Abstract
Mutations in the HSD17B10 gene were identified in two previously described mentally retarded males. A point mutation c.776G>C was found from a survivor (SV), whereas a potent mutation, c.419C>T, was identified in another deceased case (SF) with undetectable hydroxysteroid (17β) dehydrogenase 10 (HSD10) activity. Protein levels of mutant HSD10(R130C) in patient SF and HSD10(E249Q) in patient SV were about half that of HSD10 in normal controls. The E249Q mutation appears to affect HSD10 subunit interactions, resulting in an allosteric regulatory enzyme. For catalyzing the oxidation of allopregnanolone by NAD+ the Hill coefficient of the mutant enzyme is ≈1.3. HSD10(E249Q) was unable to catalyze the dehydrogenation of 2-methyl-3-hydroxybutyryl-CoA and the oxidation of allopregnanolone, a positive modulator of the γ-aminobutyric acid type A receptor, at low substrate concentrations. Neurosteroid homeostasis is critical for normal cognitive development, and there is increasing evidence that a blockade of isoleucine catabolism alone does not commonly cause developmental disabilities. The results support the theory that an imbalance in neurosteroid metabolism could be a major cause of the neurological handicap associated with hydroxysteroid (17β) dehydrogenase 10 deficiency.
Keywords: developmental disabilities, HSD10 deficiency, hydroxyacyl-CoA dehydrogenase
Hydroxysteroid (17β) dehydrogenase 10 (HSD10) is a mitochondrial multifunctional enzyme (1, 2), which catalyzes the oxidation of steroid modulators of γ-aminobutyric acid type A (GABAA) receptors (3), steroid hormones (4, 5), and xenobiotics (6). It also exhibits short-chain 3-hydroxy-2-methylacyl-CoA dehydrogenase activity such that it is essential for the degradation of isoleucine (7–9). This dehydrogenase is encoded by the HSD17B10 gene (formerly the HADH2 gene) that maps to chromosome Xp11.2 (OMIM300256) (10). Four mutations have been identified in the HSD17B10 gene (10–12). Previously, it had been proposed that the blockade in isoleucine catabolism caused the neurological disorders in patients with inherited HSD10 (formerly labeled 2-methyl-3-hydroxybutyryl-CoA dehydrogenase or MHBD) deficiency (9).
In the process of isoleucine degradation, the isoleucine metabolite 2-methyl-3-hydroxybutyryl-CoA is reversibly converted to 2-methylacetoacetyl-CoA, which is then cleaved into propionyl-CoA and acetyl-CoA. These two consecutive catabolic steps are catalyzed by HSD10 (EC 1.1.1.178/35/239/159/150) (8) and mitochondrial acetoacetyl-CoA thiolase (β-ketothiolase, EC 2.3.1.9) (13), respectively. The latter enzyme is encoded by the ACAT1 gene (OMIM607809). Loss-of-function mutations in either the HSD17B10 gene or the ACAT1 gene block the isoleucine degradation pathway such that patients with these two metabolic disorders have nearly identical urine organic acid profiles (14). However, the clinical phenotype and prognosis of these two inherited metabolic disorders, namely HSD10 deficiency and β-KT deficiency, contrast sharply. The blockade of isoleucine degradation by loss of acetoacetyl-CoA thiolase does not commonly cause developmental disabilities except for a few cases with neurological sequelae attributed to severe ketoacidotic attacks (13). A regimen consisting of the avoidance of fasting and the provision of a moderate protein-restricted diet ameliorated the developmental disabilities of patients with acetoacetyl-CoA thiolase deficiency but not those with HSD10 deficiency (15, 16). Moreover, it was recently demonstrated that the over-expression of HSD17B10 gene is associated with mental retardation (17). Therefore, we propose that an imbalance in neurosteroid metabolism (18) is likely to be a major cause of psychomotor retardation in patients suffering from hydroxy steroid (17β) dehydrogenase 10 deficiency.
Here, we report a missense mutation, c.776G>C, in the HSD17B10 gene of a male mentally retarded patient (19). This mutation results in a single amino acid substitution E249Q. The mutant enzyme displays diminished activity especially in a low substrate environment. Another male patient deceased at 10 years old (16) was identified as carrying a c.419C>T transition in his HSD17B10 gene, and this potent mutation (11) results in the complete inactivation of HSD10. Taken together, these observations suggest that, because of its multifunctionality, an appropriate level of HSD10 activity is required by the brain for normal development.
Results
Identification of HSD17B10 Mutations.
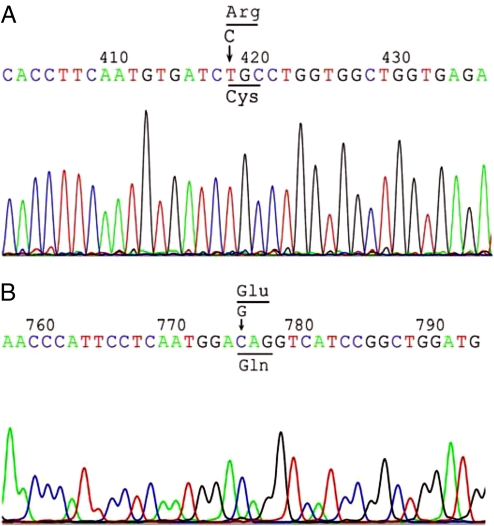
The determination of the HSD17B10 nucleotide sequence in the X chromosome of patient SF revealed that the patient carried a c.419C>T transition that resulted in mutant HSD10(R130C) (Fig. 1A). For the surviving patient SV, the only mutation identified was a c.776G>C transversion in his HSD17B10 gene. This mutation resulted in mutant HSD10(E249Q) (Fig. 1B).
Fig. 1.
Mutations on the HSD17B10 gene of patients with HSD10 deficiency. (A) Chromatogram of the forward sequence of HSD17B10 gene from patient SF showing c.419C>T transition. This mutation resulted in mutant HSD10(R130C). (B) Chromatogram of the forward sequence of the same gene from patient SV showing c.776G>C transversion. This mutation generated mutant HSD10(E249Q) with the substitution of glutamine for glutamate at residue 249 of HSD10.
Detection of HSD10 Protein in Fibroblasts.
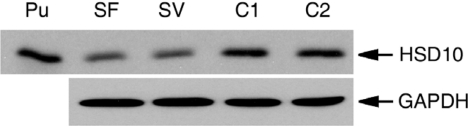
The mutations c.419C>T and c.776G>C are respectively located in exons 4 and 6 of the HSD17B10 gene (11, 20). Because the mutant proteins, HSD10(R130C) and HSD10(E249Q), possess the same C-terminal sequence as the WT HSD10, they can be detected by immunoblotting analysis with the R228 antiserum (21). As shown in Fig. 2, HSD10 protein levels in fibroblasts of HSD10 deficiency patients SF and SV were lower than in normal controls (C1 and C2). The average of four independent experiments showed that protein levels of HSD10(R130C) in patient SF and that of HSD10(E249Q) in patient SV were ≈57 ± 10% and 52 ± 9% of HSD10 in normal controls, respectively.
Fig. 2.
Immunoblotting analysis of fibroblast homogenates of patient SF, SV, and two normal controls (C1 and C2). Ten nanograms purified HSD10 (Pu) was run on the left-most lane as a 27-kDa molecular mass marker. In the other four lanes, different samples containing 10 μg of protein each were loaded and analyzed as indicated. GAPDH bands in corresponding lanes serve as an internal control for standardizing the transfer and loading of protein.
3D Structural Model of HSD10(E249Q).
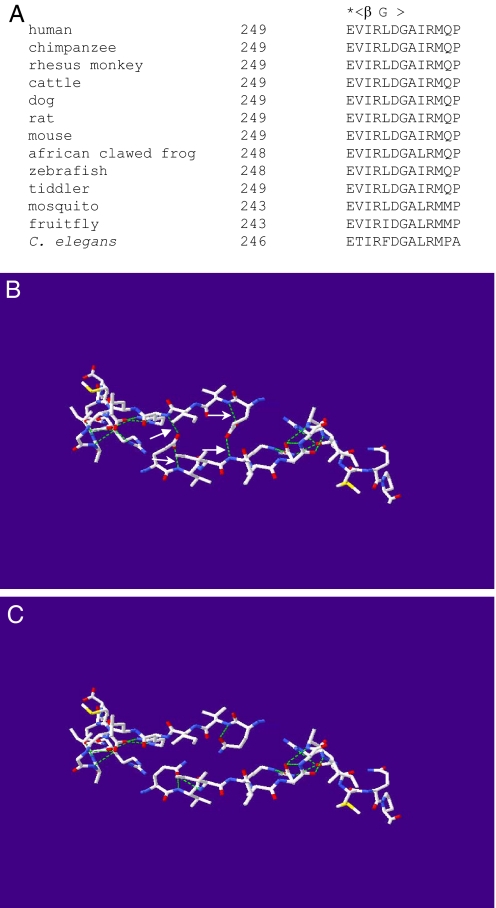
The C-terminal sequence of HSD10 is highly conserved evolutionarily, and glutamate 249 is located before the last β-strand (22) (Fig. 3A). Modeling studies, based on the X-ray crystal coordinates of the tetrameric WT HSD10, reveal that the carboxyl group of the glutamate 249 side chain forms two hydrogen bonds, that is, with the imino group of peptide bond of arginine 252 in the opposite subunit and with that of valine 250 in its own subunit (Fig. 3B). In contrast, modeling of the E249Q mutant predicts that the side chain of glutamine 249 bends toward its own subunit, and no longer contacts the opposite subunit but forms a new hydrogen bond with the imino group of peptide bond of isoleucine 251 in its own subunit (Fig. 3C). Consequently, a total of four hydrogen bonds at the interface of two dimers disappear. In addition, the pKa value of lysine 172, an ionized constituent of the reported catalytic tetrad (23), was slightly altered by the substitution of glutamine for glutamate 249 (see Fig. S1).
Fig. 3.
Alterations in hydrogen bonding of HSD10 by substitution of glutamine for glutamate 249. A comparison of C-terminal sequences of HSD10 orthologs (A) showed a conserved glutamate (*) before the last β-strand (βG). Comparison of 3-dimensional structure of C-terminals in chain D (Upper) and A (Lower) of HSD10 (B) with that of HSD10E249Q (C). The predicted hydrogen bond was displayed as a green dash line whereas oxygen in red, nitrogen in blue and sulfur in yellow. Hydrogen bonds in the subunit interface that will be broken by the E249Q mutation and other H-bonds formed by the carboxyl group of glutamate 249 side chain were indicated by arrow and small arrow, respectively.
Changes in the Catalytic Properties Because of Amination of Glutamate 249 or Replacement of Arginine 130 by Cystine.
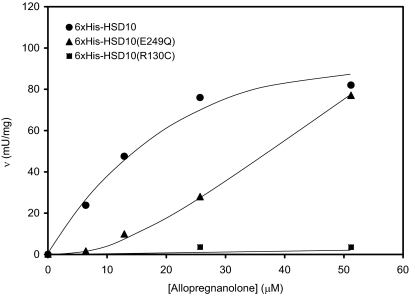
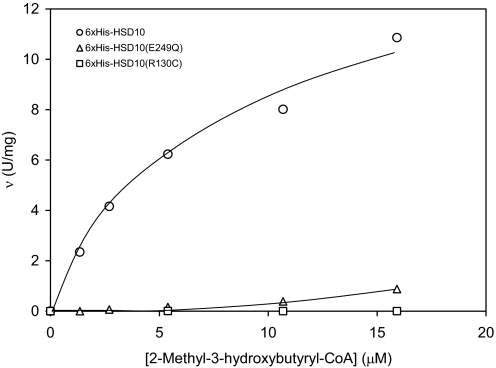
Six×His-tagged HSD10 and HSD10(E249Q) catalyzed the dehydrogenation of 2-methyl-3-hydroxybutyryl-CoA and the oxidation of allopregnanolone by NAD+, respectively (Figs. 4 and 5). Kinetic constants of 6×His-tagged HSD10 for catalyzing the oxidation of allopregnanolone (3α-hydroxy-5α-pregnane-20-one) were estimated to be Km = 30 ± 9 μM and Vmax = 150 ± 18 mU/mg, similar to those reported for HSD10 (24). The enzymatic activity of 6×His tagged-HSD10(E249Q) was proportional to the amount of enzyme added to the assay system (Fig. S2 and Fig. S3). However, the mutant enzyme appeared to adopt allosteric regulatory kinetics rather than the Michaelis–Menten Kinetics exhibited by WT HSD10. The specific activities of 6×His-tagged HSD10(E249Q) were significantly lower than those of 6×His-tagged HSD10 (P < 0.02) at low levels of allopregnanolone (Fig. 4). The Hill coefficient calculated from the sigmoid v vs. [allopregnanolone] curve (25) of the mutant HSD10(E249Q) is ≈1.3. 2-Methyl-3-hydroxybutyryl-CoA proved to be an excellent substrate for 6×His-tagged HSD10, of which kinetic constants were estimated to be Km = 7.1 ± 1.1 μM and Vmax = 14.8 ± 1.4 U/mg (Fig. 5). For catalyzing the dehydrogenation of this short-chain 3-hydroxy-2-methylacyl-CoA thioester 6×His-tagged HSD10(E249Q) behaves as an allosteric enzyme again. In contrast, 6×His-tagged HSD10(R130C) exhibited almost no catalytic activity for both substrates (Figs. 4 and 5).
Fig. 4.
Initial velocities of the oxidative reaction catalyzed by 6×His-tagged HSD10 (circle), 6×His-tagged HSD10(E249Q) (triangle), or 6×His-tagged HSD10(R130C) (square) as a function of allopregnanolone concentration.
Fig. 5.
Initial velocities of the dehydrogenation reaction catalyzed by 6×His-tagged HSD10 (circle), 6×His-tagged HSD10(E249Q) (triangle), and 6×His-tagged HSD10(R130C) (square), respectively, as a function of 2-methyl-3-hydroxybutyryl-CoA concentration.
Discussion
HSD10 deficiency is one of the diseases resulting from mutations or defective expression of the HSD17B10 gene (11, 26). Attempts were made in the present study to elucidate the basis of the pathogenesis of HSD10 deficiency, which was previously designated as 2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency (9, 19, 27, 29–31) or 3-hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency (16, 28). Its etiopathology was generally attributed to a defect of isoleucine metabolism (9, 27–31). However, MHBD or 3-hydroxyacyl-CoA dehydrogenase 2 (Hadh2) (10, 32) only represents one of the multiple functions of HSD10 (1, 8). To clarify confusions of the multiplicity of names for the gene and gene product, the Human Genome Organization (HUGO) recently announced that HSD17B10 gene and hydroxysteroid (17β) dehydrogenase 10 replaced HADH2 gene and hydroxyacyl-CoA dehydrogenase II as the official gene symbol and the designation of gene product, respectively (10, 32).
Case reports without molecular data have been published for patients SF (16) and SV (19). In the current study, a c.419C>T transition was identified in the HSD17B10 gene of patient SF. This disease-causing mutation generates a mutant HSD10(R130C), and this is a relatively common cause of HSD10 deficiency (11). The immunoblotting data indicate that there is less HSD10 protein in the patient's cells than in controls. Moreover, we found that the R130C mutation eliminated enzymatic activity, which explains why no HSD10 activity was detected (<1.4% of normal level) in his fibroblasts (16). This likely is the underlying reason for the patient's clinical presentations, which included visual loss, seizures and eventually death. This severe genotype also occurs frequently among HSD10 deficiency patients (27–31). More interestingly, a missense mutation c.776G>C was found in the HSD17B10 gene of patient SV, who has a mild phenotype. This transversion is certainly not a polymorphism because >2,500 X chromosomes have been tested thus far (12, 17). This mutation results in a mutant HSD10(E249Q). This substitution may provide a possible explanation for the observation that the HSD10, formerly short-branched-chain 3-hydroxyacyl-CoA dehydrogenase (33), activity of patient SV was greatly lower than the average of normal controls (19). Although the disease is generally believed to result from a blockade of isoleucine catabolism (9, 27–31), the catalytic properties of human HSD10 (formerly HADH2) has not yet been carefully characterized with branched-chain 3-hydroxyacyl-CoA thioesters. Data from this study clearly indicate that HSD10 represents a good example of the “flexible enzyme” originally proposed by Koshland (34).
A regimen of isoleucine-restriction diet reduced elevated levels of isoleucine metabolites to the normal level in HSD10 deficiency patients, but no improvement in their clinical manifestations or prognosis was observed (15, 16). In addition to HSD10 and mitochondrial acetoacetyl-CoA thiolase, 2-methylbutyryl-CoA dehydrogenase encoded by the ACADSB gene (OMIM600301) is also essential for the catabolic pathway of isoleucine (35). Mutations in the ACADSB gene were found to cause 2-methylbutyryl-CoA dehydrogenase deficiency (35). Nevertheless, this inborn error of isoleucine degradation has recently been summarized as a metabolic variant with a benign nature (36). Although a defect in energy metabolism may be a contributing factor to the progressive infantile neurodegeneration characteristic of HSD10 deficiency (19), metabolic acidosis is not commonly associated with this disease (11, 29). It seems no longer to be a valid explanation that the mental retardation and developmental disabilities of HSD10 deficiency patients are attributed to the blockade of isoleucine catabolism.
This enzyme may have an additional role in neuropathology. When HSD10, also known as hydroxyacyl-CoA dehydrogenase (8, 20), was bound to amyloid-β peptide, its enzymatic activity was shown to be slightly inhibited (37). However, we do not know whether amyloid-β peptide has a part to play in HSD10 deficiency because no neuropathological data for this inherited metabolic disorder is available.
The 3D structure of HSD10 was perturbed by the substitution of glutamine for glutamate before the last β-strand (Fig. 3). E249Q-mutated HSD10 showed allosteric cooperativity, as demonstrated by its decreased catalytic activity, especially at low concentrations of substrate (Fig. 4). The protein concentration of HSD10(E249Q) in patient SV's cells is less than half that of HSD10 in normal control cells (Fig. 2). The c.776G>C transversion is most likely the causative mutation because the genomic analysis failed to identify another mutation in this chromosome X region of patient SV. This was also substantiated by kinetic data of the mutant enzyme assayed with two distinct substrates. Compared with another kind of X-linked mental retardation, namely X-linked mental retardation, choreoathetosis, and abnormal behavior (MRXS10), a silent mutation (c.605C>A transversion, p.R192R), which changes an arginine codon of the HSD17B10 gene, has been identified as the causative mutation (12). Both patients SF and SV were found to have the same single nucleotide polymorphism (SNP) as the majority of normal individuals (66%) at the promoter of HSD17B10 gene.* Thus, this SNP in the promoter is unrelated to clinical presentations of patients with HSD10 deficiency.
Mentally retarded patients may carry various mutations in the HSD17B10 gene (11). For example, some cases were reported to bear only a c.771 A>G mutation in exon 6 of the HSD17B10 gene resulting in mutant HSD10(N247S) (29). The capability of this mutant enzyme to catalyze the oxidation of allopregnanolone was also found to be mostly lost. It suggests that adverse effects on cognitive development from different missense mutations of the HSD17B10 gene are probably due to an imbalance in neurosteroid metabolism.
Allopregnanolone, an important neurosteroid, can bind to the subunits of GABAA receptors (38). The 3α-hydroxyl group of this kind of neurosteroid is critical for increasing the opening frequency and duration of the receptor's chloride channel (11, 38). The biosynthesis of allopregnanolone has been widely studied (39), but its oxidation or inactivation had not. The HSD17B10 gene is expressed in various brain regions (3), and the gene product (HSD10) effectively catalyzes the oxidation of allopregnanolone to 5α-dihydroprogesterone (DHP) by NAD+ as further demonstrated in this study (Fig. 4). Also HSD10 can catalyze the oxidation of other neurosteroids such as estradiol (2, 5, 40). Sterol homeostasis was reported to be critical for brain development (41). Studies on the mouse model of Neimann-Pick C disease demonstrated that allopregnanolone significantly attenuated the progression of neurodegeneration by restoring sterol homeostasis via pregnane X receptor activation (42). Moreover, the over-expression of the HSD17B10 gene was associated with mental retardation (17). Taken together, results of the present study lead to the suggestion that an appropriate level of HSD10 is required by the brain development because of its multifunctionality.
Materials and Methods
Patients and Cell Lines.
Informed consent for molecular analyses and enzymatic studies was obtained from the parents of patients SF and SV, respectively. Case reports on these two patients have been published (16, 19). Patient SF was deceased at 10 years old because of pneumonia, whereas patient SV is currently 29 years old and his clinical manifestations are relatively stable.
Patients' fibroblasts were cultured in MEM α plus 10% FCS and 1% L-glutamine (Invitrogen–GIBCO).
Mutation Analysis.
Total DNA was purified with a DNeasy and Tissue kit (Qiagen) from cultured fibroblasts according to the instructions of the manufacturer. A fragment of X chromosomal DNA was amplified by PCR using a pair of primers, HSDF and HSDR (see Table S1). Nucleotide sequences of PCR products (≈3.7 kb) containing the HSD17B10 gene were determined by the dideoxy method using 10 primers listed in the beginning of Table S1. Then, the nucleotide sequence data were compared with the human DNA sequence from the clone RP3–339A18 on chromosome Xp11.1–11.4 (accession number Z97054) for screening mutations. Nucleotide sequence numbering according to the GenBank Ref. seq NM004493.
Construction of 6×His-Protein Expression Vectors.
The plasmid pSBET-HBHAD (20) was linearized by digestion with NdeI and EcoO109I. Oligomers 6HISF and 6HISR2 were annealed together and the resulting double-stranded DNA was subcloned into the NdeI-EcoO109I site of pSBET-HBHAD to generate pSBET-6×His-HSD10. The c.776G>C mutation and c.419C>T mutation were introduced into this 6×His-protein expression vector, respectively, with a pair of mutagenesis primers (E776C and E776G) and another pair of primers (R419T and R419A) (Table S1) using a QuikChange Site-Directed Mutagenesis kit (Stratagene) according to the instructions of the manufacturer. The mutated plasmids were designated pSBET-6×His-HSD10(E249Q) and pSBET-6×His-HSD10(R130C), respectively. The plasmid sequences were confirmed by sequencing.
Protein Expression and Purification.
Plasmids pSBET-6×His-HSD10, pSBET-6×His-HSD10(E249Q), and pSBET-6×His-HSD10(R130C) were transformed, respectively, into E. coli BL21(DE3) pLysS by the 1-step transformation method (43). The transformants were induced by 0.5 mM IPTG for 6 h. The preparation of cell extracts and the purification of 6×His-HSD10, 6×His-HSD10(E249Q), and 6×His-HSD10(R130C) were accomplished using a Ni-NTA Fast Start kit (Qiagen) according to the instructions of the manufacturer.
Tertiary Structural Model of HSD10(E249Q).
Structural differences between the mutant HSD10(E249Q) and the WT HSD10 were ascertained by bioinformatics analysis. The published crystal structure of human HSD10 was used as the template structure (22). Data were extracted from a pdb file (1U7T) of the X-ray coordinates available from Protein Data bank (www.resb.org) using DeepView/Swiss-pdb Viewer 3.7 (44). Substitution of the best rotamer mutant amino acid was made with the “Mutating Amino-Acids” function in DeepView/Swiss-pdb Viewer 3.7 (44). In addition, E249Q-induced pKa changes of the catalytic residues were analyzed with the H++ program (45).
Protein Analyses and Enzyme Assay.
Protein concentrations were determined by use of the Micro BCA protein assay kit obtained from PIERCE according to the instructions of the manufacturer. Immunoblotting was performed as described (3, 21) except that 10 micrograms of total cellular proteins were separated on a 10% SDS-polyacrylamide gel and then transferred to a nitrocellulose membrane. Protein bands were detected by use of the enhanced chemiluminescent substrate obtained from PIERCE (#32106) according to the instructions of the manufacturer. The band intensities were estimated by densitometry and normalized to the internal standard GAPDH. Then the protein level of an individual patient relative to an average of normal controls was calculated. Tiglic acid, allopregnanolone, and other chemicals were obtained from Sigma. Tiglyl-CoA was synthesized as described in ref. 7. 2-Methyl-3-hydroxybutyryl-CoA (MHB-CoA) was converted from tiglyl-CoA under the catalysis of crotonase, and effective concentrations of MHB-CoA were calculated by use of the reported equilibrium ratio of 0.33 for [MHB-CoA/tiglyl-CoA] (7). Enzymatic activity was determined as reported earlier (7, 24) but a higher concentration of NAD+ (2 mM) had been added to the assay. Kinetic data were modeled using the Michaelis–Menten equation or the Hill equation (25); all kinetic parameters were calculated with the computer program Leonora (46). One unit of activity is defined as the amount of enzyme that catalyzes the conversion of 1 μmol of substrate to product per min.
Supplementary Material
Acknowledgments.
We thank Drs. H. Schulz and D. Miller for invaluable advice and critical reading of the manuscript; Dr. C. Dobkin for his invaluable help; and Michael Fenko, Art Warman, and Shirley Clark for excellent technical assistance. This work is supported in part by the New York State Office of Mental Retardation and Developmental Disabilities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The G/C transversion has been submitted to the dbSNP of NCBI, http://www.ncbi.nlm.nih.gov/ (accession no. ss#HSD10–000595210060).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902377106/DCSupplemental.
SNP Report: rs1264014.
References
- 1.Yang SY, He XY, Schulz H. Multiple functions of type 10 17beta-hydroxysteroid dehydrogenase. Trends Endocrinol Metab. 2005;16:167–175. doi: 10.1016/j.tem.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 2.He XY, Merz G., Mehta P, Schulz H, Yang SY. Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. Characterization of a novel 17β-hydroxysteroid dehydrogenase. J Biol Chem. 1999;274:15014–15019. doi: 10.1074/jbc.274.21.15014. [DOI] [PubMed] [Google Scholar]
- 3.He XY, Wegiel J, Yang SY. Intracellular oxidation of allopregnanolone by human brain type 10 17beta-hydroxysteroid dehydrogenase. Brain Res. 2005;1040:29–35. doi: 10.1016/j.brainres.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Shafqat N, et al. Expended substrate screenings of human and Drosophila type 10 17βhydroxysteroid dehydrogenase reveal multiple specificities in bile acid and steroid hormone metabolism. Characterization of multifunctional 3α/7α/7β/20β/21-hydroxysteroid dehydrogenase. Biochem J. 2003;376:49–60. doi: 10.1042/BJ20030877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He XY, Yang YZ, Schulz H, Yang SY. Intrinsic alcohol dehydrogenase and hydroxysteroid dehydrogenase activities of human mitochondrial short chain L-3-hydroxyacyl-CoA dehydrogenase. Biochem J. 2000;345:139–143. [PMC free article] [PubMed] [Google Scholar]
- 6.Froemming MK, Sames D. Harnessing functional plasticity of enzymes: A fluorogenic probe for imaging 17beta-HSD10 dehydrogenase, an enzyme involved in Alzheimer's and Parkinson's diseases. J Am Chem Soc. 2007;129:14518–14522. doi: 10.1021/ja072601x. [DOI] [PubMed] [Google Scholar]
- 7.Luo MJ, Mao LF, Schulz H. Short-chain 3-hydroxy-2-methylacyl-CoA dehydrogenase from rat liver: Purification and characterization of a novel enzyme of isoleucine metabolism. Arch Biochem Biophys. 1995;321:214–220. doi: 10.1006/abbi.1995.1388. [DOI] [PubMed] [Google Scholar]
- 8.Yang SY, He XY, Schulz H. 3-Hydroxyacyl-CoA dehydrogenase and short-chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease. FEBS J. 2005;272:4874–4883. doi: 10.1111/j.1742-4658.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 9.Ofman R, et al. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene. Am J Hum Genet. 2003;72:1300–1307. doi: 10.1086/375116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korman SH, Yang SY. HSD17B10 replaces HADH2 as the approved designation for the gene mutated in 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency. Mol Genet Metab. 2007;91:115. doi: 10.1016/j.ymgme.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Yang SY, He XY, Miller D. HSD17B10: A gene involved in cognitive function through metabolism of isoleucine and neuroactive steroids. Mol Genet Metab. 2007;92:36–42. doi: 10.1016/j.ymgme.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Lenski C, et al. The reduced expression of the HADH2 protein causes X-linked mental retardation, choreoathetosis, and abnormal behavior. Am J Hum Genet. 2007;80:372–377. doi: 10.1086/511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukao T, Scriver CR, Kondo N. The clinical phenotype and outcome of mitochondrial acetoacetyl-CoA thiolase deficiency (β-ketothiolase or T2 deficiency) in 26 enzymatically proved and mutation-defined patients. Mol Genet Metab. 2001;72:109–114. doi: 10.1006/mgme.2000.3113. [DOI] [PubMed] [Google Scholar]
- 14.Pasquali M, Monsen G, Richardson L, Alston M, Longo N. Biochemical findings in common inborn errors of metabolism. Am J Med Genet Part C. 2006;142C:64–76. doi: 10.1002/ajmg.c.30086. [DOI] [PubMed] [Google Scholar]
- 15.Korman SH. Inborn errors of isoleucine degradation: A review. Mol Genet Metab. 2006;89:289–299. doi: 10.1016/j.ymgme.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Sutton VR, O'Brien WE, Clark GD, Kim J, Wanders RJ. 3-Hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. J Inherit Metab Dis. 2003;26:69–71. doi: 10.1023/a:1024083715568. [DOI] [PubMed] [Google Scholar]
- 17.Froyen G, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baulieu E, Robel P, Schumacher M. Neurosteroids: Beginning of the story. Int Rev Neurobiol. 2001;46:1–32. doi: 10.1016/s0074-7742(01)46057-0. [DOI] [PubMed] [Google Scholar]
- 19.Olpin SE, et al. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency in a 23-year-old man. J Inherit Metab Dis. 2002;25:477–482. doi: 10.1023/a:1021251202287. [DOI] [PubMed] [Google Scholar]
- 20.He XY, Schulz H, Yang SY. A human brain L-3-hydroxyacyl coenzyme A dehydrogenase is identical with an amyloid β-peptide binding protein involved in Alzheimer's disease. J Biol Chem. 1998;273:10741–10746. doi: 10.1074/jbc.273.17.10741. [DOI] [PubMed] [Google Scholar]
- 21.He XY, et al. Characterization and localization of human type 10 17β-hydroxysteroid dehydrogenase. Eur J Biochem. 2001;268:4899–4907. doi: 10.1046/j.0014-2956.2001.02421.2421.x. [DOI] [PubMed] [Google Scholar]
- 22.Kissinger CR, et al. Crystal structure of human ABAD/HSD10 with a bound inhibitor: Implications for design of Alzheimer's disease therapeutics. J Mol Biol. 2004;342:943–952. doi: 10.1016/j.jmb.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 23.Filling C, et al. Critical residues for structure and catalysis in short-chain dehydrogenase/reductases. J Biol Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 24.He XY, et al. Type 10 17beta-hydroxysteroid dehydrogenase catalyzing the oxidation of steroid modulators of γ-aminobutyric acid type A receptors. Mol Cell Endocrinol. 2005;229:111–117. doi: 10.1016/j.mce.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Segel IH. Enzyme Kinetics, Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme System. New York: Wiley; 1993. pp. 353–385. [Google Scholar]
- 26.He XY, Yang SY. Comments on ‘Significance of developmental expression of amphioxus Branchiostoma belcheri and zebrafish Danio rerio Hsd17b10 in biological and medical research.’. J Fish Biol. 2009;74:1689–1692. doi: 10.1111/j.1095-8649.2009.02175.x. [DOI] [PubMed] [Google Scholar]
- 27.Zschocke J, et al. Progressive infantile neurodegeneration caused by 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: A novel inborn error of branched-chain fatty acid and isoleucine metabolism. Pediatric Res. 2000;48:852–855. doi: 10.1203/00006450-200012000-00025. [DOI] [PubMed] [Google Scholar]
- 28.Ensenauer R, et al. Clinical variability in 3-hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. Ann Neurol. 2002;51:656–659. doi: 10.1002/ana.10169. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Cerda C, et al. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: An X-linked inborn error of isoleucine metabolism that may mimic a mitochondrial disease. Pediatr Res. 2005;58:488–491. doi: 10.1203/01.pdr.0000176916.94328.cd. [DOI] [PubMed] [Google Scholar]
- 30.Cazorla MR, Verdu A, Perez-Cerda C, Ribes A. Neuroimage findings in 2-methy-3-hydroxybutyryl-CoA dehydrogenase deficiency. Pediatr Neurol. 2007;36:264–267. doi: 10.1016/j.pediatrneurol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Poll-The BT, et al. Spastic Diplegia and periventricular white matter abnormalities in 2-methy-3-hydroxybutyryl-CoA dehydrogenase deficiency, a defect of isoleucine metabolism: Differential diagnosis with hypoxic-ischemic brain diseases. Mol Genet Metab. 2004;81:295–299. doi: 10.1016/j.ymgme.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Yang SY, He XY. Re: hadh2 and 3-hydroxyacyl-CoA dehydrogenase. Am J Physiol Endocrinol Metab. 2008;295:E987. doi: 10.1152/ajpendo.90521.2008. [DOI] [PubMed] [Google Scholar]
- 33.He XY, Yang SY. Roles of type 10 17beta-hydroxysteroid dehydrogenase in intracrinology and metabolism of isoleucine and fatty acids. Endocr Metab Immun Disord Drug Targets. 2006;6:95–102. doi: 10.2174/187153006776056639. [DOI] [PubMed] [Google Scholar]
- 34.Koshland DE. Crazy, but correct. Nature. 2004;432:447. doi: 10.1038/432447a. [DOI] [PubMed] [Google Scholar]
- 35.Gibson KM, et al. 2-Methylbutyryl-Coenzyme A dehydrogenase deficiency: A new inborn error of L-isoleucine metabolism. Pediatr Res. 2000;47:830–833. doi: 10.1203/00006450-200006000-00025. [DOI] [PubMed] [Google Scholar]
- 36.Sass JO, et al. 2-Methylbutyryl-coenzyme A dehydrogenase deficiency: Functional and molecular studies on a defect in isoleucine catabolism. Mol Genet Metab. 2008;93:30–35. doi: 10.1016/j.ymgme.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Oppermann UC, Salim S, Tjernberg LO, Terenius L, Jornvall H. Binding of amyloid β-peptide to mitochondrial hydroxyacyl-CoA dehydrogenase (ERAB): Regulation of an SDR enzyme activity with implication for apoptosis in Alzheimer's disease. FEBS Lett. 1999;451:238–242. doi: 10.1016/s0014-5793(99)00586-4. [DOI] [PubMed] [Google Scholar]
- 38.Hosie AM, Wilkins ME, da Silva HMA, Smart TG. Endogenous neurosteroids regulate GABAA receptors through two discrete transmembrane sites. Nature. 2006;444:486–489. doi: 10.1038/nature05324. [DOI] [PubMed] [Google Scholar]
- 39.Agís-Balboa RC, et al. Characterization of brain neurons that express enzymes mediating neurosteroid biosynthesis. Proc Natl Acad Sci USA. 2006;103:14602–14607. doi: 10.1073/pnas.0606544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McEwen BS. In: Neurosteroids: A New Regulatory Function in the Nervous System. Baulieu EE, Robel P, Schumacher M, editors. Totowa, NJ: Humana Press; 1999. pp. 233–253. [Google Scholar]
- 41.Vance JE, Karten B, Hayashi H. Lipid dynamics in neurons. Biochem Soc Trans. 2006;34:399–403. doi: 10.1042/BST0340399. [DOI] [PubMed] [Google Scholar]
- 42.Langmade SJ, et al. Pregnane X receptor (PXR) activation: A mechanism for neuroprotection in a mouse model of Neimann-Pick C disease. Proc Natl Acad Sci USA. 2006;103:13807–13812. doi: 10.1073/pnas.0606218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guex N, Peitsch MC. SWISS-MODEL and the swiss-Pdb viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 45.Gordon JC, et al. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cornish-Bowden A. Analysis of Enzyme Kinetic Data. New York: Oxford Univ Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.