Abstract
Mistic, a bacterial membrane-associating protein family, uniquely found in Bacillus species. It enhances expression of eukaryotic membrane proteins at the bacterial membrane. Mistic from B. subtilis (M110), expresses at the E. coli membrane, however its shorter orthologs have been recently shown to be mainly cytoplasmic with varying membrane affinities. Based on that, we hypfothesized that the expression level of membrane proteins fused to Mistic is correlated with the degree of membrane association of the particular Mistic protein. We compared expression levels by various Mistic proteins as fusion partners for the Aplysia californica Kv1.1 (aKv1.1) channel as a cargo membrane protein. Mistic from B. atrophaeus (M4), which has the highest membrane association among the shorter orthologs, enhanced expression of the transmembrane domain of aKv1.1 to the highest extent. In contrast, M1, which consists of the 84 C-terminal amino acids of M110 is the most soluble protein and showed the least capacity to express the channel. A chimeric Mistic, constructed with the first α-helix (H1) of M110 N-terminally fused to M4, did not increase the level of expression of aKv1.1 beyond those of either the M110 or the M4 fusions. The channel fused to M110, M4 or the aforementioned H1-M4 chimera, expresses in the highest quantity and quality among Mistic proteins, providing suitable sample for structural studies. Our data support the concept that expression levels of 'Misticated' membrane proteins are related to the independent chaperoning character of Mistic via direct membrane association, rather than related to specific sequence-dependent interaction with the E. coli translocon machinery.
Keywords: Mistic, membrane proteins, potassium channel, overexpression
Integral membrane proteins (IMPs) constitute approximately a quarter of any eukaryotic or prokaryotic proteome. Irrespective of their prevalence, structural characterization of IMPs has been very limited due to the challenges arising from the hydrophobic nature of their transmembrane (TM) domains. Although some eukaryotic IMPs, such as rhodopsin and Ca2+ ATPase, are found in high amounts in native biological membranes, most membrane proteins do not normally exist in such abundance. With the advent of the vast amount of genomic information and major advances in high-throughput structural biology techniques, there is accordingly an increasing demand for more robust techniques of expressing of IMPs to advance structural knowledge about eukaryotic IMPs.
The first major obstacle for structural studies of IMPs is the production of large amounts of properly folded protein. Unlike soluble proteins, highly hydrophobic membrane proteins require auxiliary proteins to prevent their aggregation during the protein folding process [4, 5]. It is thus likely that production of eukaryotic IMPs in bacteria is limited compared to that of soluble proteins, due to the additional complexity involved in the biogenesis of IMPs, coordinated by the ribosome and the SecYEG translocon assisted by chaperones such as SecB and the ATPase motor SecA, some of which may be needed to be co-overexpressed and/or impose additional specificity [6–10].
The next obstacle is to isolate the IMP from the membrane for further purification, which typically requires screening for the proper detergent to allow the formation of a stable mono-dispersed protein-detergent-complex (PDC). Finally, once a pure PDC is obtained (by purification techniques similar to those applied for soluble proteins), an extensive search for a detergent or a mixture of detergents with or without lipids is often needed to promote successful crystallization of the IMP in the PDC. Overcoming the above barriers for successful crystallization of IMPs requires much larger protein quantities on average, although ironically their expression yields are generally much lower than those for soluble proteins. This difference entails the challenge in structural biology of IMPs, and highlights the importance for the development of high-yield expression systems for IMPs.
Mistic is a membrane-associated protein originally discovered in Bacillus subtilis [11]. It was shown to enhance expression levels of a large number of foreign IMPs at the membrane of E. coli, when used as a fusion partner linked to the N-termini of cargo proteins [11–15]. Interestingly, Mistic lacks identified signal sequence and its overexpression, whether alone or in fusion to other IMPs, apparently lacks the toxicity issues associated with overloading the protein translation machinery, and thus is likely to bypass the E. coli translocon complex [11]. Other Mistic proteins have been identified in Bacillus species only, and they are 26 amino acids shorter than the Mistic originally identified from B. subtilis [12], referred to as M110. The discovery of shorter Mistic orthologs provided new opportunities for optimizing expression of IMPs, given that they were all shown to serve as fusion partners for expression of IMPs at the bacterial membrane [12]. Surprisingly, it was recently shown that, unlike M110, the shorter Mistic proteins are soluble in the absence of detergents and they are mainly cytoplasmic, but show significantly varying membrane association propensities [16]. In accordance with previous suggestion on the independent ability of Mistic to facilitate membrane expression of IMPs, that study indicates that isolated Mistic proteins may spontaneously insert into the membrane. Based on that, we hypothesized that the degree of membrane association displayed by the individual Mistic proteins [16], is correlated to their ability to enhance expression of IMPs at the bacterial membrane.
Despite many efforts, eukaryotic voltage-gated potassium channels have not been successfully expressed in bacteria unless fused to Mistic. To test our hypothesis, we have chosen the Aplysia californica Kv1.1 (aKv1.1) channel, which showed no basal level of expression in E. coli, as a cargo IMP for the comparison of its expression by various Mistic fusion partners. We report on the capacity of various Mistic orthologs in assisting expression of aKv1.1 in the bacterial membrane, which enabled us to achieve large quantities of pure and stable ‘Misicated’ eukaryotic IMP. Finally we discuss Mistic’s mechanistic roles in chaperoning the insertion of IMPs fused to its C-terminus into the lipid-bilayer membrane.
Materials and Methods
Cloning and Purification
The aKv1.1 channel or its six-transmembrane (6TM) domain (residues 194–444) were cloned in fusion to the C-terminus of Mistic in Gateway-adapted (Invitrogen, Carlsbad, CA) expression vectors, containing N-terminal His-tag separated from Mistic by a thrombin cleavage site and another thrombin site between Mistic and the downstream aKv1.1 channel. The various Mistic sequences tested were shuffled up stream to the channel, as described in Figure 1. Freshly transformed E. coli BL21(DE3) cells (Invitrogen, Carlsbad, CA) were grown in Terrific Broth to an approximate optical density (OD) of 1.0 in 20 ml culture. Protein production was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 18°C. Cultures were harvested by centrifugation at 3000×g for 15 min and cell pellets were resuspended in Lysis Buffer (25 mM Tris pH 8.0, 20 mM imidazole, 150 mM KCl, 1 mM phenylmethanesulphonylfluoride [PMSF], 1 mM benzamidine, 10 mM β-mercaptoethanol [BME] and 10% glycerol). Lysis was carried out by sonication, and the crude lysate was spun down by centrifugation at 100,000×g for 2 hours. The membranes-containing pellets were solubilized in Lysis Buffer containing 0.5% sodium-dodecyl-sulfate (SDS) for 5 hours at room temperature and centrifuged at 30,000×g for 30 min to pellet the inclusion bodies. The SDS-soluble fraction was then bound to Ni-NTA beads (Qiagen, Hilden, Germany), pre-washed in the solubilization buffer, and samples were eluted with the Lysis buffer containing 400 mM imidazole and 0.5% SDS.
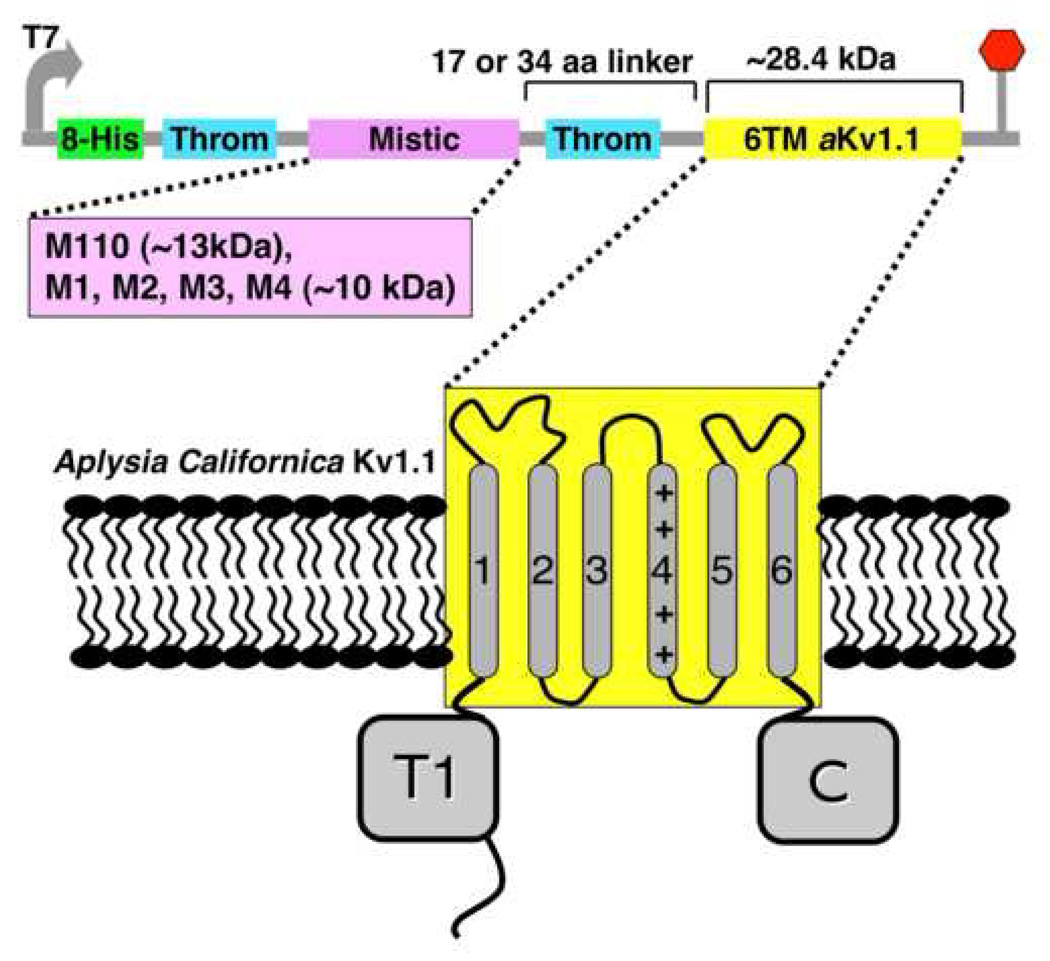
Figure 1.
Schematics of the construction of aKv1.1 fused to Mistic. The vector with the short linker is called pMis4.0 and the long linker vector is referred to as pMis3.0. The 6TM domain of aKv1.1 (yellow) is cloned downstream of Mistic. The first thrombin (Throm) cleavage site shown on the vector just downstream of the His-Tag was designed for the purpose of purifying ‘Misticated’ IMPs lacking the second site which was introduced here upstream of our channel for separation from Mistic. The short Mistic proteins were introduced into the pMis4.0 vector replacing M110 and are thus called pM1_4.0, pM2_4.0 and so on, corresponding to the M1, M2 through M4 Mistic sequences, respectively.
For the large cultures (1 L), the cells were lysed efficiently with a Microfluidizer (Microfluidics, Newton, MA USA) and purified similarly, except that the inclusion bodies were spun down prior to the detergent solublization step by centrifugation at 30,000×g for 20 min. For solubilization, we used 8 mM of the detergent lauryl-dimethylamine-N-oxide (LDAO) instead of SDS. For the affinity chromatography, only 5 mM LDAO and no PMSF or binzamidine were used. Before the size-exclusion chromatography (SEC) and light-scattering analysis, samples eluted from NiNTA column were digested with bovine thrombin (Sigma-Aldrich, Inc. St. Louis, MO) during dialysis against 20 mM Tris pH 8.0, 150 mM NaCl, 1mM DTT and 5mM LDAO. Completely cleaved proteins were concentrated using 30K MWCO Vivaspin conentrators (Vivascience AG, Hannover, Germany), and further purified by SEC on 10/30HR Superdex-200 column (Amersham Pharmacia Biotech, Piscataway, NJ), using 3 mM LDAO, 20 mM Tris pH 8.0, 200 mM NaCl and 1 mM DTT as the running buffer.
SDS-PAGE and Western Analysis
Purity and expression levels were analyzed by NuPAGE® Novex 4–12% Bis-Tris gels (Invitrogen, Carlsbad, CA), with MES or MOPS as running buffers. For the Western analysis, the gel was blotted onto a PVDF membrane by applying a constant current of 60 mA for 6 hours using, as a transfer buffer, 10 mM CAPS pH 11.0, 10% methanol and 1 mM DTT. The membrane was blocked with 3% BSA and 0.05% Tween in TBS (25 mM Tris pH 8.0, 125 mM NaCl). Mouse anti-His-tag antibody (Qiagen, Hilden, Germany) was used as the primary antibody by diluting a stock (0.2 mg/ml) 1/2000 in the blocking solution. Goat-anti-Mouse HRP conjugated secondary antibody (Bio-Rad, Hercules, California) was used similarly but with 1% gelatin instead of BSA for blocking. Colorimetric detection was performed using the Immuno-Blot HRP® (4CN) assay kit (Bio-Rad, Hercules, California) containing the substrate 4-chloro-1-naphthol with a sensitivity of 500 pg.
Light Scattering
Static light-scattering measurements were performed using Wyatt miniDAWN (Wyatt Technology, Santa Barbara, CA) coupled with a G3000Swxl (30 cm) column (Tosoh Bioscience, Montgomeryville, PA), using 20 mM Tris pH 7.0, 200 mM NaCl, 3 mM LDAO and 1 mM DTT as the running buffer. The system was also connected to a Wyatt Optilab rEX refractometer to measure the concentrations of the PDCs. By equating concentration of the PDC measured by the refractometer and of the protein in PDC (measured by UV at 280) and using respectively 0.187 and 0.148 as protein and detergent dn/dc values [17, 18], the protein and the detergent's molar masses in the PDC could be readily derived by fitting the Zimm plots using the software ASTRA V (Wyatt Technology, Santa Barbara, CA).
RESULTS AND DISCUSSION
Negative Charge and Membrane interaction
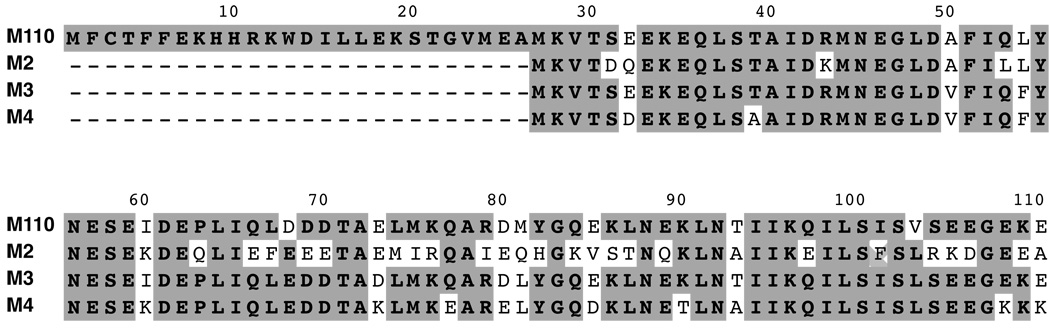
The highly acidic nature of the 110 aa-long Mistic from Bacillus subtilis (M110) with a net charge of −12.0 at pH 7.0 (based on pKa values of the isolated amino acids of its sequence), had been suggested to be central for its tight association with the lipid-bilayer membrane, whether as an isolated protein or as a fusion partner expressed recombinantly in E. coli [12]. The shorter Mistic proteins corresponding to only the C-terminal 84 aa of M110 are also highly acidic. The protein produced by the C-terminal 84 aa sequence of M110 (referred to as M1); and Mistic from B. leicheniformis (referred to as M2), B. mojavensis (referred to as M3) and from B. atrophaeus (referred to as M4) have the following approximate charge values (based on pK values of isolated aa residues), −12.0, −10.0, −12.0 and −7.0 at pH=7.0, respectively. The alignment of these Mistic sequences indicates their high similarity, with 63% identity between M1 and M2, 93% identity between M1 and M3 and 83% identity between M1 and M4 (Box 1).
Box 1.
Sequence alignment of Mistic proteins. Identical residues are shaded.
Remarkably, the shorter Mistic proteins are significantly more soluble than M110 when expressed independently in E. coli, all of which are mainly localized in the cytoplasm and do not require detergent for solubility [16]. The rank order of solubility is as follows, M1~M3 > M2 >> M4 (>M110) such that M4 interacts the most strongly with the membrane similar to the insoluble M110 except that it exists in the cytoplasm as well. In fact, the tendency of the short Mistic proteins to interact with the membrane is negatively correlated to their estimated net charge, thus, it is unlikely that the high acidic nature of Mistic is responsible for its tight interaction with the membrane as was previously suggested. These observations raised a question whether the membrane affinity expressed by intact (non-fused) Mistic proteins is correlated to their ability to aid overexpression of IMP at the bacterial membrane, as fusion partners.
Full-length versus the Transmembrane Domain of aKv1.1
To compare the capacity of the various Mistic orthologs to enhance expression of IMPs, we have searched for a eukaryotic IMP that does not express in bacteria. We found the potassium channel aKv1.1 suitable for this purpose, since it could not be detected in the membrane fraction or in the inclusion bodies when expressed recombinantly in E. coli, but expressed in large amounts (~2mg/L culture) in fusion to M110 (Figure 2). Expression of the full-length aKv1.1 fused to M110 was detectable previously by Coomassie-blue stained gel, but its six-transmembrane (6TM) domain (referred to as aKv1.1-6TM) fused to M110 appears as more pure and stable protein (Figure 1, Figure 2). Unlike the full-length aKv1.1, the 6TM domain fusion may have more intimate interaction with the C-terminus of M110, since it is resistant to cleavage by thrombin. This observation is unique to the fusion with aKv1.1-6TM, and has not been reported for other IMPs fused to Mistic. On the basis of the high purity and stability obtained after Ni+2 and SEC purification, we chose the aKv1.1-6TM channel for comparison of expression by the various Mistic proteins. Our detergent screen (data not shown) for solubilization of 'Misticated' aKv1.1 implicated LDAO suitable for extraction of the channel from E. coli membranes.
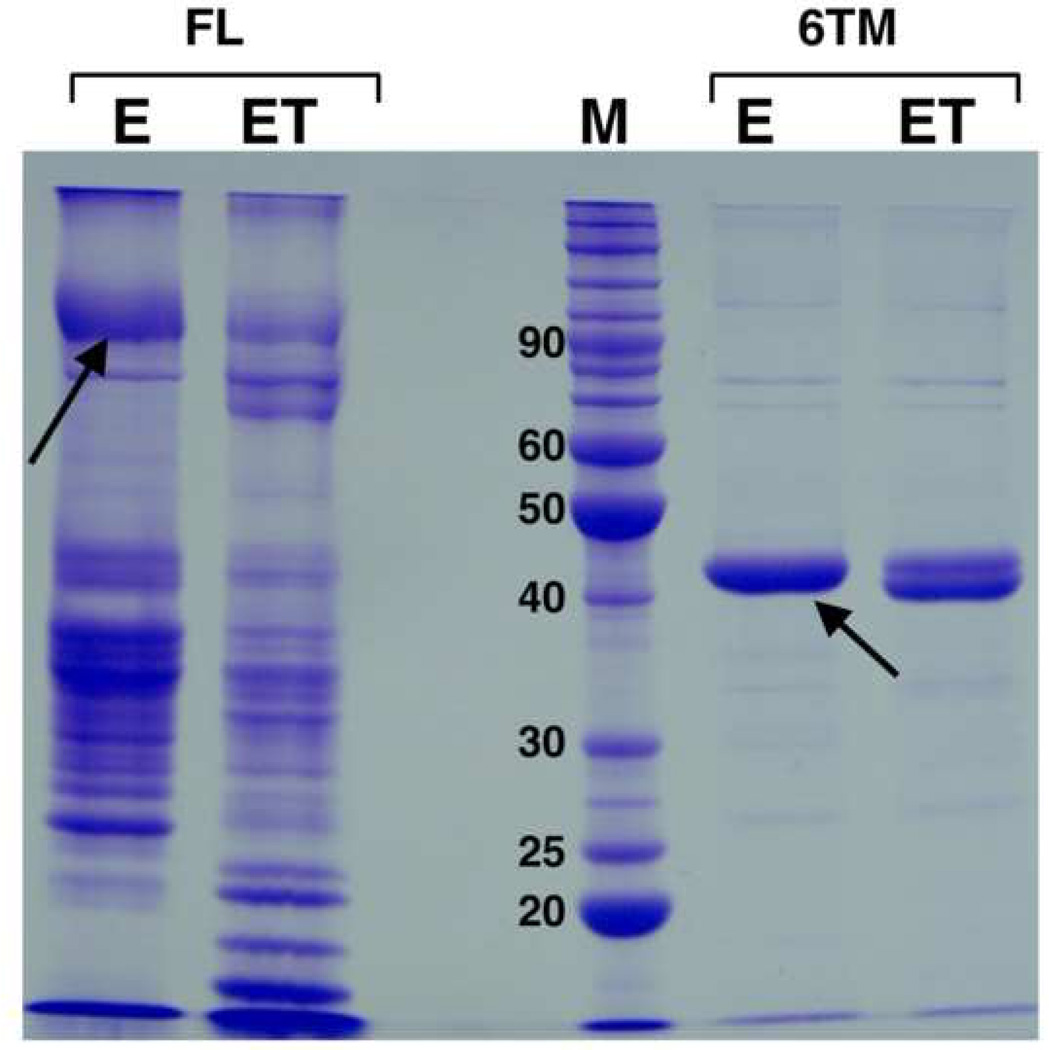
Figure 2.
Expression of aKv1.1 fused to Mistic, comparing full-length (FL) channel versus the 6TM domain only. The channel was fused to Mistic M110 (pMis4.0 vector) as shown on Figure 1. NiNTA elution (E) and thrombin cleaved elution (ET) were loaded on a 12 % SDS-PAGE gel next to a protein marker (M, numbers indicate kDa). The aKv1.1-6TM channel expresses better and is more stable and pure than the full-length aKv1.1 channel which shows aggregation and lower migrating bands. Note that only the His-tag but not Mistic could be cleaved off the aKv1.1-6TM. Arrows indicate bands corresponding to the Mistic fusion channel before cleavage.
Expression of aKv1.1 by Mistic Homologs
The interactions between Mistic and the downstream channel might play an important role for regulating the expression levels. We have, thus, shorten the length of the linker between Mistic and downstream gene to 17 amino acid residues, in the vector pMis4.0, from 34 in pMis3.0. For the expression of the aKv1.1-6TM channel, a shorter linker (17 aa residues as in pMis4.0) is preferable over the longer linker (34 residues as in the vector pMis3.0) possibly due to reduction in truncation and proteolytic degradation. Therefore, we tested the expression of the channel as a fusion with various Mistic sequences in vectors with pMis4.0 equipped with the shorter linker. Accordingly, replacing the M110 sequence in the pMis4.0 vector with M1, M2, M3 and M4 generated the vectors pM1_4.0, pM2_4.0, pM3_4.0 and pM4_4.0, respectively.
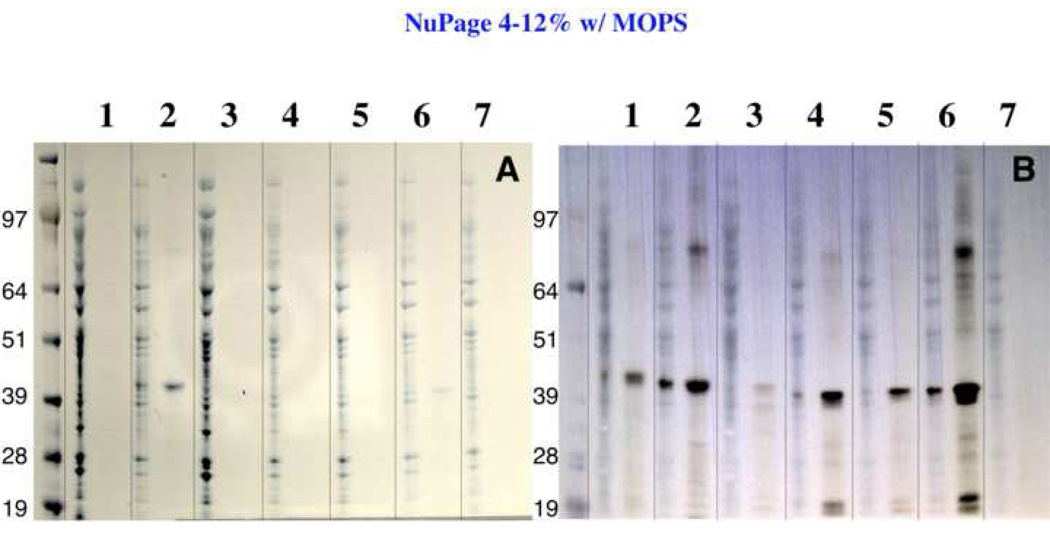
The analytical scale (20 ml bacterial culture) expression of aKv1.1-6TM fused to the various Mistic proteins shows that the only fusion products detectable by Coomassie-blue stain are from the channel fused to either M110 or M4 (Figure 3a). The anti-His Western analysis shows that there is a very low level of expression by M1 (lane 3) and some low level of expression by M2 and M3 proteins (lanes 4 and 5), while no expression is detected for the control expression in the absence of Mistic, see lane 7 (Figure 3b).
Figure 3.
Membrane expression of aKv1.1-6TM fused to various Mistic homologues. The aKv1.1-6TM channel was fused to Mistic homologues in the following vectors: 1: pMis3.0, 2: pMis4.0, 3: pM1_4.0, 4: pM2_4.0, 5: pM3_4.0, 6:pM4_4.0, 7: pHis9 (no Mistic). In Panel A, membrane associated proteins solubilized by 0.5% SDS (see M&M) were loaded on an SDS NuPAGE gel (Invitrogen, Carlsbad, CA) before and after Ni2+ purification. Panel B shows the corresponding anti-His antibody Western blot.
At first sight, the lowest expression level by M1 seemed surprising because it consists of the sequence of the high-expression level fusion partner, M110, lacking only its first α-helix H1 (amino acids 1 through 26). On one hand, the observation suggests a possible role for H1 of M110 in improving membrane integration, which we have tested subsequently and described below. On the other hand, these results can be understood in the light of the solubility studies on intact Mistic homologs, demonstrating that M1 is actually the most soluble Mistic protein, while M4 and M110 show the highest levels of membrane association propensity [16].
Solution Behavior of the aKv1.1-6TM Fused to M110 and M4
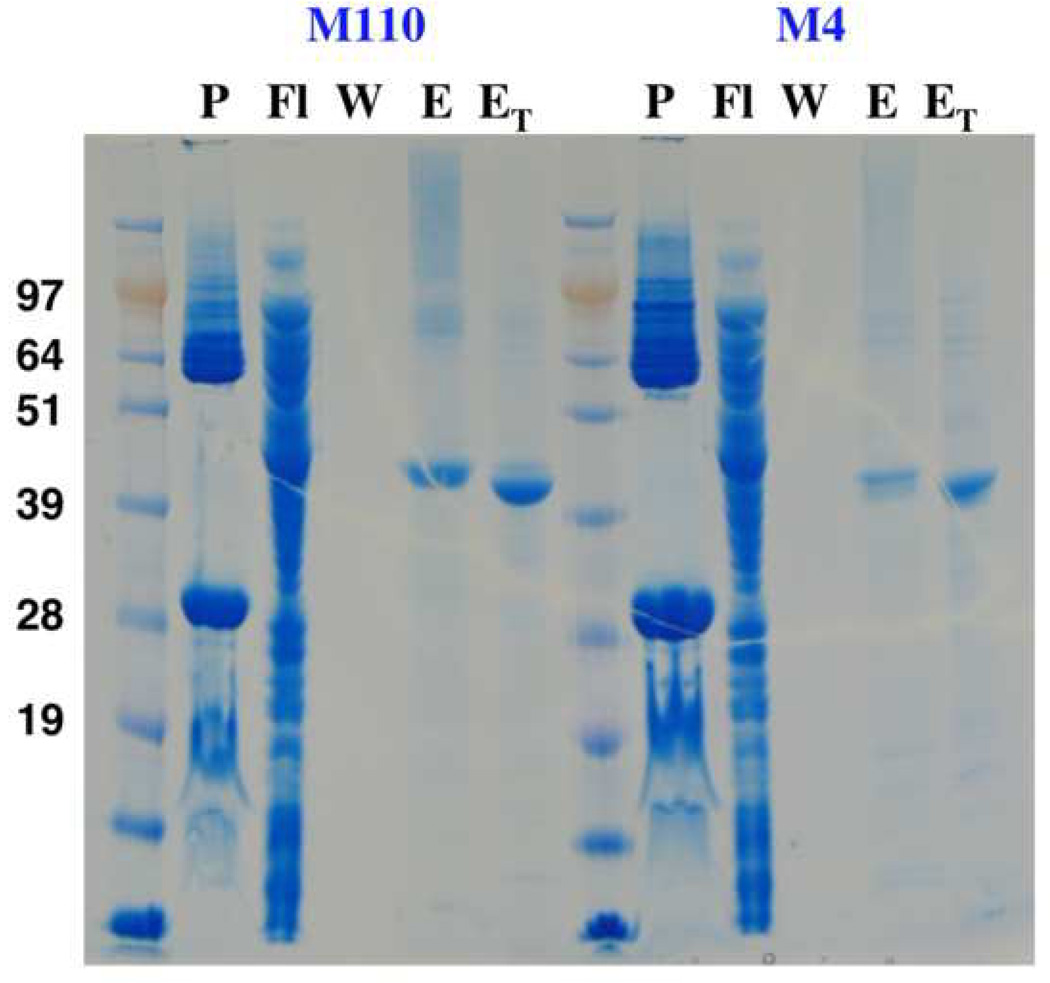
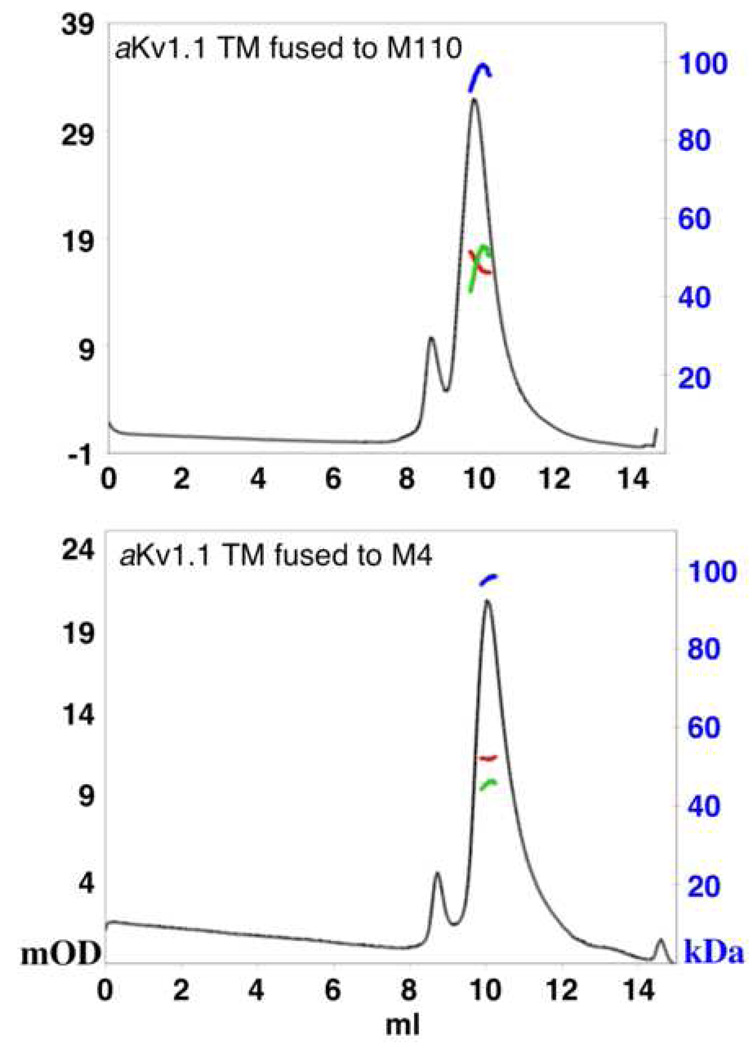
To test the stability and the oligomeric state of the ‘Misticated’ channel, we expressed it in a larger scale (1L culture) fused to either M110 or M4 for comparison. Figure 4 shows that the expression levels from both fusions are comparable. In addition, fusion to either M110 or M4 produces chimeras that are resistant for thrombin cleavage only between Mistic and aKv1.1-6TM, but both N-terminal His-tags were readily cleaved. This may suggest that both M110 and M4 associate with the N-terminus of the aKv1.1-6TM domain intimately. Further purification by SEC of the aKv1.1-6TM yielded a stable monodispersed PDC when fused to either M110 or M4 (Figure 5). Our static light-scattering data show that both exist as monomers when solubilized with LDAO. Although these fusions do not tetramerize as expected for an active Kv channel, their SEC elution profile does not indicate aggregation or misfolding, and the monomeric state maybe be preferable just due to the lack of the cytoplasmic T1 tetramerization domain [19]. For the M110 fusion, we measure a total PDC size of ~96 kDa with equal size of ~46 kDa for the protein and the detergent micelle in the PDC (47.5 kDa is calculated for the protein sequence). For the M4 fusion, the total PDC molar mass measures to ~97 kDa, with a bigger PDC detergent micelle size of ~52 kDa and ~45 kDa of protein compared to a sequence-based molecular size of ~46.5 kDa. Thus, no difference in oligomerization state is observed between the M110 and M4 fusions with aKv1.1-6TM.
Figure 4.
Comparing expression of aKv1.1-6TM by M110 and M4. All the samples are analyzed on an SDS NuPAGE gel. P: pellet of the LDAO-solubilized membranes, Fl, W and E: are the flow-through, the wash and the elution of the NiNTA purification, ET: is Ni elution cleaved by thrombin. It can be seen that the expression levels by M110 and M4 are comparable.
Figure 5.
Static light scattering analysis of aKv1.1-6TM fused to M110 or M4. Samples were injected on a G3000SWxl SEC column, in line with a light-scattering device and a refractive index instrument. UV traces (milli OD at 280 nm) are shown in black. Molar mass values were calculated for the main peaks and traces related to the blue scale in kDa are shown for the total PDC (blue), for the detergent only (red) and for the protein only (green).
Mistic chimera of M110 and M4
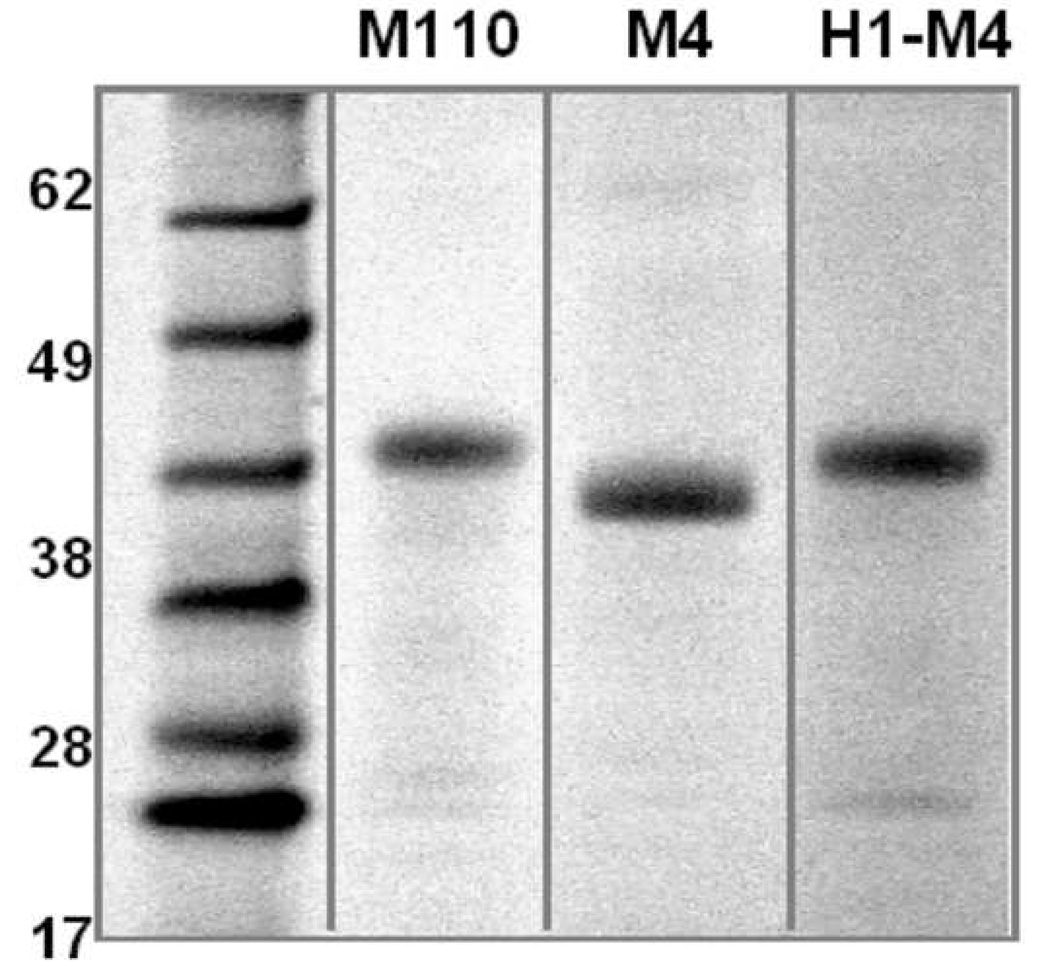
Since M110 assists expression of the aKv1.1 channel much better than its shorter form, M1, one could imagine that the additional N-terminal α-helix, residues 1–26 of M110 (H1), carries an additive quality for assisting overexpression of IMP in E. coli. In an attempt to create an 'enhanced Mistic' we have fused the H1 segment directly to the N-terminus of M4 (referred to as H1-M4) and assessed the expression levels of the aKv1.1-6TM fused to M110, M4 or H1-M4. We found that the expression levels of the channel are very similar between the three expression constructs (Figure 6) and the SEC analysis showed a monodispersed peak corresponding to the size of a monomer in the LDAO detergent micelle for all three expression constructs.
Figure 6.
Expression of aKv1.1-6TM channel by two Mistic proteins, M110, M4 and the chimera H1-M4. The two best Mistic fusion partners found for aKv1.1-6TM, M110 and M4 together with a chimeric Mistic made out of H1 of M110 and M4 (H1-M4) were fused upstream to the channel. The SDS-PAGE gel analyses the products after Ni2+ purification and cleavage of the N-terminal His-tag. Equal volumes of identical elution volumes were loaded on the gel and protein bands are identified by Coomassie stain, showing that the expression levels are similar for all three expression constructs.
Thus, for the expression of aKv1.1-6TM, we do not observe an enhancement of expression by H1-M4. It is possible that the expression by M4 is already too high for H1 to improve on, and if fused to M2 or M3 the effect would have been more significant. Regardless, the data suggests that H1 does not have an additive quality, and the enhancement with M110 is synergistic arising from the intra-interaction of H1 with the M1 sequence. Moreover, If H1 of M110 was an important sequence for interaction, more adapted with the E. coli translocon system or with the cytoplasmic membrane, we would expect to see an improvement of M4 by fusing this sequence to its N-terminus. Since this was not observed, the data further support the hypothesis that expression of IMPs by Mistic is related to the global chaperoning character of its structure to spontaneously insert into the membrane, and is likely independent of the translocon machinary. Interestingly, the recent study on polymerization and fibril formation by Mistic proteins [16], allowed the authors to propose a model for membrane insertion through polymerization of Mistic proteins.
CONCLUSION
Our data support the hypothesis that expression of foreign IMPs fused to Mistic in the membrane of E. coli depends on physical properties of the sequences that dictate their independent affinity for the membrane. We show that slightly different sequences of Mistic exhibit significantly varying capacities to enhance expression of an IMP at the E. coli membrane. The differences in expression levels are correlated with the degree of membrane association of the intact (non-fused) Mistic proteins. Thus, testing for membrane association of intact modified Mistic sequences may be useful in designing improved expression of foreign IMPs in E. coli.
Finally, since many eukaryotic IMPs could not be expressed in bacteria due to various incompatibilities, the fusion with Mistic proteins to express foreign IMPs in bacteria, offers a relatively simple and inexpensive alternative compared to other eukaryotic expression systems. This study shows means to utilize various Mistic proteins or combinations of them in bacterial expression of IMPs. It is possible that certain Mistic proteins would interact better with a particular IMP for its overexpression or for its biochemical stability, therefore, it is advisable to establish it experimentally as documented in this case study.
ACKNOWLEDGEMENT
We would like to thank Mark Vega for sharing Gateway 'Misticated' expression vectors. We acknowledge assistance of Innokentiy Maslennikov and Witek Kwiatkowski in light scattering and refractive index analysis. We thank Georgia Kefala for her critical advice. This work is supported by the NIH grant GM74821, GM74929.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Science. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd D, Schierle C, Beckwith J. How many membrane proteins are there? Protein Science. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanapin A, Batalov S, Davis MJ, Gough J, Grimmond S, Kawaji H, Magrane M, Matsuda H, Schonbach C, Teasdale RD, Yuan Z. Mouse proteome analysis. Genome Research. 2003;13:1335–1344. doi: 10.1101/gr.978703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 5.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nature Reviews Microbiology. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-ranslocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li WK, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Molecular Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 8.van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 9.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 10.Drew D, Froderberg L, Baars L, de Gier JW. Assembly and overexpression of membrane proteins in Escherichia coli. Biochimica et Biophysica Acta. 2003;1610:3–10. doi: 10.1016/s0005-2736(02)00707-1. [DOI] [PubMed] [Google Scholar]
- 11.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 12.Roosild TP, Vega M, Castronovo S, Choe S. Characterization of the family of Mistic homologues. BMC Structural Biology. 2006;6:10. doi: 10.1186/1472-6807-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KE, Kim HM, Lee JO, Jeon H, Han SS. Regulation of CD40 reconstitution into a liposome using different ratios of solubilized LDAO to lipids. Colloids and Surfaces B: Biointerfaces. 2008;62:51–57. doi: 10.1016/j.colsurfb.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Liu XJ, Wu L, Deng GS, Li N, Chu XS, Guo F, Li D. Characterization of mitochondrial trifunctional protein and its inactivation study for medicine development. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2008;1784:1742–1749. doi: 10.1016/j.bbapap.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Kefala G, Kwiatkowski W, Esquivies L, Maslennikov I, Choe S. Application of Mistic to improving the expression and membrane integration of histidine kinase receptors from Escherichia coli. Journal of Structural and Functional Genomics. 2007;8:167–172. doi: 10.1007/s10969-007-9033-4. [DOI] [PubMed] [Google Scholar]
- 16.Dvir H, Lundberg ME, Maji SK, Riek R, Choe S. Mistic: cellular localization, solution behavior, polymerization and fibril formation. Protein Science. 2009 doi: 10.1002/pro.148. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi Y, Matsui H, Takagi T. Membrane protein molecular weight determined by low-angle laser light-scattering photometry coupled with high-performance gel chromatography. Methods in Enzymology. 1989;172:514–528. doi: 10.1016/s0076-6879(89)72031-0. [DOI] [PubMed] [Google Scholar]
- 18.Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Science. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreusch A, Pfaffinger PJ, Stevens CF, Choe S. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature. 1998;392:945–948. doi: 10.1038/31978. [DOI] [PubMed] [Google Scholar]