The genetic alphabet is constrained by the four natural nucleotides and the two base pairs that they form. An unnatural base pair that is selectively replicated, transcribed, and translated would dramatically expand the information potential of the genetic alphabet.1–4 This would also increase the potential of the already ubiquitous methodologies based on DNA, RNA, and their sequence specific amplification. Moreover, the in vivo expansion of the genetic alphabet would serve as the foundation of a semi-synthetic organism with an expanded genetic code.
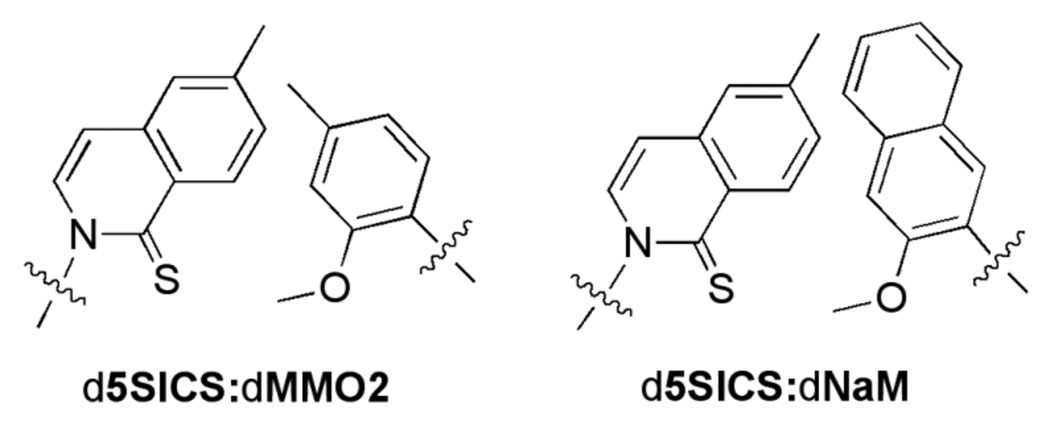
Towards this goal, we have focused on developing unnatural base pairs formed between predominantly hydrophobic unnatural nucleotides. These unnatural base pairs are stable and selectively replicated by DNA polymerases based not on complementary hydrogen-bonding, but rather on complementary shape and packing interactions. Through screening a library of nucleotide analogs, followed by hit optimization, we identified the unnatural base pair formed between dMMO2 and d5SICS (Figure 1),2a which is relatively well recognized by a variety of different replicative DNA polymerases.2b Further optimization identified dNaM which pairs with d5SICS to form an unnatural pair that is replicated with efficiencies and fidelities that are beginning to approach those of a natural base pair.2c
Figure 1.
The d5SICS:dMMO2 and d5SICS:dNaM base pairs
Expansion of the genetic code requires unnatural base pairs that are not only replicable, but also transcribed with good efficiency and selectivity in both strand contexts (i.e. dX must template YTP insertion and dY must template XTP insertion). Previous studies have examined the transcription of unnatural nucleotides bearing nucleobase analogs that pair based on either orthogonal hydrogen-bonding1 or hydrogen-bonding and hydrophobicity;3a,b however, in none of these cases was unnatural base pair transcription shown to be efficient and selective in both possible strand contexts. In addition, it remains unclear if nucleobase shape and hydrophobicity alone are sufficient for transcription.
To characterize the transcription of the unnatural base pairs formed between d5SICS and dMMO2 or dNaM, ribonucleotides were synthesized and converted to the corresponding triphosphates or deoxy-phosphoramidites, and the phosphoramidites were incorporated into DNA templates using automated DNA synthesis (Supporting Information). Transcription experiments were conducted with 100 nM DNA substrate, 1× Takara buffer (40 mM Tris-HCl pH 8.0, 8 mM MgCl2, 2 mM spermidine), DEPC treated and nuclease-free sterilized water (Fisher), T7 RNA polymerase (50 units), 20 µM each natural NTP, α-32P-ATP (0.25 mCi, MP Biomedicals), and either 5 µM 5SICSTP, 10 µM MMO2TP, or 10 µM NaMTP. After incubation for 2 hr at 37 °C, the reaction was quenched by the addition of 10 µL gel loading solution (10 M urea, 0.05% bromophenol blue), loaded onto a 20% polyacrylamide-7 M urea gel, and following electrophoresis, analyzed by phosphorimaging (Supporting Information).
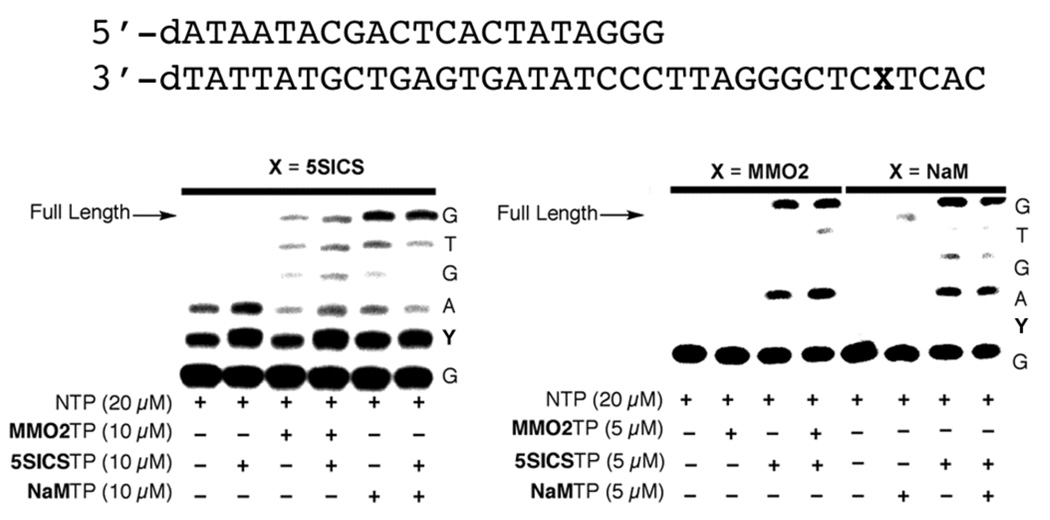
We first characterized the ability of d5SICS to template the transcription of RNA containing MMO2 or NaM (Figure 2). In the absence of the unnatural XTPs, no full length product was observed. Most of the truncated product corresponded to the termination of transcription immediately before d5SICS in the template, although a small amount corresponded to termination after mispairing, or after single nucleotide extension of a mispair. In contrast, in the presence of MMO2TP, a small amount of full length transcript was observed, although a significant amount of truncated product remained after two hours. In the presence NaMTP, significantly more full length product was observed, revealing that d5SICS templates the incorporation of NaM into RNA more efficiently than it templates the introduction of MMO2. This parallels the behavior observed with DNA polymerases,2 suggesting that at least some aspects of unnatural base pair recognition are conserved between the two classes of enzymes. In the presence of either unnatural triphosphate, the major truncation products are those corresponding to termination immediately prior to and at the unnatural nucleotide in the template, which suggests that unnatural transcription is limited by both the rate at which the unnatural base pair is synthesized and the rate with which it is extended. This is again similar to what is observed with DNA polymerases.2 Thus we tentatively conclude that recognition of the unnatural base pairs with d5SICS in the template is similar for DNA and RNA polymerases. Importantly, the addition of 5SICSTP did not alter the amount of transcript produced, suggesting that the self pair does not inhibit transcription.
Figure 2.
Full length transcription of DNA containing 5SICS, MMO, or NaM. Template sequence is shown above. X and Y correspond to the indicated unnatural base in the template and transcript, respectively.
We next characterized transcription of the unnatural base pair in the opposite strand context, by examining the ability of dMMO2 or dNaM to template the transcription of RNA containing 5SICS (Figure 2). Again, in the absence of the unnatural XTPs, virtually no full length product was observed. With either unnatural nucleotide in the template, transcription occurred up to the unnatural nucleotide and then halted, yielding only truncated product. The addition of the cognate unnatural triphosphate, in this case 5SICSTP, resulted in full length transcript with both unnatural templates. Neither the addition of MMO2TP or NaMTP interfered with transcription, again indicating that transcription is not inhibited by self pairing of the hydrophobic nucleobases. In contrast to transcription with d5SICS in the template, the observed pattern of truncated products with dMMO2 or dNaM in the template suggests that the efficiency of continued transcription after synthesis of the unnatural base pair is higher than the efficiency of unnatural base pair synthesis. This contrasts with the behavior of DNA polymerases for which synthesis of the unnatural pairs is more efficient than extension.2
We next used 2D TLC to confirm the high fidelity of unnatural base pair transcription (Table 1 and Supporting Information)3,5 Consistent with the observation that full length transcription requires the presence of the unnatural triphosphate, we found that the fidelity of transcription is not significantly different than that for a natural base pair under identical conditions, and is greater than 98% in all cases.
Table 1.
Nucleotide Composition Analysis of T7 Transcription Productsa
| Normalized composition of nucleotides incorporated 5’ to A during transcription | ||||||
|---|---|---|---|---|---|---|
| Template | ||||||
| dN | XTP | Ap | Gp | Cp | Up | Xp |
| dA | none | 1.01 ± 0.03 [1] | 1.98 ± 0.03 [2] | n.d. [0] | 0.99 ± 0.01 [1] | n.d [0] |
| d5SICS | MMO2TP | 1.07 ± 0.03 [1] | 1.96 ± 0.02 [2] | n.d. [0] | n.d. [0] | 0.97 ± 0.03 [1] |
| d5SICS | NaMTP | 0.98 ± 0.02 [1] | 2.02 ± 0.03 [2] | n.d. [0] | n.d. [0] | 0.99 ± 0.01 [1] |
| dMMO2 | 5SICSTP | 1.07 ± 0.03 [1] | 1.99 ± 0.05 [2] | n.d. [0] | n.d. [0] | 0.93 ± 0.07 [1] |
| dNaM | 5SICSTP | 1.03 ± 0.04 [1] | 2.02 ± 0.02 [2] | n.d. [0] | n.d. [0] | 0.94 ± 0.03 [1] |
Values determined by dividing the radioactivity observed for a particular radiolabeled monophosphate by the total radioactivity × the expected number of nucleotides. Predicted values assuming 100% fidelity are shown in brackets. Error reported is the standard deviation of at least three independent determinations. n.d., not detected. See Ref. 3a and 3b and Supporting information for experimental details.
Having established that each nucleotide selectively templates the incorporation of its partner into RNA, we next examined transcription efficiency by measuring, at low percent conversion, the amount of full length produced formed as a function of time (Supporting Information). Relative to the rate at which a fully natural sequence is transcribed, the incorporation of a single NaM or MMO2 opposite d5SICS reduces the rate of full length transcription by only 16-and 41-fold, respectively. The incorporation of a single 5SICS opposite either dMMO2 or dNaM reduces the rate of full length transcription by 26-and 24- fold, respectively. These relative rates of unnatural base pair transcription agree well with the qualitative gel data described above. The only ~20-fold reduction in transcription efficiency of d5SICS:dNaM in both strand contexts is remarkable and again suggests that at least some of the determinants of substrate recognition are similar with DNA and RNA polymerases, and in contrast to previously characterized unnatural base pairs, that these general determinants of recognition are possessed by d5SICS:dNaM.
Efforts to expand the genetic alphabet rely on the development of an unnatural base pair that is efficiently and selectively replicated and transcribed in both strand contexts. We have now demonstrated that dNaM:d5SICS, and to a somewhat lesser extent dMMO2:d5SICS, is not only efficiently and selectively replicated, but that it is also efficiently and selectively transcribed. This suggests that, like replication, nucleobase shape and hydrophobicity are sufficient to underlie selective and efficient transcription of an unnatural base pair into RNA. Indeed, the dNaM:d5SICS unnatural base pair seems well suited for use as part of an expanded genetic alphabet/code, with immediate in vitro applications,6,7 as well as for the long term goal of creating a semi-synthetic organism with an increased genetic code.8,9
Supplementary Material
Acknowledgment
We thank Prof. Ichiro Hirao for technical aid and the National Institutes of Health (GM060005) for funding.
Footnotes
Supporting Information Available: Details of compound synthesis and transcription analysis. This information is available free of charge via the Internet at http://www.pubs.acs.org
References
- 1.(a) Piccirilli JA, Krauch T, Moroney SE, Benner SA. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]; (b) Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hwang GT, Romesberg FE. J. Am. Chem. Soc. 2008;130:14872–14882. doi: 10.1021/ja803833h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Mitsui T, Kimoto M, Harada Y, Yokoyama S, Hirao I. J. Am. Chem. Soc. 2005;127:8652–8658. doi: 10.1021/ja0425280. [DOI] [PubMed] [Google Scholar]; (b) Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. J. Am. Chem. Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]; (c) Hirao I, Mitsui T, Kimoto M, Yokoyama S. J. Am. Chem. Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- 4.Krueger AT, Lu H, Lee AHF, Kool ET. Acc. Chem. Res. 2007;40:141–150. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Transcripts are synthesized in reactions containing α-32P ATP and then digested with RNAseI. This yields mononucleotides with 3’ phosphates that originated from the 3’ nt in the transcript. Mononucleotides are resolved by 2D TLC and quantified using phosphorimaging. Fidelity is measured as the ratio of radioactivity for each mononucleotide compared to the predicted values.
- 6.(a) Seeman NC. Trends Biochem. Sci. 2005;30:119–235. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Seeman NC. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoltenburg R, Reinemann C, Strehlitz B. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 9.Benner SA, Sismour AM. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.