another cardiac glycoside was also active, but 10-fold less potent in reducing p53 levels in multiple human cancer lines (data not shown). Our results clearly demonstrated that p53 levels in multiple human cancer cell lines are subjected to reduction or elimination by cardiac glycosides, DG or OU in a cell line-dependent, but p53 status-independent manner.
Figure 1. Cardiac glycosides, digoxin (DG) and ouabain (OU) reduces the basal p53 levels in lung cancer cell lines: Dose- and time-dependent: (A).
Structure of DG and OU. (B) DG or OU reduces p53 levels: A549 and H1355 lung cancer cells were treated with DG or OU at 100 nM for 24 hrs and subjected to western blotting. (C) Does dependent reduction of p53 levels by DG: Four lung cancer lines were treated with DG at 30 or 100 nM for 24 hrs and subjected to western blotting. (D) Time dependent reduction of p53 levels by DG or OU: H460 and H1355 cells were treated with DG or OU at 100 nM for indicated periods of time, followed by western blotting.
Cardiac glycosides blocked p53 induction and p53 upregulation of its target genes by p53 activators
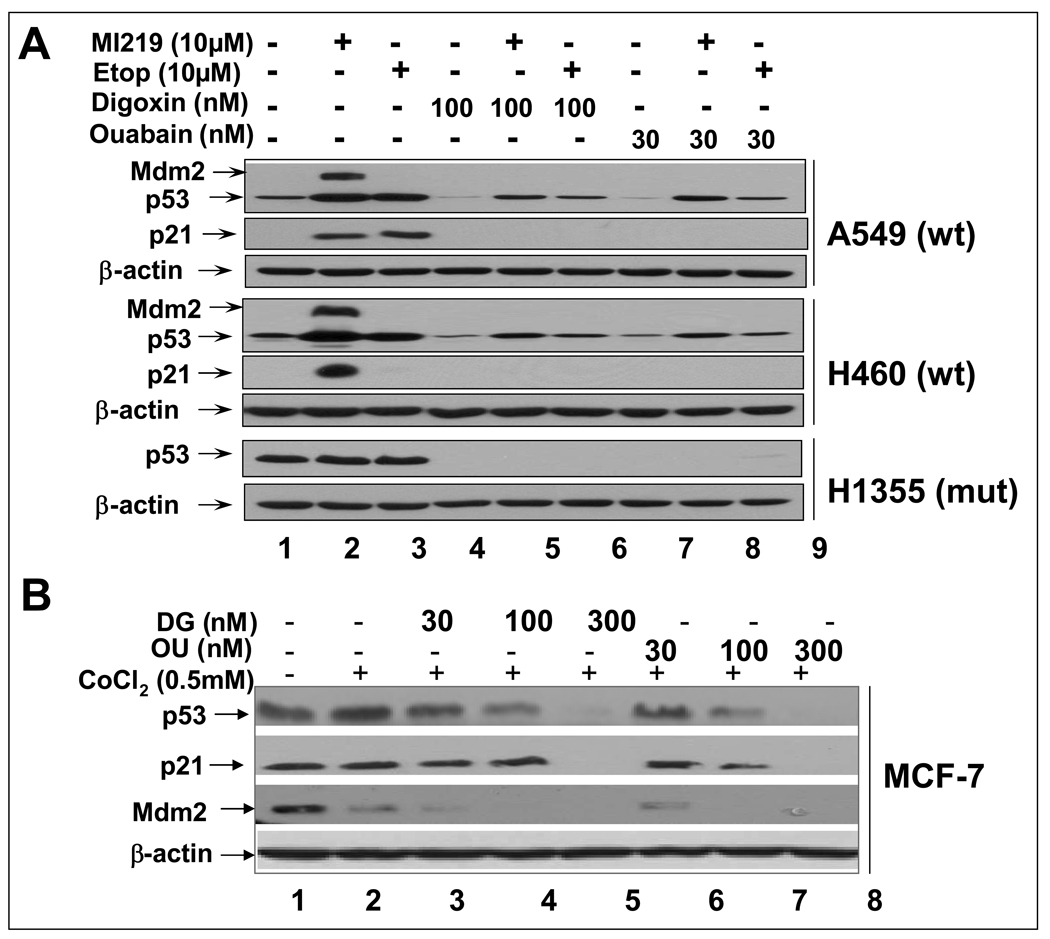
We next determined if DG or OU also reduced the levels of induced p53 and p53 transactivation of its target genes by known p53 activators. Three lung cancer cell lines were treated for 24 hr with MI-219, an Mdm2 inhibitor, which activates p53 by disrupting Mdm2-p53 binding (14) or etoposide, a DNA damaging agent, known to activate p53 in wt p53-containing lung cancer cells (16), alone or in combination with DG or OU, respectively. As shown in Figure 2A, MI-219 or etoposide treatment induced p53 levels as well as p53 target proteins, p21 or Mdm2 (by MI-219 only) in two wt p53-containing lines, A549 and H460, but not in mutant p53-containing line, H1355 (lanes 1–3). Combinational treatment with either DG or OU reduced the induced levels of p53 and completely eliminated p53 induction of its targets, p21 and Mdm2 in A549 and H460 cells (lanes 4–9). The mutant p53 in H1355 was not subjected to induction by two p53 activators, but was completely eliminated in combinational treatment (bottom panel).
Figure 2. DG or OU reduces the levels of induced or stabilized p53 and blocks p53 transactivation of its targets.
(A) Reduction of induced p53: Lung cancer cells with either wt p53 or mutant p53 were treated with p53 activator, MI219 or etoposide (Etop) alone or in combination of DG or OU. (B) Reduction of stabilized p53: MCF7 cells were treated with CoCl2 (0.5 mM) alone or in combination with DG or OU for 24 hrs. Cell lysates were prepared for western blotting.
p53 can be stabilized by hypoxia in wt p53 containing MCF7 cells by chemical hypoxic agents, cobalt chloride and desferrioxamine (24). We next determined if hypoxia-stabilized p53 is also subjected to reduction by DG or OU. As shown in Figure 2B, A 24 hr treatment of cobalt chloride stabilized p53 up to 2-fold in MCF7 cells. This stabilized p53 was reduced and eliminated by simultaneous treatment of DG or OU in a dose-dependent manner up to 300 nM (lanes 3–8). Along with p53 reduction, two p53 target proteins, p21 and Mdm2, which were detectable at the basal levels due to a relatively higher level of p53, were also reduced and eliminated. Taken together, these results indicate that both cardiac glycosides also reduced the levels of p53 and its target proteins upon p53 activation and stabilization. The results further suggest that drug-induced p53 reduction may be Mdm2-independent, since Mdm2 is free from p53 binding upon MI-219 treatment (14).
Cardiac glycosides reduced the levels of transfected p53 in cancer cells, but had no effect on endogenous wt p53 in normal cells
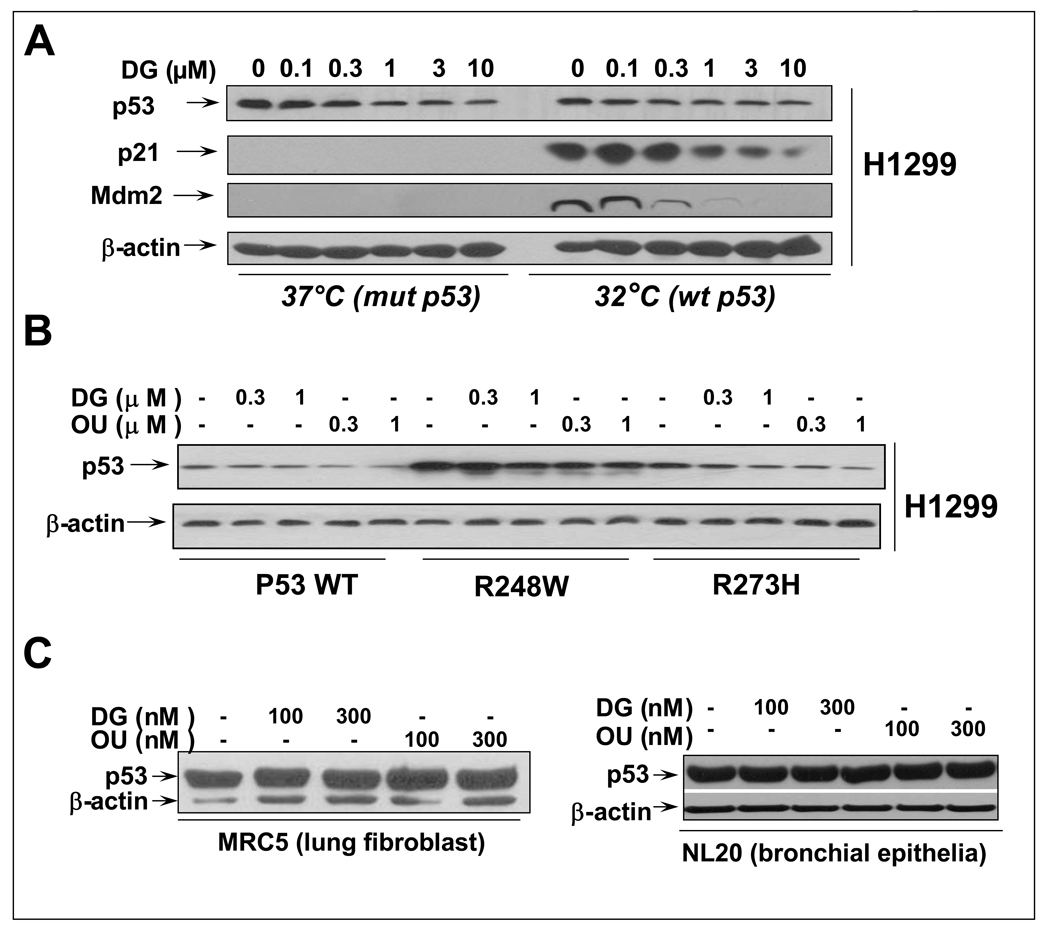
We next determined if p53 upon over-expression in a p53-null H1299 cells would also subjected to DG or OU reduction. H1299 cells over-expressing a temperature sensitive p53 mutant (codon 138) (25) were treated with DG for 24 hrs at 37°C (p53 adopts a mutant conformation) or 32°C (wt conformation). As shown in Figure 3A, DG treatment caused a dose-dependent reduction of over-expressed p53, regardless of p53 status. Upon p53 reduction, p53 target genes, p21 and Mdm2, induced at 32°C when p53 is in a wild type status, were also reduced accordingly. Likewise, DG reduced the levels of p53 transiently transfected into H1299 cells regardless of the p53 status of wild type or two mutants, R248W and R273H, most frequently found in human cancer (Figure 3B). However, the potency of drugs against over-expressed p53 was much reduced with much high drug doses up to 10 µM to achieve a moderate effect. Interestingly, DG or OU failed to reduce the endogenous wt p53 highly expressed in two immortalized lung derived cell lines, MRC5 lung fibroblast and NL20 bronchial epithelia (Figure 3C). These results indicated a tumor cell line selective and p53 status-independent reduction of p53 by DG or OU.
Figure 3. DG or OU selectively reduces the p53 levels in cancer cells, but not in immortalized normal cells: (A) p53 status-independent reduction of stably over-expressed p53.
A stable clone of H1299 lung cancer cells (endogenous p53-null), over-expressing a temperature sensitive p53 mutant (H1299-p53ts-A138V), was treated with various concentrations of DG, as indicated while grown at 37°C (adapting a mutant p53 conformation) or 32°C (adapting a wt p53 conformation). The levels of p53 and its two target proteins, p21 and Mdm2 were measured by western blotting. (B) p53 status-independent reduction of transiently over-expressed p53: p53-null H1299 cells were transiently transfected with wt p53 or two p53 mutants, most frequently found in human cancer (p53-R248W and p53-R273H). Cells were subsequently treated with the drugs for 24 hrs, followed by western blotting. (C) Lack of p53 reduction by DG or OU in two immortalized lung-derived cells: Lung fibroblast, MRC5 and bronchial epithelial NL20 cells were treated with DG or OU for 24 hrs, followed by western blotting.
Dissociation between drug cytotoxicity and drug-induced p53 reduction
To exclude the possibility that p53 reduction is the consequence of drug-induced cytotoxicity, we measured the IC50 values of a panel of cancer cell lines, either sensitive or resistant to drug-induced p53 reduction. As shown in Figure S2, DG or OU was quite potent growth inhibitor with an IC50 ranging from 50 to 100 nM in the majority of lines tested. However, the drug cytotoxicity appears not to be associated with cellular sensitivity to drug-induced p53 reduction. For examples, two sensitive lines, H1355 and DLD1 had similar IC50 values to two resistant lines, HCT116 and HT29, whereas MCF7 cells are rather resistant to DG cytotoxicity (Figure S2A), but sensitive to DG-induced p53 reduction (Figure 1S). The morphological appearances of A549 and H1355 after drug treatment for different periods of time were shown in Figure 3S, which demonstrated a moderate cytotoxicity only at 24 hr, particularly in H1355 cells. Furthermore, since two wt p53-containing lines, A549 and H460 appeared to be the most sensitivity to drug cytotoxicity with an IC50 of 20–40 nM, we determined if the cytotoxicity is p53 dependent. As shown in Figure 2S B–D, siRNA silencing of p53 did not change drug-cytotoxicity, indicating that cytotoxicity is not mediated through, nor associated with wild type p53.
Cardiac glycosides induced p53 reduction does not occur at the mRNA levels, but at the protein levels
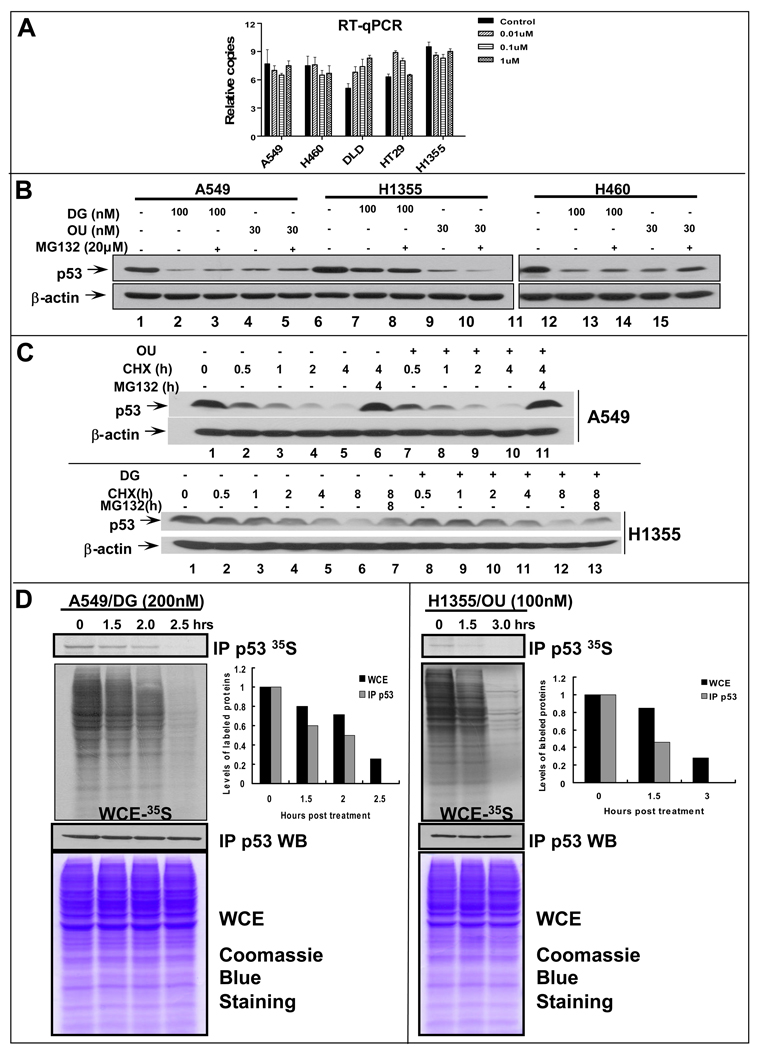
p53 regulation by DG or OU could occur at the levels of transcription, translation or post-translation, although post-translational regulation is the most common mechanism for p53 regulation. We first examined if p53 reduction occurred at the mRNA levels. Five cell lines with four sensitive (A549, H460, DLD-1, and H1355) and one resistant (HT29) were treated with DG at various doses up to 1 µM and subjected to RT-qPCR analysis. As shown in Figure 4A, the mRNA levels were rather consistent with all treatments among all tested lines, regardless of their drug sensitivity or p53 status. Thus, p53 reduction does not occur at the mRNA levels.
Figure 4. Reduction of p53 by DG or OU does not occur at the mRNA levels, but at the protein levels: (A) Lack of p53 mRNA changes upon DG treatment.
Five cancer cell lines were treated with DG for 24 hrs, followed by RNA isolation and qRT-PCR analysis. The relative p53 mRNA levels were plotted. (B). Proteasome inhibitor failed to rescue p53 reduction: Cells were treated with DG or OU for 24 hrs, alone or in combination with MG132 (added in last 6 hrs), followed by western blotting. (C) DG or OU did not shorten p53 protein half-life: Cells were treated with cycloheximide (CHX) to block new protein synthesis in the absence or presence of DG (100 nM) or OU (30 nM). Cells were harvested at various time points and subjected to western blot analysis. (D). DG or OU inhibited de novo p53 synthesis: Cells were treated with DG or OU for indicated periods of time with MG132 addition at the last hour. Cells were pulse-labeled with 35S-methionine for 5 min, followed by p53 immunoprecipitation and autoradiography (top panel). Total 35S-methionine incorporation into cellular proteins was assessed by autoradiography (second panel). The steady-state levels of p53 was measured by p53 immunoprecipitation, followed by Western blotting (third panel), whereas the total amounts of cellular proteins were measured by Coomassie blue staining (bottom panel).
We next determined if DG or OU-induced p53 reduction was due to enhanced degradation. Three sensitive lung cancer lines were treated with DG or OU, alone or in combination with proteasome inhibitor, MG132 in last 6 hrs prior to cell harvesting. As shown in Figure 4B, MG132 treatment only slightly, if any, blocked DG- or OU-induced reduction of p53 levels in A549 and H460 cells (lanes 3 vs. 2; 5 vs. 4; and 13 vs. 12; 15 vs. 14), but had no effect on H1355 cells (lanes 7–10). Similar results were obtained when PS341, a potent proteasome inhibitor, was used (data not shown). A minor blockade of p53 reduction by MG132 was also observed in sensitive DLD-1 and MCF-7 cells (data not shown).
We then measured the effect of DG or OU on p53 protein half-life using A549 or H1355 cells. Cells were treated with cycloheximide (CHX) to block new protein synthesis in the absence or presence of DG or OU and harvested at various time points for p53 level measurement by western blotting. As shown in Figure 4C, wt p53 has a protein half-life of ∼0.5–1 hr in A549 cells (top panel), whereas mutant p53 has a half-life of 2–4 hrs in H1355 cells (bottom panel). In both cases, however, DG or OU treatment did not change the p53 half-life. As expected, p53 degradation can be blocked by MG132, independent of drug treatment (lanes 6 & 11 vs. 5 and 10; 7&13 vs. 6 and 12). Taken together, these results indicate that DG or OU-induced p53 reduction is not due to enhanced degradation.
We finally determined if DG or OU could inhibit p53 protein synthesis. Two drug sensitive lines A549 and H1355 were used and newly synthesized p53 was measured by 35S-methionine labeling in the presence of MG132 to block p53 degradation. As shown in Figure 4D, in both lines DG or OU treatment induced a time-dependent inhibition of de novo p53 protein synthesis with a complete elimination of p53 synthesis at 2.5 or 3 hrs post treatment, respectively (top panel). The drugs also caused a time dependent inhibition of overall de novo protein synthesis but to less extent (second panel with quantification data shown on the right). In contrast, the same treatment did not change the steady-state levels of p53 (third panel), nor the total cellular proteins (bottom panel). The drug also inhibited de novo synthesis of p53 in an additional sensitive line, MCF7 (data not shown). Thus, DG or OU-induced p53 reduction is not due to enhanced degradation, rather due to inhibited protein synthesis.
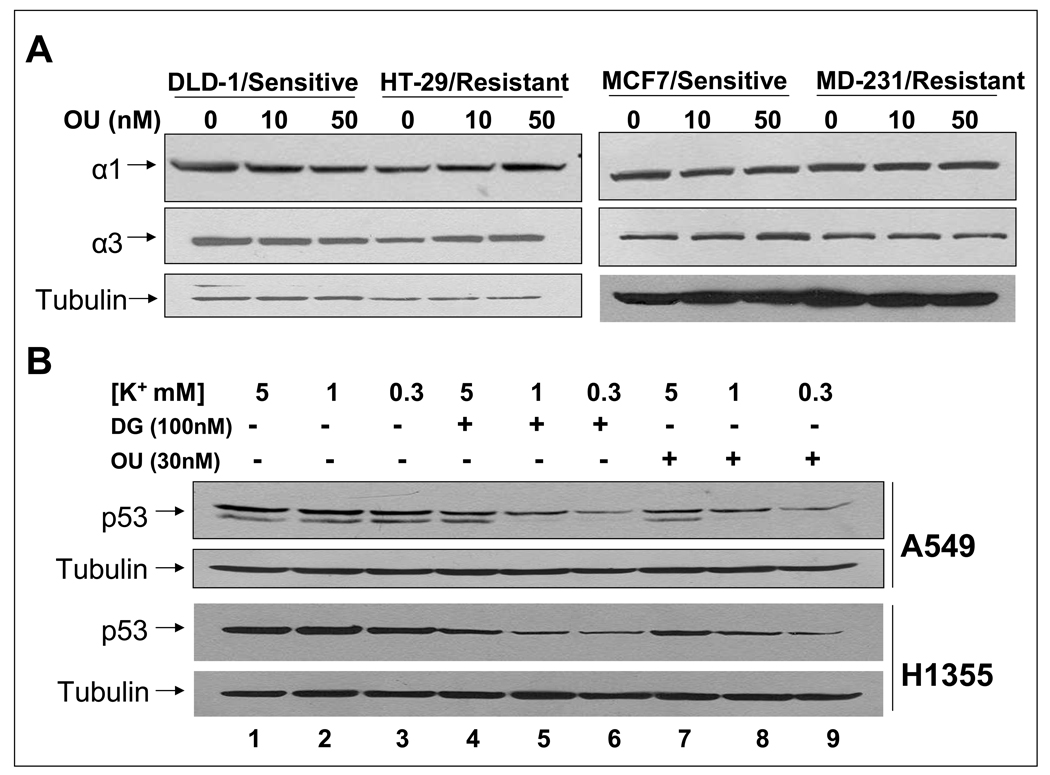
Cardiac glycosides-induced p53 reduction is independent of the levels of α subunits of Na+/K+-ATPase, but is enhanced by lowering extracellular K+
The cardiac glycosides target for treatment of congestive heart failure is the Na+/K+-ATPase (26). We determine if the levels of Na+/K+-ATPase α subunits are associated with cellular sensitivity to OU-induced p53 reduction. As depicted in Figure 5A, Western blot analyses reveal that both OU-sensitive and resistant cells express α1 and α3. Apparently, the cellular sensitivity of OU-induced p53 reduction did not correlate with the basal levels of Na+/K+-ATPase in these cells. To test if inhibition of Na+/K+-ATPase by means other than cardiac glycosides is sufficient to reduce p53, we lowered extracellular K+ from 5 mM to 1 and 0.3 mM (27), and measured for p53. As shown in Figure 5B, reduction of potassium concentration had no effect on the basal levels of p53 (lanes 1–3). Interestingly, DG- or OU-induced p53 reduction was more pronounced progressively, consistent with the fact that lowering extracellular K+ increases DG- and OU-binding to the Na+/K+-ATPase. These results clearly showed that DG- or OU-induced p53 reduction is promoted by the binding of these drugs to the Na+/K+-ATPase.
Figure 5. Effect of Na+/K+-ATPase on p53 reduction by DG or OU: (A). The levels of α subunits of the pump did not correlate with cellular sensitivity to p53 reduction.
Two sensitive and two resistant colon and breast cancer cell lines were treated with OU and the levels of α1 and α3 subunits were measured with western blotting. pSrc/pERK: phosphorylated form of Src/Erk; tSrc/tERK (total Src/ERK). (B). Lowering the pump activity enhanced p53 reduction: Cells were cultured in regular medium containing 5 mM potassium or special media with potassium concentration reduced to 1 or 0.3 mM. Cells were left untreated or treated with DG or OU for 24 hrs, followed by western blotting. Note that two bands (p53 and p47) were visualized in A549 cells after prolonged electrophoresis, but both are subjected to reduction by drugs.
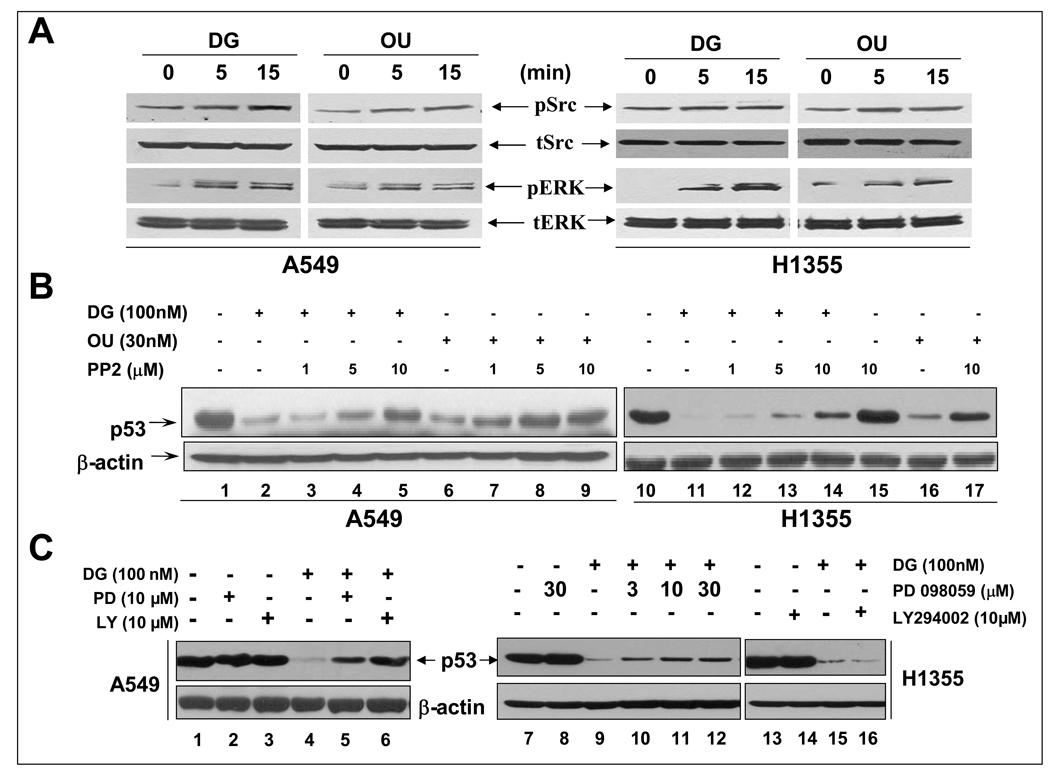
Cardiac glycosides reduced p53 levels via triggering and activating Src/MAPK signaling pathways
It has been proposed that the Na+/K+-ATPase is preassembled with its partners in caveolae; the binding of OU or DG to the pump activates the signalosome to transduce the signals via multiple pathways, including Src, FAK, MAPK and PI-3K (28–30). We first determined if DG or OU treatment would activate Src or MAPK in sensitive A549 and H1355 cells. As shown in Figure 6A, a short treatment of cells with DG or OU for 5 or 15 min caused Src and MAPK activation, as demonstrated by increased phosphorylation at the activation sites (pSrc-Y418 and pERKs-T183/Y185). We then determined the effect of Src and FAK inhibition on drug-induced p53 reduction, since Src and FAK are two upstream molecules activated upon OU- or DG-pump binding (29). In both A549 and H1355 cells, a potent Src tyrosine kinase inhibitor PP2 (31) blocked the p53 reduction by DG or OU in a dose dependent manner (Figure 6B, lanes 3–5 vs. 1; 7–9 vs. 6 and 12–15 vs. 10; 17 vs. 16, respectively), while the inhibitor itself had no effect on the p53 level (lane 15 and data not shown). On the other hand, siRNA silencing of FAK in either A549 or H1355 cells had no effect on p53 reduction by DG or OU (Supplemental Figure S4). We, therefore, focused our attention on Src downstream pathways, particularly MAPK and PI3K pathways using specific inhibitors to determine if they blocked p53 reduction. Indeed, while PD098059, a MEK inhibitor that blocks MAPK pathway or LY294002, a PI3K inhibitor had no effect on the levels of p53 by drug itself (Figure 6C, lanes 2&3 vs. 1 and lanes 8 vs. 7 and 14 vs. 13), both inhibitors were able to rescue the p53 reduction by DG in A549 cells (Figure 6C, lanes 5&6 vs. 4). Only MEK inhibitor but not PI3K inhibitor, partially rescued p53 reduction in H1355 cells (Figure 6C, 10–12 vs. 9 and 16 vs. 5). Taken together, these results demonstrated that activation of SRC/MAPK/PI3K signaling pathways triggered by the binding of cardiac glycosides to the pump is responsible for p53 reduction. While the SRC activation mediates p53 reduction in both cell lines, MAPK and PI3K are involved in A549 cells, whereas MAPK, but not PI3K, is partially involved in H1355 cells.
Figure 6. p53 reduction by OU or DG was mediated by Src/MAPK/PI3K signaling pathways: (A) Activation of Src and MAPK upon OU/DG exposure.
Cells were serum starved for 36 hrs, followed by exposed to OU (30 nM) or DG (100 nM) for indicated periods of time, and analyzed by western blotting. (B) p53 reduction rescued by Src inhibitor, PP2: Cells were left untreated or treated with DG or OU alone or in combination with increasing concentrations of PP2 for 24 hrs. (C). p53 reduction rescued by MAPK or PI3K inhibitor: Cells were left untreated or treated with DG alone or in combination with MEK inhibitor, PD098059 or PI3K inhibitor, LY294002 for 24 hrs. Cell lysates were prepared for western blotting.
Discussion
Cardiac glycosides are a class of natural products that have been used for medical purposes since ancient time. Three well-known cardiac glycosides, digoxin (DG), ouabain (OU) and digitoxin were used for the treatment of congestive heart failure and atrial fibrillation via binding and inhibiting Na+/K+ ATPase to increase intracellular calcium concentrations (28). In addition to benefit heart failure patients, the drugs were also found to be beneficial to breast cancer patients (32), and were associated with a lower risk for leukemia, lymphoma as well as kidney and urinary tract cancer (33). Accumulated data in past few years have shown that cardiac glycosides selectively inhibited proliferation and induced apoptosis and autophagy in cancer cells, but not normal cells, suggesting their utility in anticancer therapy [(34) and for review, see (28)]. Mechanistically, DG or OU inhibited catalytic activity of topoisomerase II (35) and stabilized DNA-topoisomerase II complexes to suppress growth. DG, OU or other cardiac glycosides up-regulated death receptor 4 and 5 to sensitize lung cancer cells to apoptosis induced by Apo2L/TRAIL (36). DG or OU also remarkably inhibit protein synthesis of HIF-1α to block HIF1 transcription factor activity and to inhibit tumor cell growth both in vitro and in vivo (37). Furthermore, DG or OU could modulate signaling pathways of MAPK/AKT, PKC/AP-1, NF-κB, and reactive oxygen species to regulate cell growth and survival [for reviews, see (26, 28)]. Finally, globe level reduction of protein synthesis upon drug exposure, as shown in our study (Fig. 4D), could also contribute to their cytotoxicity. However, the role of p53 in the action of cardiac glycosides is totally unknown, although a recent study showed an observation that OU slightly reduced p53 level in a breast cancer line, MDA-MB-435s without providing any mechanistic insight (38).
DG was identified in our chemical library screening for the drugs that selectively kill cancer cells with mutant p53 via synthetic lethal mechanism (21). Subsequent analysis serendipitously found that the drug effectively reduces p53 levels in a cell line-dependent, but p53 status independent manner. It is well-known that p53 is a short-lived protein whose expression was regulated mainly at the post-translational levels, including phosphorylation, acetylation, ubiquitination, sumolyation and methylation (39–41). Recently, p53 is shown to be regulated at the translational level on protein synthesis by ribosomal protein L26 (42), mTOR (43) or Mdm2. Interestingly, Mdm2 could either stimulate p53 synthesis via a direct binding to p53 mRNA (44) or inhibit p53 synthesis by promoting the degradation of L26 (45). We found that the drug-induced p53 reduction did not occur at the mRNA level, as demonstrated by RT-qPCR analysis, but at the protein level. Failure to rescue drug-induced p53 reduction by proteasome inhibitors and failure of drugs to shorten the p53 protein half-life indicate that p53 reduction by the drugs is not due to enhanced p53 degradation. The drug inhibition on metabolic labeling of newly synthesized p53 suggests that the change occur at the level of protein synthesis. Thus, DG and OU are potent inhibitors of p53 protein synthesis.
What is then the mechanism by which DG or OU inhibits the de novo p53 protein synthesis? Although the details are still unknown at the present time, it appears to involve Src/MAPK signaling pathways, triggered and activated by OU/DG binding to Na+/K+ ATPase, since 1) OU or DG activates Src and MAPK, 2) their inhibitors are able to abrogate the OU/DG-induced p53 reduction, whereas 3) inhibition of Na+/K+-ATPase activity by lowering extracellular K+ has no effect. Thus, a simplest explanation for these observations is that upon DG or OU binding to Na+/K+-ATPase, Src/MAPK signaling pathways are activated particularly in drug-sensitive cancer cells, leading to activation of their down-stream effectors, eventually the inhibition of p53 protein synthesis. Future effort will be directed 1) to characterize which type of p53 synthesis; cap-dependent and/or cap-independent, also known as internal ribosome entry site (IRES) element dependent (46), is actually inhibited by cardiac glycosides and 2) to elucidate the mechanism of action.
Another interesting observation reported here is that cardiac glycosides, DG and OU, are potent cytotoxic agents in a panel of tumor cell lines with IC50s ranging from 50 to 100 nM. However, the cytotoxicity is tumor cell line dependent, but independent of wild type p53, dissociating cancer cell killing from wild type p53 elimination. Although wild type p53 induces growth arrest and apoptosis in most cases (1, 6, 7), and acts as a survival protein in some particular cases [for review, see (47)], it is unlikely that wild type p53 plays any significant role in DG or OU-induced cell killing in few cancer cell lines tested. Nevertheless, the fact that cardiac glycosides are inactive against wild type p53 in normal cells, but potently active in elimination of mutant p53 in some cancer cells, suggests that these drugs could have utility in the treatment of human cancer harboring a gain-of-function p53 mutant (48, 49).
Supplementary Material
Acknowledgements
We would like to thank Dr. Jun-Lin Guan at the University of Michigan for providing us the Ad-sh-RNA virus silencing FAK as well as FAK antibody (50). This work was supported by the NCI grants (CA111554 and CA118762) and DOD concept Award (W81XWH-08-1-0539) to YS.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Chipuk JE, Kuwana T, Bouchier-Hayes L, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 3.Mihara M, Erster S, Zaika A, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 4.Bouchet BP, de Fromentel CC, Puisieux A, Galmarini CM. p53 as a target for anti-cancer drug development. Crit Rev Oncol Hematol. 2006;58:190–207. doi: 10.1016/j.critrevonc.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y. p53 and its downstream proteins as molecular targets of cancer. Mol Carcinog. 2006;45:409–415. doi: 10.1002/mc.20231. [DOI] [PubMed] [Google Scholar]
- 6.Bassett EA, Wang W, Rastinejad F, El-Deiry WS. Structural and functional basis for therapeutic modulation of p53 signaling. Clin Cancer Res. 2008;14:6376–6386. doi: 10.1158/1078-0432.CCR-08-1526. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 8.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 9.Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low- molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 10.Peng Y, Li C, Chen L, Sebti S, Chen J. Rescue of mutant p53 transcription function by ellipticine. Oncogene. 2003;22:4478–4487. doi: 10.1038/sj.onc.1206777. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Ludwig RL, Jensen JP, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 12.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 14.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang K, Guo M, Sun Y, Yang D, Wang S. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition PNAS. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shangary S, Ding K, Qiu S, et al. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol Cancer Ther. 2008;7:1533–1542. doi: 10.1158/1535-7163.MCT-08-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun SH, Zheng M, Ding K, Wang S, Sun Y. A small molecule that disrupts Mdm2-p53 binding activates p53, induces apoptosis, and sensitizes lung cancer cells to chemotherapy. Cancer Biol Ther. 2008;7:845–852. doi: 10.4161/cbt.7.6.5841. [DOI] [PubMed] [Google Scholar]
- 17.Komarov PG, Komarova EA, Kondratov RV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 18.Strom E, Sathe S, Komarov PG, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 19.Blagosklonny MV, Toretsky J, Neckers L. Geldanamycin selectively destabilizes and conformationally alters mutated p53. Oncogene. 1995;11:933–939. [PubMed] [Google Scholar]
- 20.Blagosklonny MV, Trostel S, Kayastha G, et al. Depletion of mutant p53 and cytotoxicity of histone deacetylase inhibitors. Cancer Res. 2005;65:7386–7392. doi: 10.1158/0008-5472.CAN-04-3433. [DOI] [PubMed] [Google Scholar]
- 21.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 22.Zheng M, Morgan-Lappe SE, Yang J, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570–7578. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowen JW, McDonough A. Pretranslational regulation of Na-K-ATPase in cultured canine kidney cells by low K+ Am J Physiol. 1987;252:C179–C189. doi: 10.1152/ajpcell.1987.252.2.C179. [DOI] [PubMed] [Google Scholar]
- 24.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 25.Robinson M, Jiang P, Cui J, Li J, Wang Y, Swaroop M, Madore S, Lawrence TS, Sun Y. Global Genechip profiling to identify genes responsive to p53-induced growth arrest and apoptosis in human lung carcinomas. Cancer Biol Therapy. 2003;2:406–415. doi: 10.4161/cbt.2.4.437. [DOI] [PubMed] [Google Scholar]
- 26.Schoner W, Scheiner-Bobis G. Endogenous and exogenous cardiac glycosides: their roles in hypertension, salt metabolism, and cell growth. Am J Physiol Cell Physiol. 2007;293:C509–C536. doi: 10.1152/ajpcell.00098.2007. [DOI] [PubMed] [Google Scholar]
- 27.Tepperman K, Millette LA, Johnson CL, Jewell-Motz EA, Lingrel JB, Wallick ET. Mutational analysis of Glu-327 of Na(+)-K(+)-ATPase reveals stimulation of 86Rb+ uptake by external K+ Am J Physiol. 1997;273:C2065–C2079. doi: 10.1152/ajpcell.1997.273.6.C2065. [DOI] [PubMed] [Google Scholar]
- 28.Newman RA, Yang P, Pawlus AD, Block KI. Cardiac glycosides as novel cancer therapeutic agents. Mol Interv. 2008;8:36–49. doi: 10.1124/mi.8.1.8. [DOI] [PubMed] [Google Scholar]
- 29.Xie Z, Cai T. Na+-K+--ATPase-mediated signal transduction: from protein interaction to cellular function. Mol Interv. 2003;3:157–168. doi: 10.1124/mi.3.3.157. [DOI] [PubMed] [Google Scholar]
- 30.Liang M, Cai T, Tian J, Qu W, Xie ZJ. Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. J Biol Chem. 2006;281:19709–19719. doi: 10.1074/jbc.M512240200. [DOI] [PubMed] [Google Scholar]
- 31.Hanke JH, Gardner JP, Dow RL, et al. Discovery of a novel, potent, and Src familyselective tyrosine kinase inhibitor Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 32.Stenkvist B, Bengtsson E, Eriksson O, Holmquist J, Nordin B, Westman-Naeser S. Cardiac glycosides and breast cancer. Lancet. 1979;1:563. doi: 10.1016/s0140-6736(79)90996-6. [DOI] [PubMed] [Google Scholar]
- 33.Haux J, Klepp O, Spigset O, Tretli S. Digitoxin medication and cancer; case control and internal dose-response studies. BMC Cancer. 2001;1:11. doi: 10.1186/1471-2407-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman RA, Kondo Y, Yokoyama T, et al. Autophagic cell death of human pancreatic tumor cells mediated by oleandrin, a lipid-soluble cardiac glycoside. Integr Cancer Ther. 2007;6:354–364. doi: 10.1177/1534735407309623. [DOI] [PubMed] [Google Scholar]
- 35.Bielawski K, Winnicka K, Bielawska A. Inhibition of DNA topoisomerases I and II, and growth inhibition of breast cancer MCF-7 cells by ouabain, digoxin and proscillaridin A. Biol Pharm Bull. 2006;29:1493–1497. doi: 10.1248/bpb.29.1493. [DOI] [PubMed] [Google Scholar]
- 36.Frese S, Frese-Schaper M, Andres AC, Miescher D, Zumkehr B, Schmid RA. Cardiac glycosides initiate Apo2L/TRAIL-induced apoptosis in non-small cell lung cancer cells by up-regulation of death receptors 4 and 5. Cancer Res. 2006;66:5867–5874. doi: 10.1158/0008-5472.CAN-05-3544. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Qian DZ, Tan YS, et al. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kometiani P, Liu L, Askari A. Digitalis-induced signaling by Na+/K+-ATPase in human breast cancer cells. Mol Pharmacol. 2005;67:929–936. doi: 10.1124/mol.104.007302. [DOI] [PubMed] [Google Scholar]
- 39.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 40.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 41.Scoumanne A, Chen X. Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23:1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 43.Lee CH, Inoki K, Karbowniczek M, et al. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 2007;26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Candeias MM, Malbert-Colas L, Powell DJ, et al. p53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008 doi: 10.1038/ncb1770. PubMed ID18690233. [DOI] [PubMed] [Google Scholar]
- 45.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halaby MJ, Yang DQ. p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene. 2007;395:1–7. doi: 10.1016/j.gene.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 47.Janicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15:959–976. doi: 10.1038/cdd.2008.33. [DOI] [PubMed] [Google Scholar]
- 48.Di Agostino S, Strano S, Emiliozzi V, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 50.Peng X, Wu X, Druso JE, et al. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. Proc Natl Acad Sci U S A. 2008;105:6638–6643. doi: 10.1073/pnas.0802319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.