Summary
Cockayne syndrome group B protein (CSB) is a member of the SWI/SNF2 subgroup of Superfamily 2 ATPases/nucleic acid translocases/helicases and is defective in the autosomal recessive segmental progeroid disorder Cockayne syndrome. We examine herein ATP dependent and ATP independent biochemical functions of human CSB. We report that Ca2+ is a novel metal cofactor of CSB for ATP hydrolysis, mainly through the enhancement of kcat, and that a variety of biologically relevant model nucleic acid substrates can function to activate CSB ATPase activity with either Mg2+ or Ca2+ present. However, CSB lacked detectable ATP dependent helicase and single- or double-stranded nucleic acid translocase activities in the presence of either divalent metal. CSB was found to support ATP independent complementary strand annealing of not only DNA/DNA duplexes, but DNA/RNA and RNA/RNA duplexes, with Ca2+ again promoting optimal activity. CSB formed a stable protein:DNA complex with a 34mer double-stranded DNA in electrophoretic mobility shift assays, independent of divalent metal or nucleotide (e.g. ATP). Moreover, CSB was able to form a stable complex with a range of nucleic acid substrates, including bubble and “pseudo-triplex” double-stranded DNAs that resemble replication and transcription intermediates, as well as forked duplexes of DNA/DNA, DNA/RNA, and RNA/RNA composition, the latter two of which do not promote CSB ATPase activity. Association of CSB with DNA, independent of ATP binding or hydrolysis, was seemingly sufficient to displace or rearrange a stable pre-bound protein:DNA complex, a property potentially important for its roles in transcription and DNA repair.
Keywords: CSB/ERCC6, SWI/SNF2, Calcium, DNA-dependent ATPase, Nucleic acid binding
Introduction
Cockayne syndrome (CS) is a rare autosomal recessive segmental premature aging disorder characterized by cachectic dwarfism, cutaneous photosensitivity, sensorinural hearing loss, cataracts, loss of subcutaneous fat and neuropathologies; including white matter hypomyelination, central nervous system calcification, and microcephaly1, 2. In contrast to the photosensitive disease xeroderma pigmentosum (XP), patients with CS do not display any increased risk of cancer. CS has two strict complementation groups, CSA (ERCC8, CKN1) and CSB (ERCC6), while patients with combined XP-CS harbor mutations in XPB, XPD or XPG3. Mutations in the CSB gene account for ~80% of CS cases4 and have also been found in DeSantis-Cacchione syndrome (DSC)5, Cerebro-oculo-facio-skeletal syndrome (COFS)6, and UV sensitive syndrome (UVSS)7.
The ERCC6 gene is located on chromosome 10q21 and encodes a 4479 nt mRNA consisting of twenty-one exons8. The CSB protein is 1493 amino acids (~168 kDa) and has two putative nuclear localization signals, an acidic region, a glycine rich region, and seven sequence motifs characteristic of Superfamily 2 (SF2) ATPases/helicases/nucleic acid translocases9. CSB has been found to be essential for transcription coupled nucleotide excision repair (TC-NER) – a sub-pathway of NER that operates to remove RNA polymerase blocking lesions10 – and is considered the human transcription coupled repair factor, similar in nature to the Escherichia coli protein Mfd11. CSB also appears to play a role in general transcription12 and as an auxiliary factor in the base excision repair (BER) pathway2. CSB interacts with proteins involved in transcription, i.e. RNA polymerases (RNAP) I, and II13, 14, the p44 and p62 subunits of TFIIH15, and p5316; in NER, i.e. XPB, XPD, XPG15, XAB217 and the p34 subunit of TFIIE11; and in BER, including PARP118, OGG119, and APE120.
A hallmark of CSB deficiency is sensitivity to ultraviolet (UV) light and a failure to recover RNA synthesis after UV exposure21. This latter defect is presumably reflective of inoperable TC-NER and/or faulty chromatin reorganization, typically mediated by CSB to facilitate transcription re-initiation22. CSB deficient cells are also sensitive to agents, such as 4-nitroquinoline (4-NQO) and N-acetoxy-2 aminofluorene (NA-AAF), which form bulky helix-distorting lesions repaired primarily by NER23–25. Moreover, CSB deficient cells are sensitive to ionizing radiation (IR)26, paraquat27, methyl methanesulfonate (MMS), and 5-hydroxymethyl-2’-deoxyuridine20, suggesting that CSB plays a role in protection against or removal of BER substrates (e.g. base lesions, abasic sites, or single-strand breaks).
Previous biochemical work on CSB has shown that the protein possesses the ability to hydrolyze ATP to ADP plus inorganic phosphate (Pi) in a manner that is dependent on structured or double-stranded (ds) DNA, with DNA/RNA and RNA/RNA only weakly stimulating this function11, 28, 29. In addition, CSB has been found to change the nicked plasmid linking number by introducing negative supercoils in a seemingly ATP binding dependent manner30, 31 and to pair complementary DNA strands in the absence of ATP32. CSB has also been reported to modify mononucleosome positioning, alter nucleosome arrays, and interact with histone tails30, properties that are similar to other SWI/SNF2 chromatin remodeling enzymes33. Unlike SWI/SNF2 remodeling proteins, however, CSB was not able to change the topology of nucleosomal plasmid DNA30. Finally, CSB, in the presence of ATP or the non-hydrolyzable ATP analog (AMP-PNP), has been found by atomic force microscopy to wrap dsDNA around itself31, perhaps explaining the ability of CSB to perturb nucleosomes.
Emerging evidence suggests that CSB is able to act in two modes, one that requires its ATPase activity, presumably in some remodeling aspect, and one that operates in an ATP independent fashion. Consistent with the ATP hydrolysis activity of CSB serving some function, in vivo complementation experiments indicate that an intact ATPase/SF2 core domain is required for rescue of 4-NQO sensitivity, UV-sensitivity, RNA synthesis recovery after UV, and prevention of UV induced apoptosis of CSB deficient cells23, 34. Additionally, CSB SF2 motif V/VI mutant cells have lower repair activity for 8-hydroxyguanine in DNA than comparable wild type complemented cells35. On the other hand, the CSB E646Q ATPase mutant only partially complements the MMS sensitivity of CSB deficient cells, suggesting that CSB functions, at least in BER, in two capacities: one involving its core ATPase-dependent activity and one invoking an ATP independent function that may engage the other regions of the protein, which perhaps mediate specific physical associations20. Furthermore, CSB is able to stimulate Ape1 AP site incision20 and elongation of RNA polymerase I36 in an ATP independent manner, indicating ATP independent functions of CSB in DNA repair and transcription. We have explored further the ATP dependent and ATP independent biochemical properties of CSB, focusing on its interactions with and manipulation of nucleic acid.
Results
Influence of divalent metals on DNA dependent ATP hydrolysis by CSB
Before proceeding with further biochemical characterization of CSB, we determined the optimal reaction parameters for the human enzyme. Specifically, we assayed the double stranded DNA (dsDNA) stimulated ATPase activity of CSB under two buffer conditions (Tris-Cl and HEPES-OH) at varying pH values. We found that CSB dsDNA ATPase activity was active over a wide pH range (6.5 to 8.5), under both buffer conditions (unpublished observations). With HEPES-OH pH 8.0, CSB consistently hydrolyzed the highest level of ATP to ADP and Pi, and thus, this platform was used for all subsequent experiments.
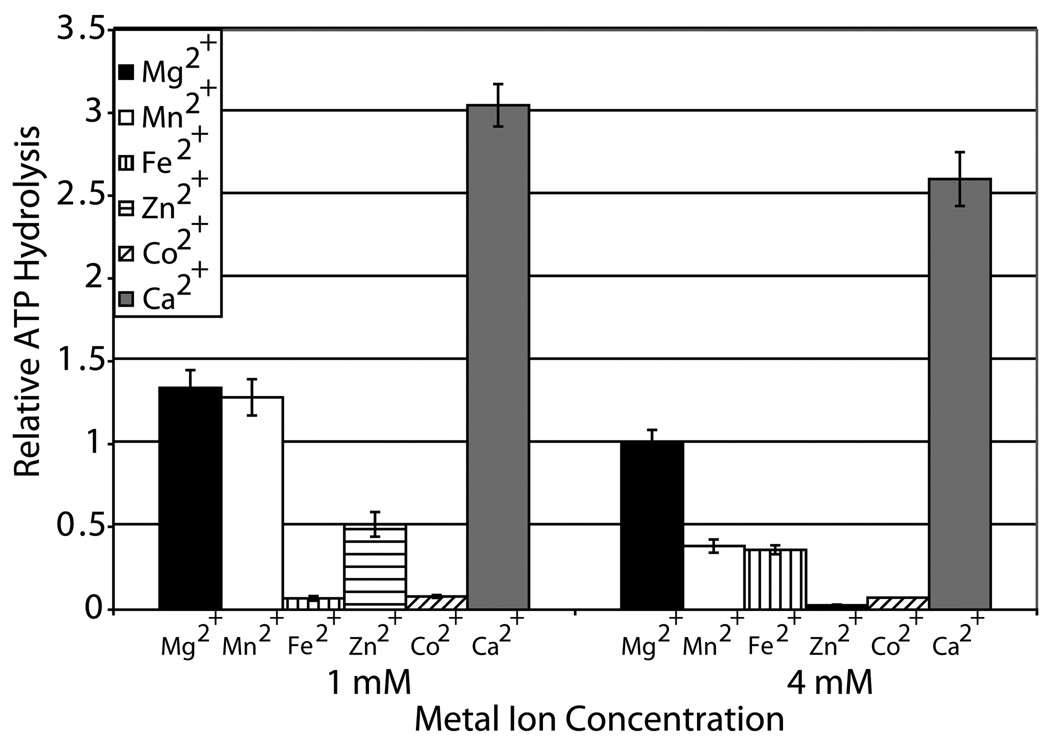
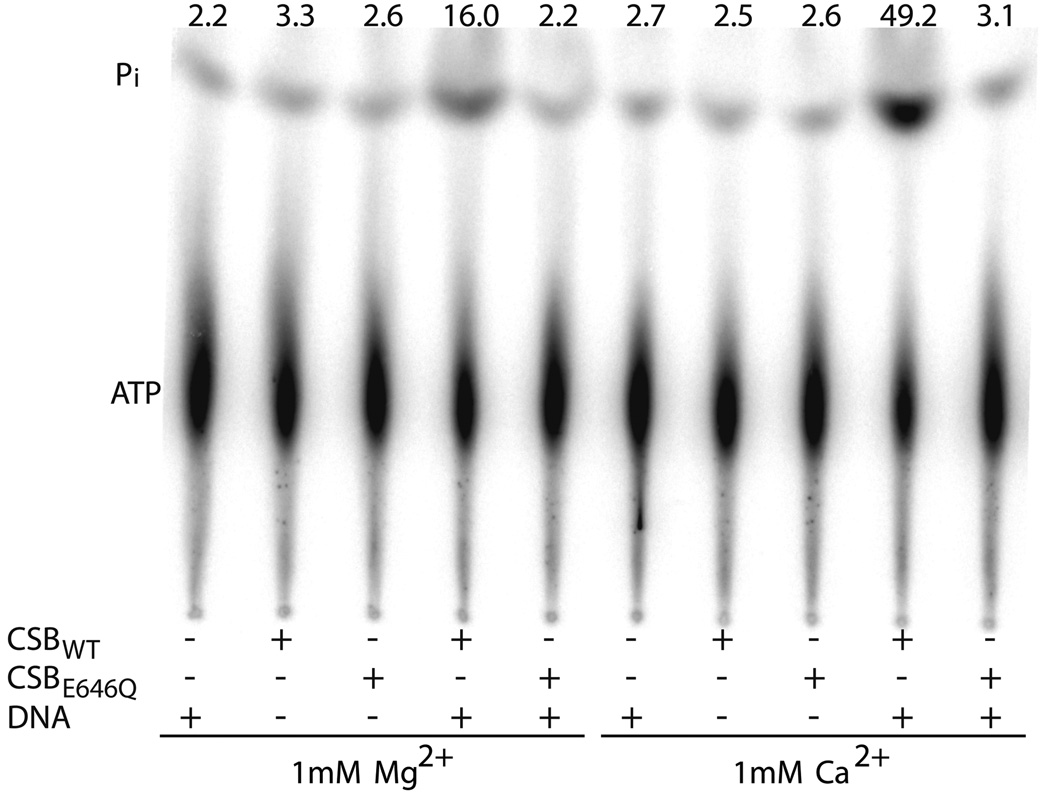
We also examined at two concentrations (1 and 4 mM) the effect of various divalent cations on the ATP hydrolytic activity of CSB. Ca2+ > Mg2+ ≥ Mn2+ > Zn2+, Fe2+ were found to activate dsDNA dependent ATPase activity – in some cases in a concentration specific manner – whereas Co2+ was not (Fig. 1a). To our surprise, Ca2+, at both concentrations tested, was able to promote the greatest CSB dsDNA dependent ATP hydrolysis, ~ 2.5 – 3 fold over Mg2+. An ATPase motif II mutant protein, E646Q, was unable to hydrolyze ATP in the presence of either Mg2+ or Ca2+ and dsDNA, indicating that intact SF2 motifs are required for DNA dependent ATP hydrolysis and that the activity with either metal is intrinsic to CSB (Fig. 1b).
Fig. 1. Ca2+ is a novel metal cofactor for DNA dependent ATP hydrolysis by CSB.
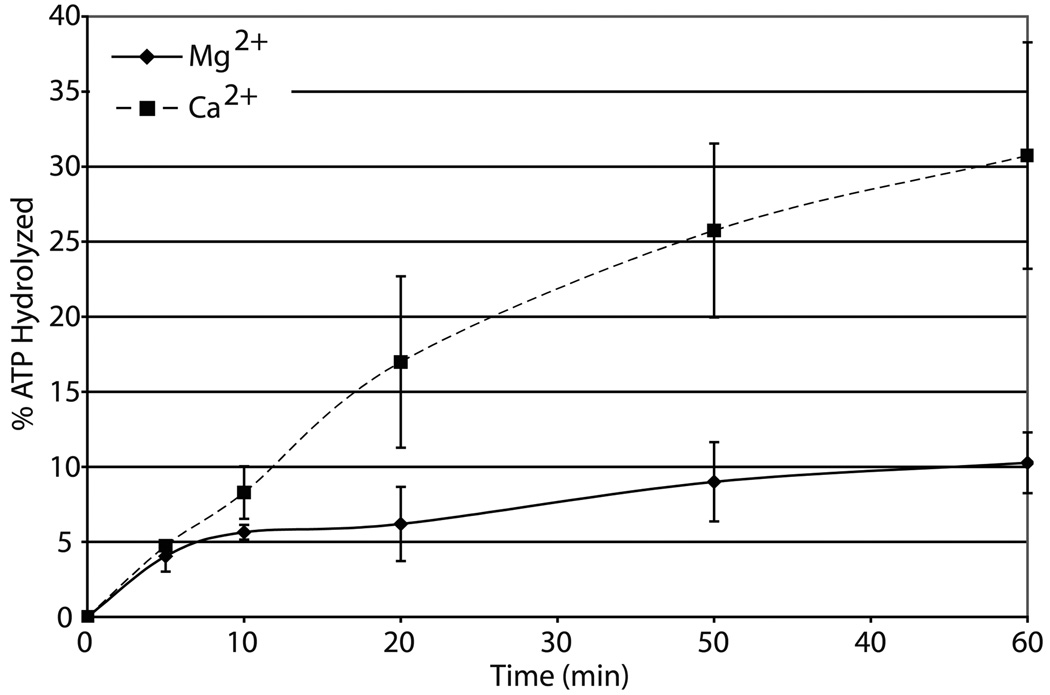
(a) Graph of relative ATP hydrolysis by CSB in the presence of various divalent metal ions at two concentrations (1 mM or 4 mM). Average values are plotted with standard deviations of at least 3 independent experiments. All values are relative to the 4 mM Mg2+ reaction, the metal condition typically used to assess CSB ATPase function 28. (b) CSB ATP hydrolysis is dependent on active SF2 motif and the presence of dsDNA for both 4 mM Mg2+ and 1 mM Ca2+. PEI-TLC plate of ATP hydrolysis by wild-type CSB or Motif II mutant (E646Q) in the presence or absence of dsDNA (pUC19) and divalent metal (Mg2+ or Ca2+). Shown above is the average percent ATP hydrolyzed, calculated from at least 3 independent experiments. (c) Graph of CSB ATPase time course kinetics in the presence of Mg2+ or Ca2+. Average values are plotted with standard deviations of at least 3 independent experiments.
Time course studies indicated that Ca2+ promoted ATP hydrolysis by CSB more effectively throughout the reaction period (Fig. 1c). To further characterize the mechanism by which Ca2+ stimulates CSB dsDNA dependent ATPase activity, we determined the kinetic parameters for ATP hydrolysis in the presence of either metal ion. Using pUC19 as the effector DNA, we found that the Km values for ATP were similar: 16.5 µM with Mg2+ and 27.6 µM with Ca2+. Vmax values were 0.06 µM product min−1 with Mg2+ and 0.2 µM product min−1 with Ca2+, indicating a ~ 3 fold increase in kcat specifically.
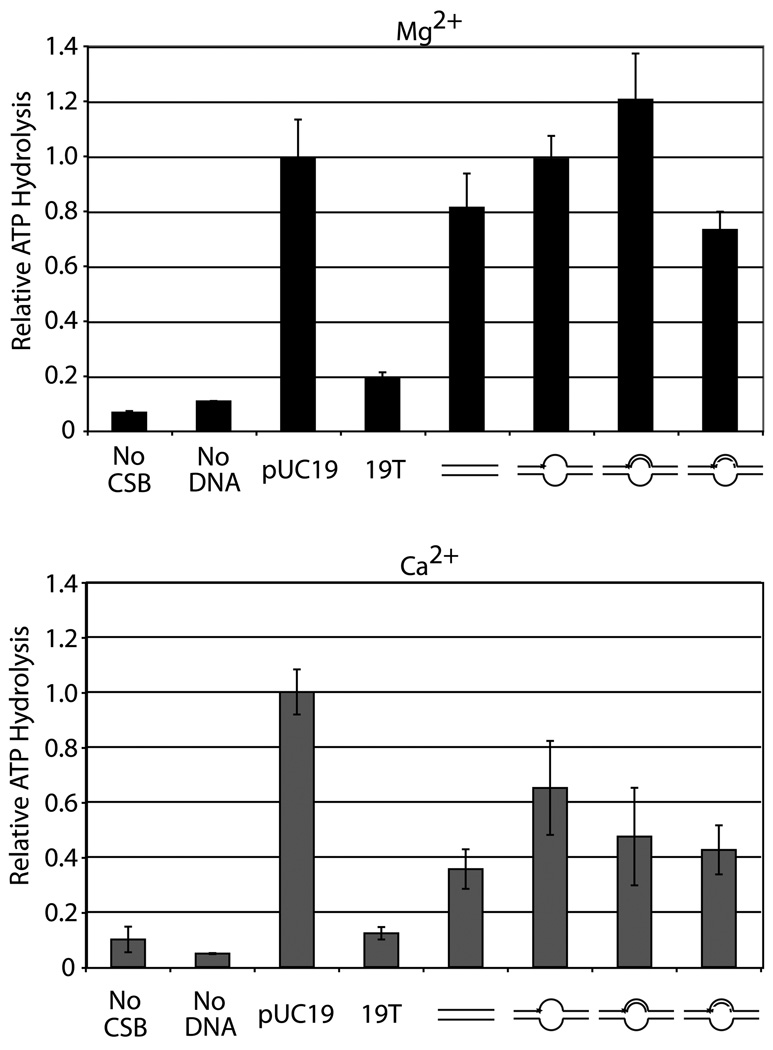
It has been shown that CSB requires DNA with some double-stranded nature to stimulate its ATP hydrolysis activity in the presence of Mg2+11, 28, 29, 37. We examined CSB ATPase function when presented with a variety of model substrates in the presence of Mg2+ or Ca2+ (see Experimental Procedures). Substrates used were a true ssDNA (19T), a supercoiled plasmid (pUC19), a short dsDNA duplex (34G/34C), a dsDNA containing an 18 nucleotide bubble (54FE/54C), and two “pseudo-triplex” substrates (54FE/54C/18D and 54FE/54C/18R) designed to mimic a DNA replication bubble or a transcription bubble, respectively (see Table 1 for oligonucleotide sequences). Consistent with prior results11, 28, the single-stranded oligonucleotide (19T) poorly stimulated the ATPase activity of CSB under either metal condition (Fig. 2). Conversely, with each metal, supercoiled plasmid DNA stimulated CSB catalyzed ATP hydrolysis most significantly, with little difference seen in comparison to the other duplex-containing substrates (e.g. compare pUC19 to 34G/34C) in the case of Mg2+ (Fig. 2, top) or with a <3-fold difference in the case of Ca2+ (Fig. 2, bottom). No unique substrate preference was uncovered when Ca2+ was the divalent cation instead of Mg2+, although a slightly different profile was observed.
Table 1.
Oligonucleotides used in this study
| Name | Sequence 5'-3' | Reference |
|---|---|---|
| 19T | TTTTTTTTTTTTTTTTTTT | This work |
| 34G | GTACCCGGGGATCCGTACGGCGCATCAGCTGCAG | 62 |
| 34C | CTGCAGCTGATGCGCCGTACGGATCCCCGGGTAC | 62 |
| 54FE | CCGCTGCCGCTGAATTGCFCCCTCGATCTAGGTCGATGATCCTAAGCATAAGCA | 63 |
| 54C | TGCTTATGCTTAGGATCAAGCTGGATCAAGCTCCCAGCAATTCAGCGGCAGCGG | 63 |
| 18D | TCGACCTAGATCGAGGGT | 63 |
| 18R | rUrCrGrArCrCrUrArGrArUrCrGrArGrGrGrU | 63 |
| DNA50 | GGGACGCGTCGGCCTGGCACGTCGGCCGCTGCGGCCAGGCACCCGATGGC | 61 |
| DNA49 | TTTGTTTGTTTGTTTGTTTGTTTGCCGACGTGCCAGGCCGACGCGTCCC rGrGrGrArCrGrCrGrUrCrGrGrCrCrUrGrGrCrArCrGrUrCrGrGrCrCrGrCrUrGrCrGrGrCrCr |
61 |
| RNA50 | ArGrGrCrArCrCrCrGrArUrGrGrC rUrUrUrGrUrUrUrGrUrUrUrGrUrUrUrGrUrUrUrGrUrUrUrGrCrCrGrArCrGrUrGrCrCrArG |
61 |
| RNA49 | rGrCrCrGrArCrGrCrGrUrCrCrC | 61 |
| DNA25F | CCGACGTGCCAGGCCGACGCGTCCC | 61 |
| DNA25 | GCCATCGGGTGCCTGGCCGCAGCGG | 61 |
| RNA25F | rCrCrGrArCrGrUrGrCrCrArGrGrCrCrGrArCrGrCrGrUrCrCrC | 61 |
| RNA25 | rGrCrCrArUrCrGrGrGrUrGrCrCrUrGrGrCrCrGrCrArGrCrGrG GGGACGCGTCGGCCTGGCACGTCGGrCrCrGrArCrGrUrGrCrCrArGrGrCrCrGrArCrGr |
61 |
| D25R25 | CrGrUrCrCrC rUrUrUrGrUrUrUrGrUrUrUrGrUrUrUrGrUrUrUrGrUrUrUrGCCGACGTGCCAGGCCGA |
61 |
| R24D25 | CGCGTCCC rGrCrCrArUrCrGrGrGrUrGrCrCrUrGrGrCrCrGrCrArGrCrGrGCCGCTGCGGCCAGGCAC |
61 |
| R25D25 | CCGATGGC | 61 |
| (TC)20 | TCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTC | 52 |
| 42BamHI | CCGCTGAATTGCACCGGATCCCTAGGTCGATGATCCTAAGCA | This work |
| 42Comp | TGCTTAGGATCATCGACCTAGGGATCCGGTGCAATTCAGCGG | This work |
Fig. 2. Alternate model nucleic acid substrates serve as cofactors for CSB dependent ATP hydrolysis in the presence of Mg2+ (top) or Ca2+ (bottom).
CSB ATPase assays were performed as described in Experimental Procedures. Average relative values (to pUC19 reaction under each metal condition) are plotted with standard deviations of at least 3 independent experiments. Substrate depictions are displayed underneath each graph: from left to right pUC19, 19T, 34G/34C, 54FE/54C, 54FE/54C/18D, and 54FE/54C/18R. Solid lines represent DNA and dashed lines represent RNA. (see Table 1 for oligonucleotide details).
The effect of divalent metals on nucleic acid complementary strand annealing by CSB
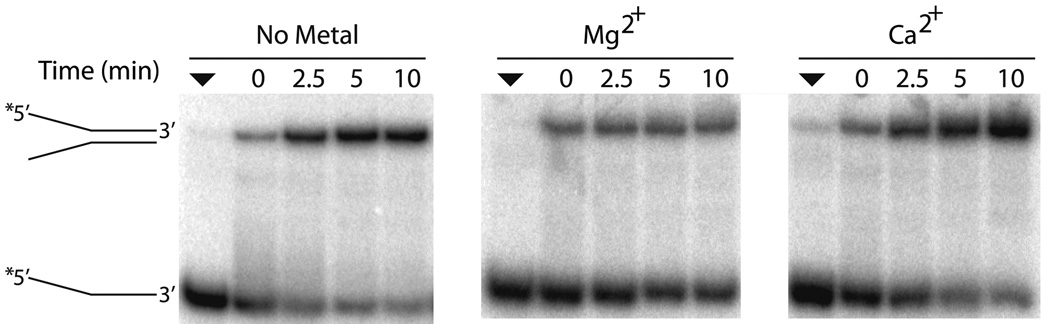
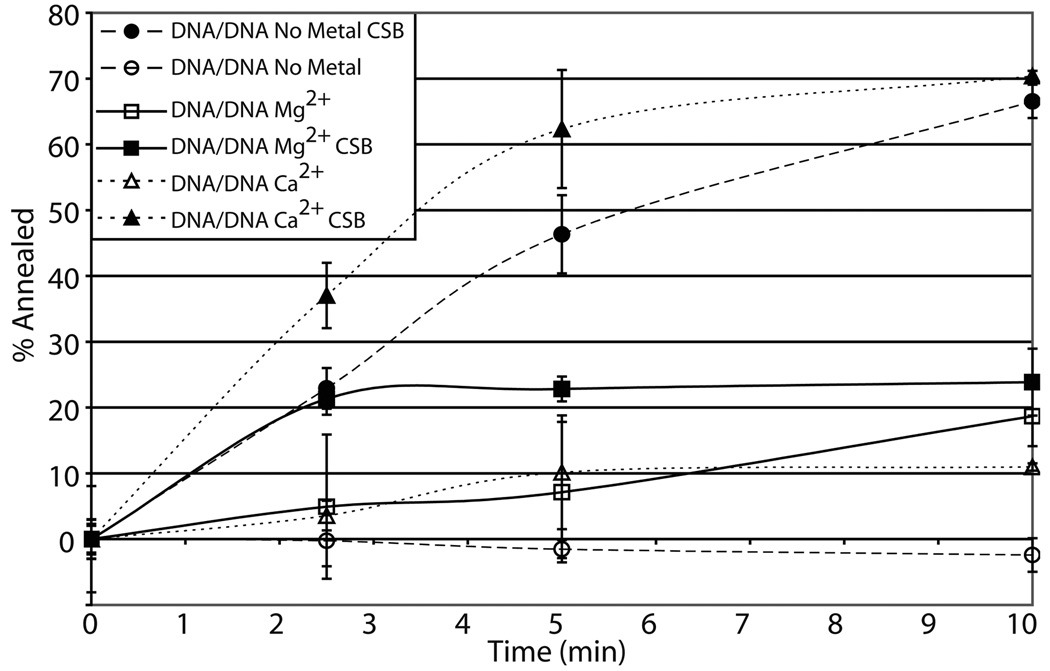
Previously, it had been demonstrated that CSB was able to pair two partially complementary DNA strands that form a fork structure when annealed in the presence of Mg2+, a function that may be relevant to its participation in various DNA transactions32. We sought to examine if divalent metal was required and whether substitution of Ca2+ for Mg2+ would affect, in any way, the ability of CSB to pair two partially complementary DNA oligonucleotides, DNA50 and DNA49 (Table 1). We found that (i) CSB does not require divalent metal to hybridize complimentary DNA/DNA strands, (ii) Ca2+ stimulates DNA/DNA strand pairing by CSB, and (iii) Mg2+ negatively impacts the annealing function relative to the no metal control (Fig. 3a and b). Similar to the ATP hydrolysis reactions, substitution of Ca2+ for Mg2+ increased the DNA/DNA pairing activity of CSB maximally ~ 3 fold.
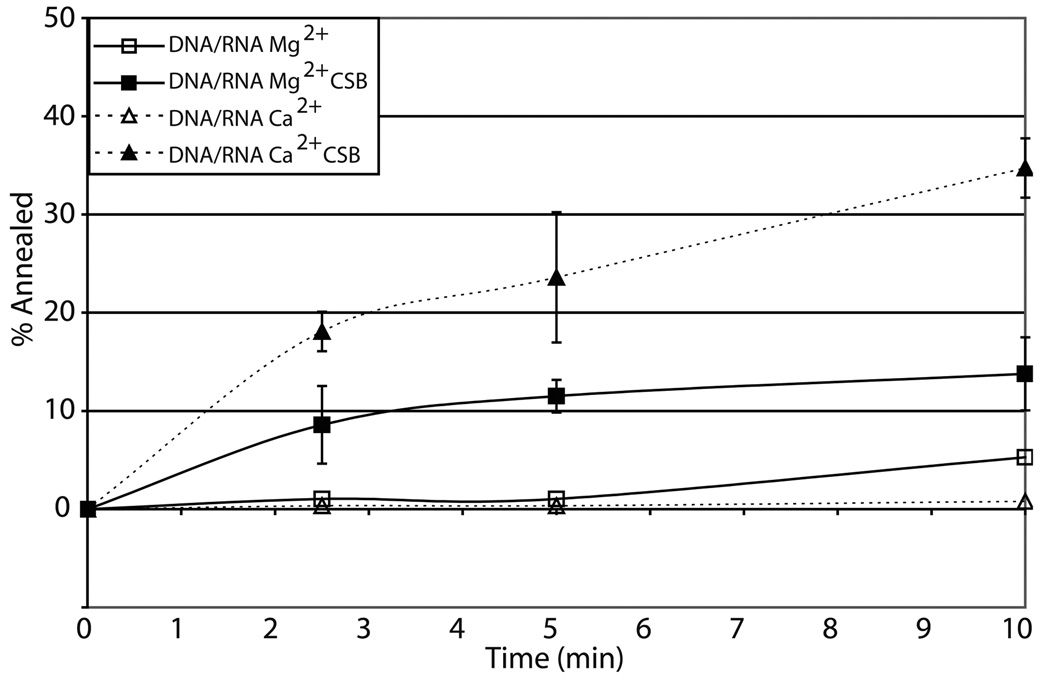
Fig. 3. Effects of Mg2+ or Ca2+ on CSB nucleic acid strand pairing.
(a) Representative gel of CSB strand pairing activity for DNA50/DNA49 in the presence of no metal, 4 mM Mg2+, or 1 mM Ca2+ (see Experimental Procedures). The positions of the initial single-stranded substrate and the duplex product are shown. The filled triangle represents labeled single-strand nucleic acid alone. (b) Graph of CSB dependent DNA/DNA strand pairing time course kinetics. (c) DNA/RNA strand pairing time course kinetics. In panels b and c, shown are the annealing reactions with and without CSB (see inset). Reactions were carried out with 450 fmol CSB protein with 100 fmol labeled and 100 fmol unlabeled oligonucleotides for the time indicated. Average values are plotted with standard deviations of at least 3 independent experiments.
We next asked whether this strand annealing function of CSB extended to DNA/RNA complementary hybrids (see Materials and Methods), which would be expected to arise during processes such as transcription. Similar to the DNA/DNA strand pairing results above, we found that CSB was able to promote the formation of double stranded DNA/RNA hybrids (DNA50 and RNA49; Table 1) more efficiently (~2-fold) in the presence of Ca2+ relative to Mg2+ (Fig. 3c). It is interesting to note that Mg2+ alone appeared to promote more efficient strand annealing than Ca2+ alone (i.e. without CSB), particularly in the case of the DNA/DNA and, to a lesser extent, the DNA/RNA reactions, making the Ca2+ stimulation of the CSB strand pairing activity even more dramatic (Fig. 3). Finally, we observed that CSB was able to promote strand annealing of partially complementary RNA oligonucleotides (RNA50 and RNA49; Table 1) in the presence of Mg2+ or Ca2+; in this case, either divalent cation supported CSB facilitated strand pairing to similar levels (data not shown).
CSB binds a variety of nucleic acid structures independent of divalent metals or nucleotide
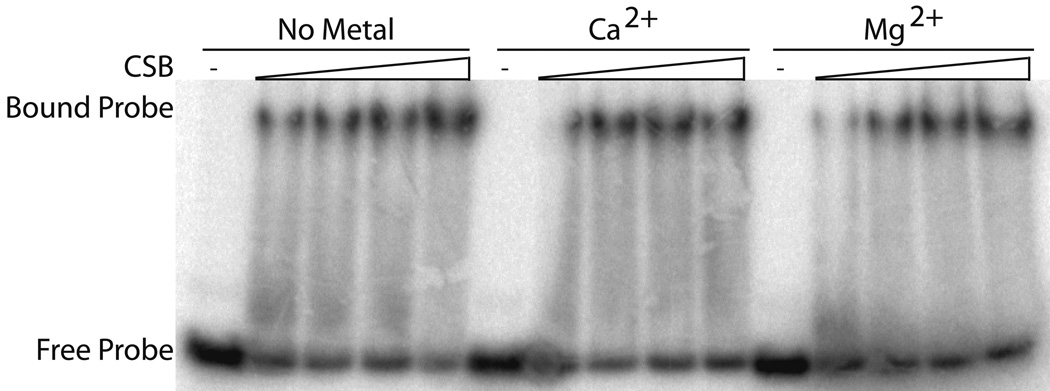
We next sought to elucidate whether CSB could form a stable complex with oligonucleotide duplexes and whether divalent metal or nucleotide status would affect complex formation. Previous DNA binding by CSB was shown using a 90mer and Mg2+ containing buffer11. We found that CSB DNA binding did not require any divalent metal and that binding to the short 34mer duplex substrate (34G/34C) was consistently, albeit only slightly, better in buffer lacking divalent metal (Ca2+ or Mg2+) (see representative binding gel in Fig. 4a). Apparent dissociation constants (KD) for CSB DNA binding were 10.7±0.6 nM without metal, 13.7±1.1 nM with Ca2+, and 20.5±5.2 nM with Mg2+. In addition, nucleotide status (i.e. addition of ATP, ADP, or AMP-PNP) did not dramatically affect DNA binding by CSB and, consistent with previous observations11, nucleotide was not required for CSB:DNA binary complex formation (data not shown).
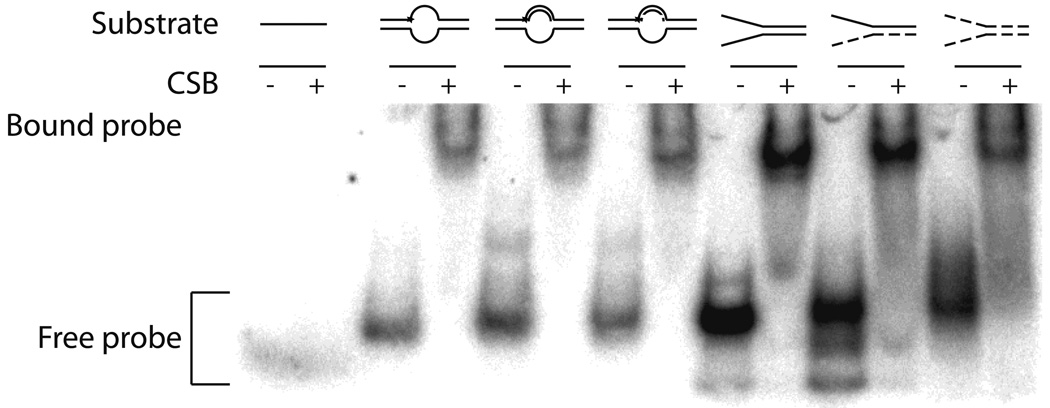
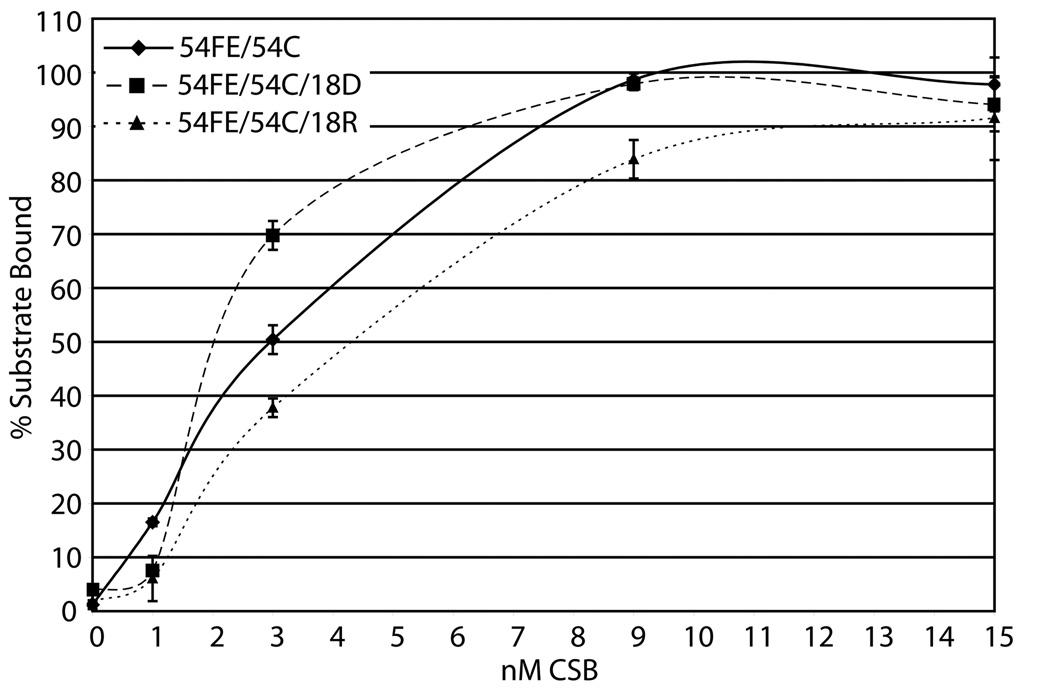
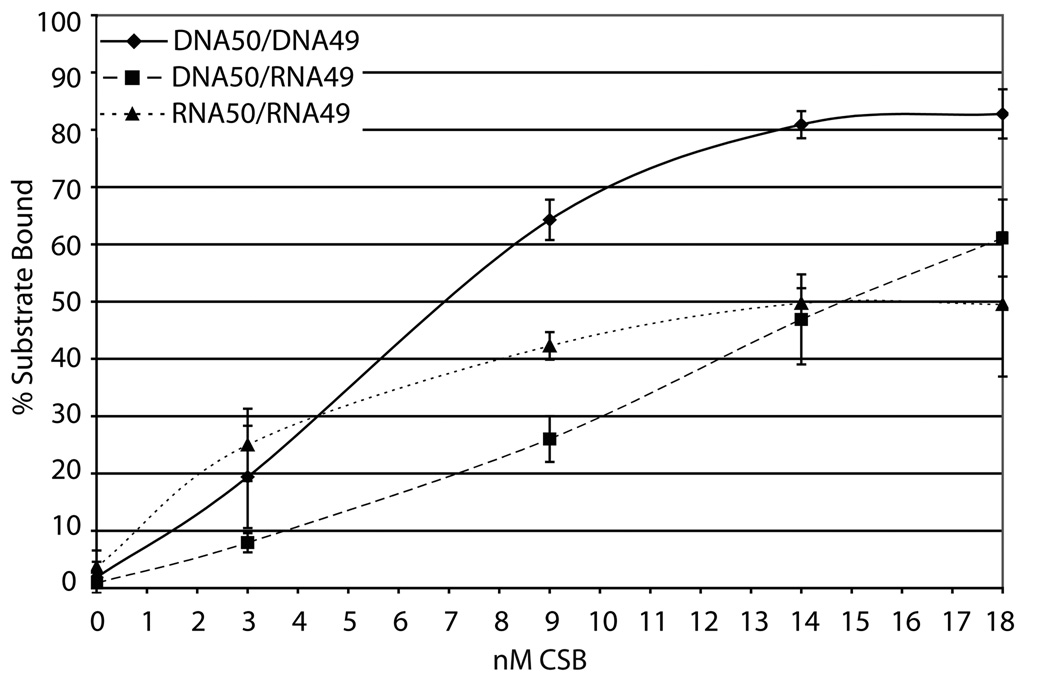
Fig. 4. Nucleic acid binding by CSB.
(a) Mg2+ and Ca2+ have little effect on CSB dsDNA binding. Reactions were carried out with 50 fmol labeled duplex 34G/34C DNA (probe) and 150 to 500 fmol CSB in the conditions indicated (see Experimental Procedures). Shown is a representative native polyacrylamide gel. (b) CSB binds to a wide variety of alternate model nucleic acid substrates. Substrate depictions are displayed on top: from left to right, 19T, 54FE/54C, 54FE/54C/18D, 54FE/54C/18R, DNA50/DNA49, DNA50/RNA49, and RNA50/RNA49. Solid lines represent DNA and dashed lines represent RNA. Reactions with (+) or without (−) CSB are shown. Note: free probe migrates differently depending on nucleic acid substrate. (c) Graph of CSB bubble substrate and pseudo-triplex nucleic acid binding. (d) Graph of CSB forked duplex nucleic acid substrate binding. Saturating binding of DNA50/RNA49 was observed at 25 nM CSB concentration with 67.5 ± 3.6% substrate binding (data not shown). Average values are plotted with standard deviations of at least 3 independent experiments.
In addition to 34mer dsDNA binding by CSB (Fig. 4a), we examined whether CSB could bind stably to alternate nucleic acid structures that might arise in vivo. We utilized duplex substrates that stimulated CSB ATP hydrolysis activity (54FE/54C, 54FE/54C/18D, 54FE/54C/18R; Table 1) and that were hybridized in the complementary strand annealing assays (DNA50/DNA49, DNA50/RNA49, RNA50/RNA49; Table 1). Interestingly, CSB was able to bind all six substrates (Fig. 4b), although the DNA/RNA hybrid (DNA50/RNA49) and the RNA/RNA duplex (RNA50/RNA49) did not promote significant ATP hydrolysis by CSB (data not shown). Apparent KD values for each substrate are as follows: 3.2±0.6 nM for 54FE/54C, 2.2±0.2 nM for 54FE/54C/18D, 3.8±0.2 nM for 54FE/54C/18R, 5.9±0.6 nM for DNA50/DNA49, 13.4±3.3 nM for DNA50/RNA49, and 3.6±1.5 nM for RNA50/RNA49 (Fig. 4c and d). The data indicate that CSB has slightly higher DNA binding affinity or stability for bubble containing DNA structures over small (34mer) dsDNAs (see for instance 54FE/54C/18D vs. 34G/34C without metal, ~ 5 fold difference in KD) and that CSB shows little difference in preference for the forked duplexes of varying nucleic acid composition (compare DNA50/DNA49, DNA50/RNA49, and RNA50/RNA49).
CSB is neither a nucleic acid helicase nor a single- or double -strand nucleic acid translocase
CSB belongs to the SWI/SNF2 subgroup of SF2 helicases and nucleic acid translocases38, 39. As such, we examined whether CSB possessed any ATP dependent nucleic acid unwinding or nucleic acid translocation activities similar to other characterized SF2 enzymes40.
First, we tested a large number of nucleic acid substrates for ATP dependent CSB catalyzed strand separation under the same buffer conditions where CSB was able to hydrolyze ATP in the presence of dsDNA, examining reactions that contained either Mg2+ or Ca2+ (see Materials and Methods). We examined DNA/DNA and DNA/RNA substrates that contained a free 5’ or a free 3’ end, as well as substrates that formed DNA/DNA, DNA/RNA, RNA/DNA, and RNA/RNA forked duplexes. In other words, all combinations of single-strand nucleic acid loading platforms and double-stranded nucleic acid duplexes were explored (Supplemental Fig. 1, top two tiers). CSB was found to have no ATP dependent unwinding activity on these substrates (see examples in Supplemental Fig. 2). We also tested DNA:RNA hybrid oligonucleotides where there was a nucleic acid switch at a single-strand:double-strand junction (Supplemental Fig. 1, third tier). CSB, again, displayed no unwinding activity on any of these substrates (data not shown). Finally, we examined a set of bubble and “pseudo-triplex” substrates (Supplemental Fig. 1, bottom tier) for CSB unwinding activity and observed no strand separation by the protein (data not shown).
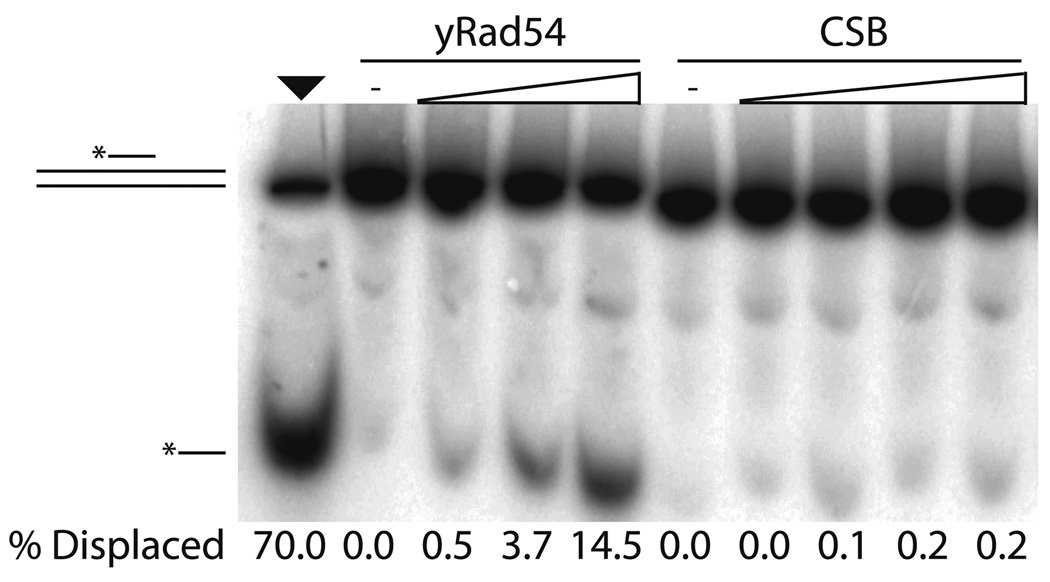
Second, we asked whether CSB could translocate along either single-stranded or double-stranded nucleic acid, again under conditions in which CSB was active as a DNA dependent ATPase. Using 50mer DNA or RNA oligonucleotide strands that harbor a centrally located biotinylated nucleotide pre-bound to streptavidin, we found no evidence that CSB was able to translocate along these ssDNA or ssRNA molecules to release the stretpavidin (Supplemental Fig. 3). We also employed a standard triplex DNA displacement assay to determine whether CSB was able to translocate along dsDNA (see Materials and Methods). Briefly, the dsDNA substrate consists of a polypurine tract to which a shorter third polypyrimidine DNA strand is bound via Hoogsteen base pairing. Many, but not all, SWI/SNF2 family proteins have been demonstrated to translocate along dsDNA and release a triplex strand in an ATP dependent manner41. When CSB and ATP were incubated with the triplex substrate, no CSB dependent release of the third radiolabeled strand was observed in the presence of either Mg2+ (Fig. 5) or Ca2+ (data not shown), although in the presence of Ca2+ alone, the triplex was somewhat destabilized. In parallel reactions with Mg2+, we incubated a known dsDNA translocase Sacchromyces cerevisiae Rad54 (yRad54) with the triplex substrate plus ATP and were able to observe strand displacement, supporting the conclusion that human CSB is unable to translocate along dsDNA (Fig. 5). We note that the translocase assay with CSB was performed under conditions found to be optimal for CSB ATPase activity, and that CSB ATPase function was less robust under the conditions used for yRAD54.
Fig. 5. CSB is unable to use ATP hydrolysis for dsDNA translocation.
Representative gel of triplex displacement assay with yRad54 or CSB in the presence of 4 mM Mg2+. Triplex displacement by increasing concentrations of yRad54 (180 fmol to 1.8 pmol) is shown as a control (left). Reactions with increasing CSB (60 fmol to 1.8 pmol) were performed as described in Experimental Procedures. % triplex strand displacement is indicated below each lane.
dsDNA binding by CSB promotes removal or rearrangement of a protein pre-bound to DNA
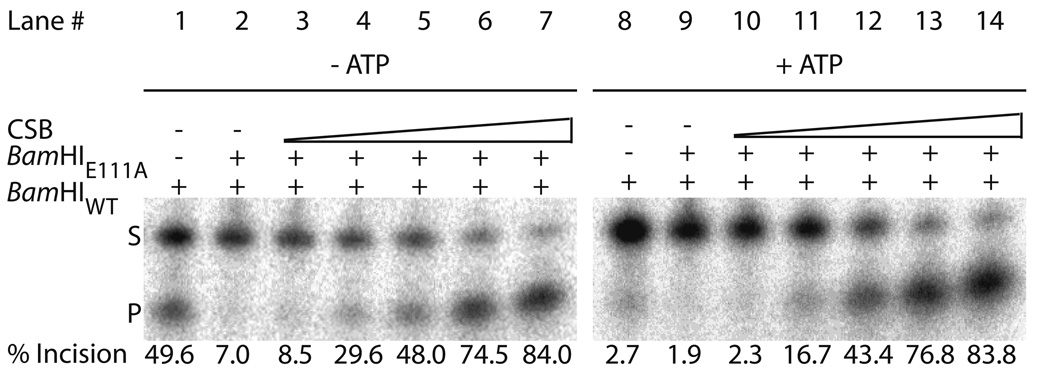
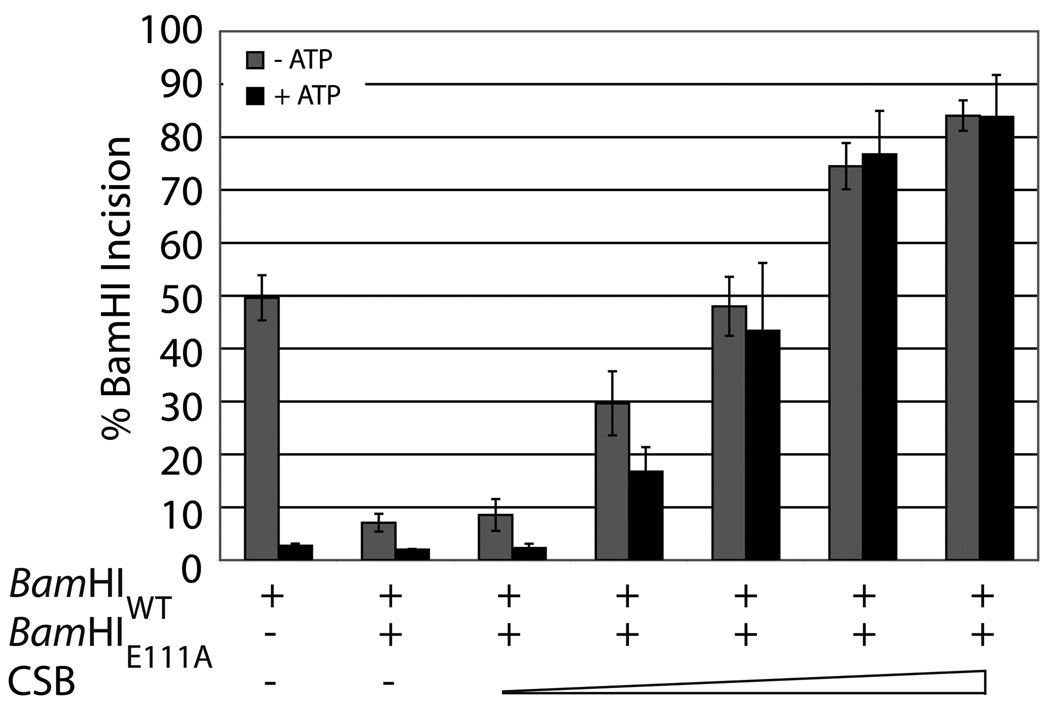
We next asked, in a heterologous system, whether DNA binding by CSB could perturb a stable protein:DNA complex. Prior studies on stand-alone SWI/SNF2 proteins (i.e. not large chromatin remodeling complexes) have indicated that they can remove specific proteins (TBP by Mot1 and Rad51 by Rdh54) from DNA in an ATP dependent manner42, 43. Towards this end, we obtained a mutant BamHI protein, BamHIE111A, which can bind its recognition sequence (5’-GGATCC-3’), yet lacks endonucleolytic activity44. In our experimental design, BamHIE111A was prebound in excess to a 42mer dsDNA duplex containing a single BamHI restriction site. When BamHI wild-type (BamHIWT) enzyme was added, endonuclease activity of BamHIWT was reduced to <10% in the presence of the prebound mutant protein (Fig. 6a, lanes 1 and 8, and Fig6b). When wild-type CSB was added in increasing amounts simultaneously with BamHIWT enzyme, increasing incision activity by the wild-type endonuclease was observed, indicating that CSB was able to remove or rearrange the binding interface of the prebound BamHIE111A to allow for cleavage (Fig. 6a, lanes 3–7). Notably, addition of ATP to the reaction did not have any appreciable effect, either positively or negatively, on the ability of CSB to activate BamHIWT endonuclease cleavage (Fig. 6a, lanes10–14). As ATP alone was found to inhibit BamHIWT activity (Fig 6a, lane 8) restoration of the cleavage event was likely mediated both by CSB dsDNA dependent ATP hydrolysis (i.e. removal of ATP) and by BamHIE111A protein displacement/rearrangement. The lack of a direct effect of ATP on CSB stimulation is seemingly consistent with the results obtained above, where DNA binding by CSB of the 34mer duplex was not affected by the presence of nucleotide (e.g. ATP, ADP, AMP-PNP).
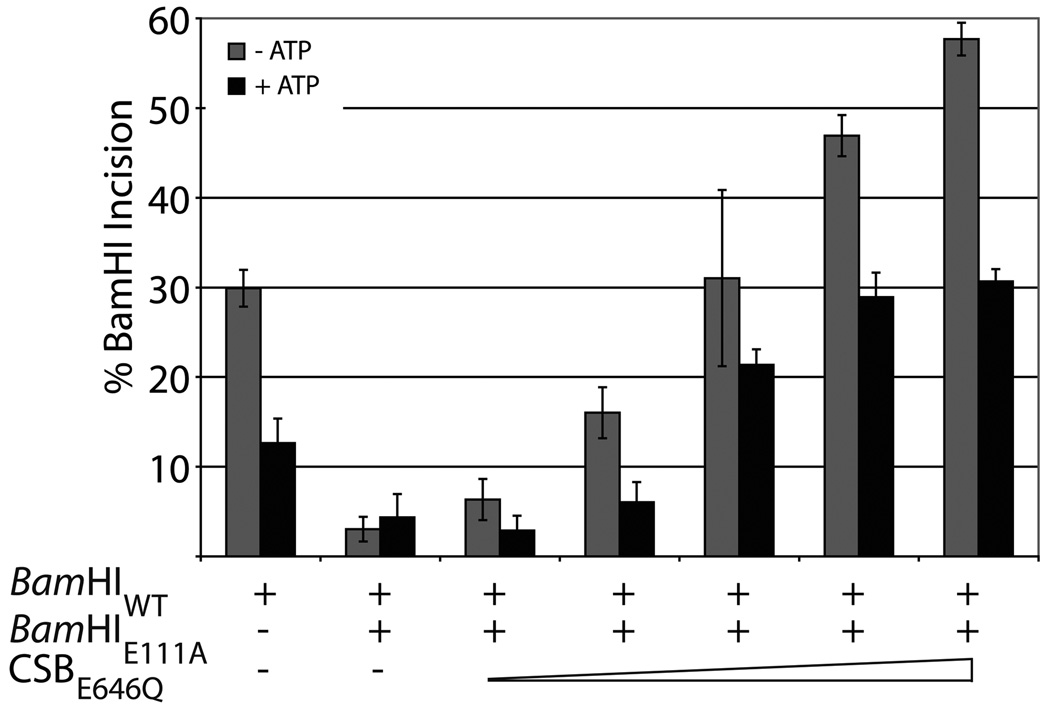
Fig. 6. CSB is able to displace or rearrange a pre-bound protein:dsDNA complex in an ATP independent manner.
(a) Representative gels of CSB concentration-dependent protein displacement/rearrangement monitored by BamHI digestion (Left panel, 0 mM ATP; Right panel, 2.5 mM ATP). See reaction condition details in Experimental Procedures. Shown above are respective lane numbers. Shown below is the average percent BamHIWT incision, following pre-binding of BamHIE111A, calculated from at least 3 independent experiments. (b) Graph quantitating percent BamHIWT incision in the presence of BamHIE111A and increasing concentrations of CSB (Grey, 0 mM ATP; Black, 2.5 mM ATP). (c) Graph quantitating percent BamHIWT incision in the presence of BamHIE111A and increasing concentrations of CSBE646Q (Grey, 0 mM ATP; Black, 2.5 mM ATP). Average values are plotted with standard deviations of at least 3 independent experiments. Note inhibition of BamHIWT activity upon ATP addition alone.
Utilizing the SF2 motif II mutant CSBE646Q protein, which can bind but not hydrolyze ATP29, we found that, in the absence of ATP, increasing concentrations of CSBE646Q were also able to restore BamHIWT cleavage activity (to levels similar to those seen with wild-type CSB), supporting that this property of CSB functions independent of ATP (Fig. 6c). In the presence of ATP, however, CSBE646Q was able to only partially relieve the inhibition by mutant BamHIE111A, suggesting that ATP binding (i.e. sequestration) in the absence of hydrolysis incompletely alleviated the inhibition of ATP on BamHIWT endonuclease activity. In all, the ability of CSB to remove or rearrange the binding interface of the prebound BamHIE111A does not depend on ATP binding or hydrolysis, as both wild-type CSB and CSBE646Q were able to promote BamHIE111A displacement/rearrangement in the absence and presence of ATP.
We also observed that, independent of ATP, CSB was able to stimulate BamHIWT restriction endonuclease activity (compare lane 1 to 7 or 8 to 14), somewhat reminiscent of the ability of CSB to stimulate Ape1 AP site incision activity20. We sought to determine whether this stimulation, as well as whether the CSB protein removal/rearrangement activity, was due to a direct CSB:BamHI physical interaction. Towards this end, we designed an in vitro pull down assay. Specifically, an α-His6 antibody was coupled and crosslinked to Protein A/G agarose, which allowed for capture of His6-tagged CSB protein. When Ape1 protein was incubated with the crosslinked α-His6 antibody Protein A/G agarose and CSB, we were able to detect a direct physical interaction between these two proteins, as evidenced by CSB-dependent pull-down (Supplementary Fig. 4, lane 12), consistent with previous findings20; Ape1 alone did not interact with the crosslinked α-His6 antibody Protein A/G agarose beads (Supplementary Fig. 4, lane 9). When BamHI was incubated with crosslinked a-His6 antibody Protein A/G agarose and CSB, we did not observe a direct physical interaction between the two proteins (Supplementary Fig. 3, lane 6), and BamHI did not interact with the crosslinked α-His6 antibody Protein A/G agarose (Supplementary Fig. 4, lane 3). These results strongly support that the CSB protein removal/rearrangement activity and its ability to stimulate BamHI restriction endonuclease activity is independent of a direct protein:protein interaction and likely the result of a CSB:DNA interaction.
To address whether CSB induces structural changes (i.e. bending or kinking) in DNA upon binding, and whether these structural changes, if observed, are dependent on the “motor” ATPase function of CSB, we carried out footprinting studies with the 34G/34C duplex substrate. Unfortunately, attempts to perform DNA footprinting in solution failed, as addition of chemical footprinting (e.g. iron-EDTA, H2O2, ascorbic acid) or crosslinking (gluteraldehyde, even as low as 0.01%) reagents disrupted the CSB:DNA complex (data not shown). This fact suggests that, at least in vitro, CSB DNA binding is sensitive to fluxes in metal ions, oxidation and/or charge, and limited our ability to utilize different solution-based footprinting strategies. We therefore employed an in situ footprinting technique. In these experiments, CSB-DNA binding reactions were resolved on a native polyacrylamide gel, and free and CSB bound nucleic acid was subjected to hydroxyl radical attack using 1,10-phenanthroline-copper [(OP)2Cu] footprinting reagents. These studies, however, did not uncover a difference in the digestion pattern of bound and unbound DNA, regardless of whether ATP or AMP-PNP was added to the binding reactions (Supplemental Fig. 5). We presume that the lack of a CSB binding footprint on DNA and the absence of (OP)2Cu hypersensitive/resistant sites is due to sequence independent binding by CSB and the fact that the protein does not induce a detectable structural change in dsDNA. This conclusion is consistent with previous experiments using DNaseI to interrogate the interaction of CSB with DNA, where no footprint or hypersensitive/resistant sites were observed30.
Discussion
CSB is a member of the SWI/SNF2 subgroup of SF2 ATPases/nucleic acid translocases/helicases based on sequence homology of its core ATP binding and hydrolysis motifs38, 39. It is a dsDNA dependent ATPase that operates over a wide physiological pH range and is operational in the presence of several divalent metal ions (Fig. 1). We examined herein ATP dependent and ATP independent biochemical activities of CSB, focusing on its interactions with and manipulation of nucleic acid.
Despite the fact that Mg2+ is generally considered to be the relevant metal cofactor for many ATPases in vivo, CSB ATPase function is significantly more active in the presence of Ca2+, regardless of the DNA cofactor (Fig. 2). To our knowledge, this is the first instance of Ca2+ promoting ATP hydrolysis in a Superfamily 1 or 2 (RecA-like domain containing) enzyme. In fact, the yeast homolog of CSB, Rad26, was found to require Mg2+ for ATP hydrolysis, but was unable to utilize Mn2+, Zn2+, Co2+, or Ca2+ as cofactors45. Characterization of the kinetic parameters of CSB dsDNA dependent ATPase activity indicates that Ca2+ exerts an effect on the enzyme turnover, increasing kcat ~ 3 fold. The significant effect of Ca2+ on human CSB ATPase function is noteworthy given that this divalent metal plays a prominent role in neuronal function and that CS patients characteristically exhibit neuroabnormalities1, 46. It is tempting to speculate that loss of CSB function would be most evident in cells where Ca2+ concentration is highest.
Previous analysis of nucleic acid cofactors for CSB catalyzed ATP hydrolysis indicated that dsDNA was a prerequisite and that CSB displayed a slight (2-fold) preference for cofactors containing a small (10 nt) bubble37. We report herein that alternate model substrates, e.g. DNA replication and transcription mimics, serve as cofactors for ATP hydrolysis by CSB, in the presence of either Ca2+ or Mg2+ (Fig. 2). Furthermore, while CSB had been shown to exhibit Mg2+ dependent, ATP independent DNA/DNA strand annealing activity32, we report that (i) this activity can occur independent of metal, yet is most efficient with Ca2+ present and slightly impaired by the addition of Mg2+ relative to the no metal control, and (ii) CSB can hybridize DNA/RNA (as well as RNA/RNA) duplexes, most efficiently in the presence of Ca2+. Although complementary DNA/DNA strand annealing activities have been ascribed to many human RecQ family bona fide helicases (RecQ147, WRN48, BLM49, and RecQ550), no in vivo functions for this activity – antagonistic to their DNA unwinding function – have been defined, and alternate nucleic acid preferences for these enzymes have not been reported. For CSB, it is conceivable that the DNA/DNA or DNA/RNA strand annealing activity plays a role: (a) in promoting the formation of duplex substrates during repair; (b) in facilitating the movement of the active bubble structure during transcription, akin to the transcription bubble rewinding by E. coli Mfd51; or (c) during homologous recombination, which requires complementary strand pairing at several phases.
Since nucleic acid binding requirements and the substrate range for SWI/SNF2 proteins have not been analyzed extensively, we evaluated the effect of metal cofactors, nucleotides, and substrate composition on CSB:DNA complex formation. EMSAs revealed that CSB bound stably to a diverse collection of nucleic acid substrates, and that inclusion of either Mg2+ or Ca2+ only slightly affected the affinity of CSB for dsDNA (Fig. 4). Moreover, addition of ATP, ADP, or AMP-PNP did not overtly affect the DNA binding properties of CSB, indicating that neither ATP binding nor hydrolysis are required for the stable interaction of CSB with DNA. Our data also indicate that CSB has a slight nucleic acid binding preference for dsDNA substrates that contain both single-stranded and double-stranded regions, such as the model replication and transcription bubbles (54FE/54C/18D and 54FE/54C/18R), implying that CSB is active, both for complex formation and ATP hydrolysis, at sites where these conformations are formed in vivo. Furthermore, it had been shown previously that DNA/RNA hybrid duplexes and RNA molecules are poor cofactors for ATP hydrolysis by CSB28. We demonstrate here that CSB is able to stably interact with DNA/DNA, DNA/RNA, and RNA/RNA forked duplexes in EMSAs, suggesting that CSB may impart function, independent of ATP hydrolysis, via binding to nucleic acid substrates. The indiscriminate manner in which CSB is able to interact with a diverse array of nucleic acids implies a wide range of activities in vivo and is consistent with CSB being implicated in a variety of cellular processes (e.g. TC-NER, BER, and transcription).
It is clear that under the conditions examined in the studies herein, CSB is unable to separate nucleic acid strands. Whereas previous studies tested simple DNA/DNA substrates for CSB helicase activity11, 28, we examined many nucleic acid combinations/structures for potential ATP dependent helicase and nucleic acid translocase functions (see examples in Supplemental Fig. 1–3, and Fig. 5). Given that the CSB ATPase core (i.e. Motifs I through VI) primary amino acid sequence, secondary structure, and predicted tertiary structure falls under the SF2 SWI/SNF2 subgroup and that, to date, no SWI/SNF2 family member has been shown to possess ATP dependent unwinding or single-strand translocase activities, the absence of these properties for CSB was not surprising. What was unexpected, however, was that CSB apparently lacks dsDNA translocase activity under conditions in which it is active as an ATPase. Indeed, many SWI/SNF2 members possess the ability to translocate along dsDNA in an ATP dependent manner; including FancM (mutated in Fanconi Anemia)52, Rad54 (involved in recombination)53, Rdh54 (a yeast Rad54 paralog)54, and SSO1653 (an archaeal SWI/SNF2 protein)55; and the chromatin remodeling motors SWI/SNF56, RSC57, and ISWI58. That said, other SWI/SNF members appear to lack classic translocase activity. For example, S. cerevisiae Mot1p (a homolog of human BTAF1) uses energy from ATP hydrolysis to “grip” DNA proximal to promoters, causing dissociation of TBP, consequently inhibiting transcription initiation42. Thus, it is not unprecedented that SWI/SNF2 members can use the energy from ATP hydrolysis for activities other then movement along dsDNA59. Moreover, modeling of the CSB core using the Sulfolobus solfataricus SWI/SNF2 co-crystal structure with DNA (PDB ID: 1Z63) indicates that the amino acid side chain contacts seen between the SSO1653 ATPase domain and DNA are likely not the same for a CSB-DNA complex (Supplemental Fig. 6). This observation implies that, at least for the ATPase core, these two proteins interact with nucleic acid differently and may in part explain why the biochemical properties of CSB differ from this and other SWI/SNF2 family members.
We explored herein if CSB possessed the ability to remove or rearrange a protein bound to DNA. CSB was indeed able to dissociate or reorganize BamHIE111A that was pre-bound to its cognate site, most likely through an interaction with DNA, as no direct physical association could be detected between CSB and the restriction enzyme (Fig. 6 and Supplementary Fig. 4). Intriguingly, we found that this property of CSB was ATP independent, and not affected by ATP binding or hydrolysis. Other SWI/SNF2 proteins that remodel chromatin (SWI/SNF, RSC, ISWI, and Rad54 among others) and those that remove specific proteins from DNA (Mot1p (BTAF)) require ATP for their protein displacement activity. Nonetheless, the ATP independent protein removal/rearrangement activity of CSB is likely important in (i) DNA repair to uncover occluded lesions and allow for the repair machinery to recognize and excise the target damage, (ii) transcription to clear a path for RNAP elongation, and (iii) transcription initiation to promote binding of trans acting initiation factors to their regulatory elements. Studies are underway to delineate the precise ATP dependent and independent tasks of CSB, with a focus on the potential impact of post-translational modifications, other protein binding partners, and the unique N-(amino acids 1 to 501) and C-terminal (amino acids 1013 to 1493) regions of CSB in modulating or directing protein function in vivo and in vitro. Such studies are imperative given the recent evidence that a normally suppressed triplex displacement activity of full-length E. coli Mfd is activated upon binding to RNAP or following deliberate removal of an auto-inhibitory domain60.
Materials and Methods
Proteins
Recombinant wild-type and E646Q CSB proteins were expressed in Sf9 insect cells via a baculoviral system as previously described28. Purification was performed by Ni2+ affinity, heparin, and typically anion exchange (source Q) chromatography, which allowed for isolation of protein to near homogeneity. The BamHI mutant protein E111A (BamHIE111A)44 was graciously provided by Drs. Michael Dalton and Lydia Dorner of New England Biolabs (Ipswitch, MA). Saccharomyces cerevisiae Rad54 (yRad54) was kindly provided by Dr. Patrick Sung (Yale University, New Haven, CT). Wild-type BamHI and T4 polynucleotide kinase were purchased from New England Biolabs.
Oligonucleotides
Oligonucleotides for complementary strand annealing, helicase, and single-strand nucleic acid translocase assays 61 were a gift from Dr. Zvi Kelman (University of Maryland Biotechnology Institute, Rockville, MD) and/or purchased from IDT (Coralville, IA). (TC)20 oligonucleotide was a kind gift from Dr. Weidong Wang (National Institute on Aging, Baltimore, MD), as was plasmid pBSCR52.
ATPase Assay
Buffers for standard ATPase reactions contained 20 mM HEPES-OH pH 8.0, 0.05 mM ATP, 40 µg/mL BSA, and 1 mM DTT. Reactions were supplemented with 1 or 4 mM divalent metal as indicated. ATPase reactions employed CSB and 12.5 µCi γ-32P ATP. Reactions were incubated for 1 hr at 30°C with 150 ng of nucleic acid substrate. Reactions were stopped by the addition of 0.5 M EDTA. ATP hydrolysis was analyzed by Polyethyleneimine-thin layer chromatography (PEI-TLC) using 1 M formic acid/0.8 M LiCl. PEI-TLC plates were analyzed and quantitated on a Typhoon phosphoimager using OptiQuant TL software. Reactions for time course kinetics were performed as above, except that 10 µL aliquots were taken at the time points specified. For kinetic parameter determinations, CSB (150 ng, 900 fmol, 90 nM) was incubated with varying amounts of unlabeled ATP (500, 100, 50, 10, 5, 1 µM) for 1 hour at 30°C and processed as above. Km and Vmax parameters were calculated from Lineweaver-Burk plots.
Complementary strand annealing assay
Oligonucleotides (DNA50 or RNA50) were labeled at the 5’ end with 32P. Unincorporated γ-32P ATP was removed by centrifugation through a Bio-Rad micro bio spin P30 column. Labeled oligos were stored in 20 mM HEPES-OH pH 8.0. Buffers for strand annealing contained 20 mM HEPES-OH pH 8.0, 40 µg/mL BSA, 1 mM DTT and either no metal, 1 mM Ca2+ or 4 mM Mg2+. DNA/DNA, DNA/RNA, and RNA/RNA strand pairing reactions contained 100 fmol each DNA50 plus DNA49, DNA50 plus RNA49, or RNA50 plus RNA49 oligonucleotides, respectively, and 75 ng (450 fmol, 20 nM) CSB. Reactions were kept on ice and initiated by the addition of CSB and incubation at 30°C for the times specified. Reactions were stopped by the addition of Stop buffer containing 35 mM EDTA, 0.6% SDS, 25% glycerol, 0.05% bromphenol blue and 0.05% xylene cyanol. Reactions were run on 12% native polyacrylamide-TBE gels at 160 V for 2 hr. Substrates and products of strand pairing reactions were visualized and quantitated by phosphoimager analysis.
EMSA
Buffers for EMSA reactions contained 20 mM HEPES-OH pH 8.0, 50 mM KCl, 200 µg/mL BSA, 5 mM DTT, and 5% (v/v) glycerol. Where mentioned, 1 mM Ca2+ or 4 mM Mg2+, and 2.5 mM ATP, ADP, or AMP-PNP final concentration, were added. Binding reactions contained 50 fmol of substrate and 15, 50, 150, 250, 350, or 500 fmol CSB. Binding reactions were incubated at 30°C for 30 min, then immediately loaded onto a native 4% polyacrylamide-TBE gel and run at 15 mA for 90 min at 4°C. Substrates and products were visualized and quantitated by phosphoimager analysis as above. Nucleic acid saturation binding curves were plotted (% substrate bound vs. nM CSB concentration) and KD values were calculated by non-linear curve fitting using the Hill equation in Origin 7.0 (Northampton, MA).
dsDNA translocation assay
(TC)20 was labeled at the 5’end with 32P. Plasmid pBSCR was linearized by digesting with ScaI. Triplex substrate was created by incubating 10 pmol labeled (TC)20 oligonucleotide with 15 pmol linearized pBSCR plasmid in Triplex buffer (33 mM Tris-Acetate pH 5.5, 66 mM Potassium acetate, 100 mM NaCl, 10 mM MgCl2, and 0.4 mM Spermine), and incubated for 1 hr at 56°C and then overnight at room temperature. Unincorporated γ-32P ATP and unannealed (TC)20 were removed via centrifugation as above. Labeled substrate was stored in 33 mM Tris-Acetate pH 5.5, 5 mM MgCl2. Buffers for CSB triplex displacement assays contained 20 mM HEPES-OH pH 8.0, 40 µg/mL BSA, 1 mM DTT, 2.5 mM ATP and either 4 mM Mg2+ or 1 mM Ca2+. Buffer for yRad54 triplex displacement contained 40 mM Tris-HCl pH 7.4, 25 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 2% glycerol, and 100 ng/µL bovine serum albumin. Increasing concentrations of CSB [10 ng (60 fmol, 3 nM), 30 ng (180 fmol, 9 nM), 100 ng (600 fmol, 30 nM), and 300 ng (1.8 pmol, 90 nM)] or yRad54 [18 ng (180 fmol, 9 nM), 60 ng (600 fmol, 30 nM) and 180 ng (1.8 pmol, 90 nM) were incubated with 100 fmol triplex substrate at 30°C for 1 hr. Reactions were stopped by the addition of Stop buffer containing 0.9% SDS, 40% glycerol, 0.1% bromphenol blue, and 0.1% xylene cyanol. Reactions were run on 12% polyacrylamide-Tris-acetate-MgCl2 gel containing 25% glycerol at 160 V for 4–5 hr. Substrate and product were visualized and quantitated by phosphoimager analysis as above.
Restriction enzyme site accessibility assay
Buffers for restriction enzyme site accessibility assays contained 20 mM HEPES-OH, pH 8.0, 50 mM KCl, 200 µg/mL BSA, 5 mM DTT, and 5% (v/v) glycerol and 4 mM Mg2+. 50 fmol of substrate (42BamHI/42Comp) was incubated with 400 ng (20 pmol, 850 nM) BamHIE111A at 30°C for 15 min with or without 2.5 mM ATP; we note that ATP addition had no effect on the final pH of the reaction mixture. Subsequently, CSB or the CSB SF2 motif II mutant CSBE646Q (50, 150, 250, 350, or 500 fmol) were added to the reactions, followed immediately by addition of 1 U BamHI wild-type (BamHIWT) enzyme and incubation at 30°C for 30 min. Stop buffer (95% formamide, 20 mM EDTA, 0.5% bromphenol blue, and 0.5% xylene cyanol) was then added and samples were heated at 95°C for 3 min and then loaded onto a 15% polyacrylamide-urea denaturing gel and electrophoresed at 225 V for 90 min. Substrates and products were visualized and quantitated by phosphoimager analysis using OptiQuant TL.
Supplementary Material
Acknowledgements
We would like to thank Dr. Vilhelm Bohr (NIA/NIH) for support, comments, and helpful discussions; Dr. Wei Yang (NIDDK/NIH) for assisting with homology modeling and helpful comments on the manuscript; Dr. Zvi Kelman (CARB/UMBI) for providing oligonucleotides for helicase, ss-nucleic acid translocase, and complementary strand pairing assays; Drs. Weidong Wang and Yutong Xue (NIA/NIH) for providing DNAs for triplex displacement assays; Drs. Michael Dalton and Lydia Dorner (NEB) for supplying BamHIE111A protein; and Dr. Patrick Sung (Yale) for providing yRad54 for triplex displacement assays. We thank Jason Piotrowski (NIA/NIH) for initial help with CSB protein purification. We thank Dr. P.J. Brooks (NIDDK/NIH), Dr. Robert Brosh Jr. (NIA/NIH) and Maria Aamann (NIA/NIH) for critical input on the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7:834–848. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vermeulen W, van Vuuren AJ, Chipoulet M, Schaeffer L, Appeldoorn E, Weeda G, Jaspers NG, Priestley A, Arlett CF, Lehmann AR. Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH): evidence for the existence of a transcription syndrome. Cold Spring Harb. Symp. Quant. Biol. 1994;59:317–329. doi: 10.1101/sqb.1994.059.01.036. [DOI] [PubMed] [Google Scholar]
- 4.Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AM. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colella S, Nardo T, Botta E, Lehmann AR, Stefanini M. Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-cacchione variant of xeroderma pigmentosum. Hum. Mol. Genet. 2000;9:1171–1175. doi: 10.1093/hmg/9.8.1171. [DOI] [PubMed] [Google Scholar]
- 6.Meira LB, Graham JM, Jr, Greenberg CR, Busch DB, Doughty AT, Ziffer DW, Coleman DM, Savre-Train I, Friedberg EC. Manitoba aboriginal kindred with original cerebro-oculo- facio-skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am. J. Hum. Genet. 2000;66:1221–1228. doi: 10.1086/302867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M, Tanaka K. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Troelstra C, van GA, de WJ, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 9.Troelstra C, Hesen W, Bootsma D, Hoeijmakers JH. Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res. 1993;21:419–426. doi: 10.1093/nar/21.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orren DK, Dianov GL, Bohr VA. The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res. 1996;24:3317–3322. doi: 10.1093/nar/24.17.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 12.Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradsher J, Auriol J, Proietti De SL, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 14.van Gool AJ, Citterio E, Rademakers S, van OR, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer N, Reagan MS, Wu KJ, Canagarajah B, Friedberg EC. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 16.Wang XW, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly JM, Wang Z, Freidberg EC, Evans MK, Taffe BG. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat. Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 17.Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo JP, Khaw MC, Saijo M, Kodo N, Matsuda T, Hoeijmakers JH, Tanaka K. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J. Biol. Chem. 2000;275:34931–34937. doi: 10.1074/jbc.M004936200. [DOI] [PubMed] [Google Scholar]
- 18.Thorslund T, von KC, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuo J, Chen C, Zeng X, Christiansen M, Bohr VA. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 2002;1:913–927. doi: 10.1016/s1568-7864(02)00116-7. Ref Type: Journal. [DOI] [PubMed] [Google Scholar]
- 20.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., III Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 22.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 23.Brosh RM, Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De SL, Bohr VA. The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol. Biol. Cell. 1999;10:3583–3594. doi: 10.1091/mbc.10.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunesen M, Selzer RR, Brosh RM, Jr, Balajee AS, Stevnsner T, Bohr VA. Molecular characterization of an acidic region deletion mutant of Cockayne syndrome group B protein. Nucleic Acids Res. 2000;28:3151–3159. doi: 10.1093/nar/28.16.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade MH, Chu EH. Effects of DNA damaging agents on cultured fibroblasts derived from patients with Cockayne syndrome. Mutat. Res. 1979;59:49–60. doi: 10.1016/0027-5107(79)90194-5. [DOI] [PubMed] [Google Scholar]
- 26.Rainbow AJ, Howes M. A deficiency in the repair of UV and gamma-ray damaged DNA in fibroblasts from Cockayne’s syndrome. Mutat. Res. 1982;93:235–247. doi: 10.1016/0027-5107(82)90138-5. [DOI] [PubMed] [Google Scholar]
- 27.de WH, de WJ, Gorgels TG, van den AG, Andressoo JO, Vermeij M, van SH, Hoeijmakers JH, van der Horst GT. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2003;2:13–25. doi: 10.1016/s1568-7864(02)00188-x. [DOI] [PubMed] [Google Scholar]
- 28.Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 29.Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM, Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Citterio E, van dBV, Schnitzler G, Kanaar R, Kanaar E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J. Biol. Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 32.Muftuoglu M, Sharma S, Thorslund T, Stevnsner T, Soerensen MM, Brosh RM, Jr, Bohr VA. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006;34:295–304. doi: 10.1093/nar/gkj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muftuoglu M, Selzer R, Tuo J, Brosh RM, Jr, Bohr VA. Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene. 2002;283:27–40. doi: 10.1016/s0378-1119(01)00870-8. [DOI] [PubMed] [Google Scholar]
- 35.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 36.Lebedev A, Scharffetter-Kochanek K, Iben S. Truncated Cockayne Syndrome B Protein Represses Elongation by RNA Polymerase I. J. Mol. Biol. 2008 doi: 10.1016/j.jmb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 38.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 41.Durr H, Hopfner KP. Structure-function analysis of SWI2/SNF2 enzymes. Methods Enzymol. 2006;409:375–388. doi: 10.1016/S0076-6879(05)09022-1. [DOI] [PubMed] [Google Scholar]
- 42.Sprouse RO, Sprouse M, Auble DT. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J. 2006;25:1492–1504. doi: 10.1038/sj.emboj.7601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi P, Kwon Y, Seong C, Epshtein A, Lam I, Sung P, Klein HL. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J. Biol. Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- 44.Dorner LF, Schildkraut I. Direct selection of binding proficient/catalytic deficient variants of BamHI endonuclease. Nucleic Acids Res. 1994;22:1068–1074. doi: 10.1093/nar/22.6.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. RAD26, the yeast homolog of human Cockayne’s syndrome group B gene, encodes a DNA-dependent ATPase. J. Biol. Chem. 1996;271:18314–18317. doi: 10.1074/jbc.271.31.18314. [DOI] [PubMed] [Google Scholar]
- 46.Mattson MP. Calcium and neurodegeneration. Aging Cell. 2007;6:337–350. doi: 10.1111/j.1474-9726.2007.00275.x. [DOI] [PubMed] [Google Scholar]
- 47.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 48.Machwe A, Lozada EM, Xiao L, Orren DK. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC. Mol. Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J. Biol. Chem. 2007;282:30776–30784. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- 55.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auble DT, Steggerda SM. Testing for DNA tracking by MOT1, a SNF2/SWI2 protein family member. Mol. Cell Biol. 1999;19:412–423. doi: 10.1128/mcb.19.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin JH, Kelman Z. The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates. J. Biol. Chem. 2006;281:26914–26921. doi: 10.1074/jbc.M605518200. [DOI] [PubMed] [Google Scholar]
- 62.Wilson DM., III Ape1 abasic endonuclease activity is regulated by magnesium and potassium concentrations and is robust on alternative DNA structures. J. Mol. Biol. 2005;345:1003–1014. doi: 10.1016/j.jmb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 63.Berquist BR, McNeill DR, Wilson DM., III Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J. Mol. Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.