Summary
In vitro biochemical reactions are most often studied in dilute solution, a poor mimic of the intracellular space of eukaryotic cells which are crowded with mobile and immobile macromolecules. Such crowded conditions exert volume exclusion and other entropic forces that have the potential to impact chemical equilibria and reaction rates. In this article, we used the well characterized and ubiquitous molecule calmodulin (CaM) and a combination of theoretical and experimental approaches to address how crowding impacts CaM’s conformational plasticity. CaM is a dumbbell shaped molecule that contains four EF hands (two in the N-lobe and two in the C-lobe) that each could bind Ca2+ leading to stabilization of certain substates that favor interactions with other target proteins. Using coarse-grained molecular simulations, we explored the distribution of CaM conformations in the presence of crowding agents. These predictions in which crowding effects enhances the population of compact structures were then confirmed in experimental measurements using fluorescence resonance energy transfer techniques of donor/acceptor labeled CaM under normal and crowded conditions. We further explored the folding energy landscape and examined the structural characteristics of CaM at free energy basins using protein reconstruction methods. We discovered that crowding stabilizes several different compact conformations, which reflects the inherent plasticity in CaM’s structure. From these results, we suggest that the EF hands in the C-lobe are flexible and can be thought of as a switch, while those in the N-lobe are stiff as analogous to a rheostat. New combinatorial signaling properties may arise from the product of the differential plasticity of the two distinct lobes of CaM in the presence of crowding. We discuss the implications of these results for modulating CaM’s ability to bind Ca2+ and target proteins.
Keywords: Calmodulin, EF-hands, macromolecular crowding, coarse-grained molecular simulations, FRET, Ca2+-signaling
I. Introduction
Thermodynamic analyses to investigate the effect of volume interactions on proteins have been applied since the pioneering work of Ogston and Laurent1. Most non-ideal thermodynamic behavior of biomolecules in biochemical reactions that depend on available space can be explained by modeling crowding agents as impenetrable hard-core models2; 3. The role of density, geometry and amount of crowding agents in the displacement of thermodynamic equilibrium can be analyzed through the theoretical framework of the scaled particle theory4. These studies, however, assumed that the native folded proteins are hard-core objects, disregarding their inherent conformational flexibility. In this regard, molecular simulation provides an excellent approach to study the dynamics of proteins by probing their statistical information5. For example, certain proteins that exhibit structural plasticity in the folded state were shown to change their structural characteristics in the presence of crowding agents6. As such, the conditions imposed by the surrounding environment of a protein could be critical to its function as some characteristics may only be induced under crowded cell-like conditions when only rarely present in dilute solutions7; 8; 9; 10.
Here, we apply experimental and theoretical approaches to address how macromolecular crowding impacts the structural characteristics of the essential and ubiquitous protein calmodulin (CaM). CaM is the predominant Ca2+ sensing protein in cells responsible for regulating a wide variety of biological functions including, muscle contraction, cell division, and neuronal signaling among many others (for reviews 11; 12; 13). CaM is a dumbbell shaped protein composed of 148 amino acids that contain four EF-hands; two in each half of the dumbbell, and two Ca2+ ions bind cooperatively to each lobe. What is critical to the present study is that there is a wealth of structural and kinetic information available for both the Ca2+ free and Ca2+ saturated states 14; 15 16 17; 18 on which simulation and experiments can be based to probe the impact of crowding. Interestingly, the structures of CaM solved by different methods and under different conditions vary drastically implying the molecule exhibits a range of metastable conformations. Results from NMR show that, in the Ca2+-free state, the C-lobe of CaM exhibits significantly greater backbone flexibility than the N-lobe14. The inter-lobe linker also plays an important role in CaM function. Even though in some crystal structures it is resolved as an extended alpha helix, NMR studies have shown this central helix to be highly flexible19. This flexibility is critical in allowing the lobes of CaM to adjust in the recognition of over 300 target proteins. The remarkable promiscuity in molecular recognition is due to the structural plasticity exhibited by CaM in its capacity to sample a wide range of conformational substates20.
Inherent flexibility of CaM has been investigated by using computer modeling and simulations. Fluctuations in lobes and helices associated with conformational changes were investigated by all-atomistic molecular dynamics simulations21; 22; 23. Differentiable intrinsic flexibility of the C-lobe compared with that of the N-lobe was suggested by using the normal mode analysis24 and low-resolution protein models25; 26. How such inherent flexibility in CaM’s globular lobes relates to its ability in target-binding are discussed for various substrates27; 28. Nevertheless, the plasticity of CaM has not been investigated under crowded conditions that are most relevant to the determination of the functional states of CaM inside cells.
A fundamental challenge to studying protein dynamics in vivo is to quantitatively dissect protein structure and dynamics29 within a complex environment. The surrounding macromolecules, such as proteins, nucleic acids, lipids, and cytoskeletons, exert volume exclusions and the volume fractions can reach as high as 40% of the total cellular volume30. To circumvent the physical and chemical complexities presented by polydisperse macromolecules, experimentalists mimic crowding effects by adding inert synthetic polymers such as polyethylene glycol, dextran, or Ficoll as crowding agents31; 32. Using this framework, studies of protein folding33; 34; 35; 36; 37, stability35; 38; 39; 40; 41 and interaction42 under crowded conditions suggested that reactions in a crowded milieu are indeed different from that in dilute solution. The effects of excluded volume of crowding agents on the kinetics of protein folding mechanism and stability have been also studied by computer modeling and simulations5; 6; 10; 43; 44; 45; 46; 47 and analytical calculations using different polymer models3; 48; 49.
Our approach consisted on one hand the use of Fluorescence spectroscopy to measure the distance between N- and C-lobes of CaM in the absence and presence of Ficoll 70. On the other hand we use coarse-grained molecular simulations that adequately probe large-scale structural fluctuations of CaM in a spatiotemporally complex system providing complementary analysis to the experimental findings. Together with the reconstruction of protein models, we found an increased probability of compacted conformations with an enhanced flexibility of the EF-hands when CaM is placed in a crowded environment. Our interpretation of these results is that crowding changes the free energy barrier between the basins corresponding to different compact CaM conformations. As a consequence, there is a shift in the distribution of compact substates to those more likely to bind Ca2+ and/or target enzymes. This suggestion is consistent with recent results describing that the conformational entropy of CaM contributes in a significant way to the free energy of binding50.
II. Results
Simulations indicate that compact states of CaM are favored in the presence of Ficoll
The thermodynamic properties of CaM were investigated by using the coarse-grained molecular simulation strategy51. To reflect the plasticity in the conformations of CaM observed in the protein database bank (PDBID: 1CFD), we use the Betancourt-Thirumalai (BT) statistical potential52 that incorporates side chain interactions depending on their amino acid types. As a result, this kind of interaction potential dictated by protein sequences allows us to sample all possible collapsed conformations7; 53. In contrast, other protein models that adopt a double Go-type25; 54 interaction can only sample a few folded states determined by a priori known structures.
We then investigated the thermodynamic characteristics of CaM in the absence and presence of crowding agents at different volume fractions (φc=0, 25% and 40%). CaM is represented in a side chain – Cα model (SCM, see Methods) and crowding agents are hard-core spheres with a radius of 55Å55 that represents Ficoll 70 molecules (Fig. 1(a)). Fig. 1(b) shows a cartoon representation of CaM, labeled with Alexa488 and QY9 fluorophores at position 34 at the N-lobe and position 110 at the C-lobe, respectively, to illustrate the comparative distance measurements between computer simulation and experiments in the presence of Ficoll 70.
Figure 1.
(a) A schematic representation of a coarse-grained CaM in the presence of Ficoll 70. CaM is represented by a side-chain Cα model. The N-lobe of CaM is shown in blue, while the C-lobe is shown in red. Ficoll is displayed as hard-spheres with a radius of 55 Å, shown in gold. (b) A ribbon representation of all-atom CaM (PDB: 1CFD) with attached fluorophores. The color code of the lobes is the same as in (a). Alexa488 (in green) is the donor fluorophore used in the FRET studies and is shown attached to position 34 while QSY9 (in magenta) is the quencher fluorophore and is shown attached to position 110. CaM is 148 amino acid residues in length.
First we plot the folding energy landscape 56; 57 of CaM as a function of the radius of gyration (Rg) and the overlap function (χ)58 at T=0.96Tf (Fig. S1) and T=1.2Tf (Fig. S2) in the supplement material. χ measures the similarity to the crystal structure of CaM. It is shown in Fig. S1(a) that there are two dominant free energy basins at φc=0 (crowder-free condition) at a temperature lower than Tf. Ensemble structures of the two basins are compact and similar in sizes (Rg ~ 15–17Å). However, they are structurally different. The ensemble structure with χ~0 is similar to the crystal structure that resembles a dumbbell. The other one (pointed by an arrow in Fig. S1), with χ~0.1 and a slightly lower Rg, represents a collapsed conformation of CaM different from the crystal structure. As φc increases, the area of the basin centered at χ~0.1 (pointed by an arrow) grows while the area of the basin centered at χ~0 shrinks. We speculate that at high levels of crowding the compact structures of CaM may not be similar to the crystal structure. At temperatures greater than Tf, (Fig. S2 (a) φc =0%, (b) 25% and (c) 40%, respectively) the sizes of the unfolded conformations decreases, indicated by a reduced population of extended conformations in the presence of Ficoll 70 with respect to those under crowder-free conditions. This agrees with the destabilization of unfolded conformations by crowding effects captured by theories3; 59 as well as computer simulations5.
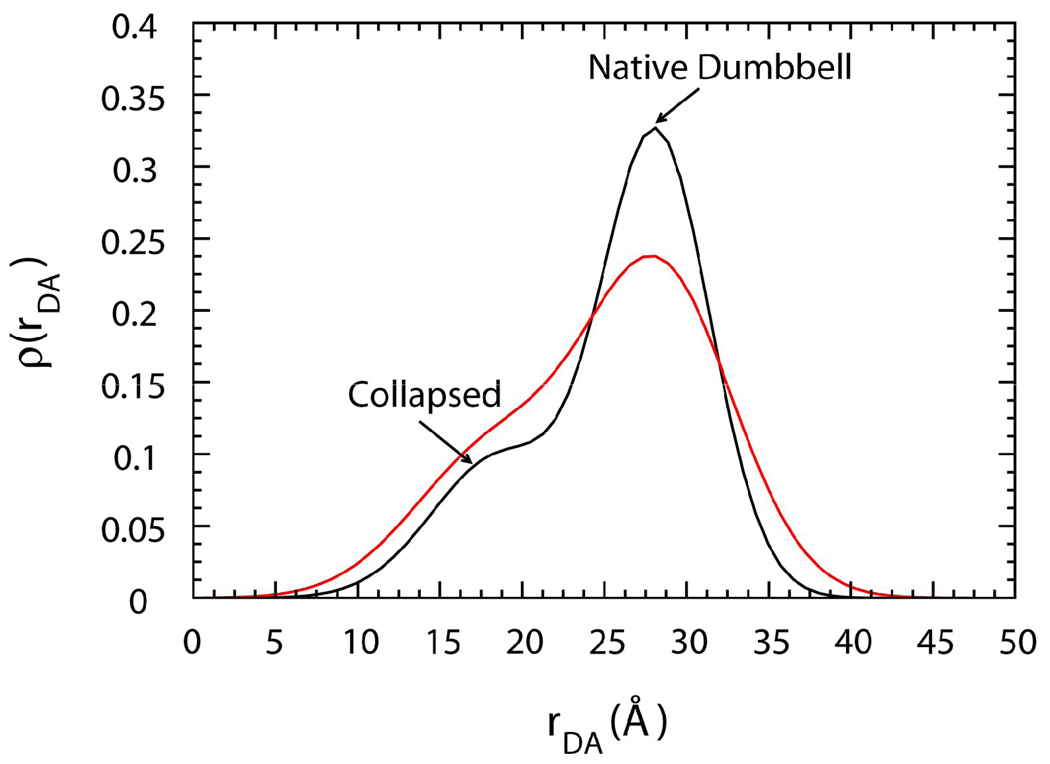
To compare these results with an experimental readout, we analyzed the distribution ρ(rDA) of the average distance of side chain beads of residue 34 and 110 (rDA) in Fig. 2. In the absence of crowding (φc=0), the profile of ρ(rDA) is broad and there are two peaks found at rDA=19Å and rDA=28Å. The ensemble conformations at rDA=28Å resemble the crystal structures. It is labeled ND (Native-Dumbbell) for its resemblance to a dumbbell. The ensemble conformations at rDA=19Å represent two collapsed lobes connected by a linker and the overall sizes are smaller than ND. In the presence of a high level of crowding (φc =40%), the profile of ρ(rDA) shifts toward the left to populate a more compact ensemble with the most probable rDA=19Å.
Figure 2.
Computer simulation of the distribution of the donor-acceptor distance, rDA, of CaM in the bulk case (black) and in the presence of 40% volume fraction of Ficoll70 (red) at 0.96Tf.
FRET experiments confirm that CaM assumes a more collapsed conformation in the presence of crowding agents
The predictions of the impact of crowding on the conformational states of CaM were tested with steady state fluorimetry. To accomplish these experiments we engineered a mutant of CaM to place Cys residues at amino acids 34 and 110. This protein was subsequently labeled with Alexa 488 as the donor fluorophore and QSY-9 as the acceptor quencher (termed CaMDA) and purified. Steady state measurements representing an ensemble average of the conformations of CaMDA were used to monitor the distances between the N and C-lobes of CaM in different conditions. A representative spectrum of the CaMDA in the absence and presence of Ficoll 70 at 40 % (w/v) is shown (Fig. 3(a)) that indicates a substantial increase in FRET (decrease in distance of the probes) in the presence of Ficoll. The potential impact of Ficoll 70 on the fluorescent properties of the donor molecule on CaM were controlled for by evaluating the fluorescence spectrum of CaM labeled with donor alone (CaMD). As seen in the inset of Fig. 3(a), the spectra are almost identical for CaMD in the presence of absence of Ficoll. The results of these experiments are summarized in Fig. 3(b). The distance between donor-acceptor, rDA, and its standard deviation, calculated as described in materials and methods, is shown for various conditions without crowding agents and at 20 % and 40% (w/v) Ficoll 70. Ficoll 70 at 40% w/v exerted a change in distance of ΔR = 7.1 Å from rDA relative to apo-CaM. In Ficoll 70 at 20% (w/v) rDA does not change significantly. For comparison, Ca2+ saturated CaM showed a change in separation distance of ΔR = 8.2 Å from the apo-CaM lobe-lobe distance, which is comparable to the separation distance (7.1 Å) observed at 40% (w/v) Ficoll 70.
Figure 3.
(a) Emission spectra of CaMDA in the absence (0% w/v) and presence of Ficoll 70 (40 % w/v). Inset shows the normalized fluorescence of donor-alone labeled CaM. (b) rDA was calculated for CaMDA in the various buffer conditions (−Ca2+, +Ca2+ and with Ca2+ and target peptide) from emission spectra similar to the one shown in panel (a) as described in Materials and Methods. On the same bar plot the separation distance, rDA, shows a decrease in distance from Ficoll 70 at 20% (w/v) to 40% (w/v).
As an additional control, the impact on the FRET reported by CaMDA of a target peptide that binds to CaM with high affinity (Kd < fM)60 was evaluated. In the presence of Ca2+, the target peptide was found to drive CaM to a collapsed conformation with the distance between the N- and C-lobes of 14.8 Å. This extensive collapse was never reached even at the maximal concentration of Ficoll 70 (40% w/v), but served as an anchor point to the extreme where all CaM is locked into a collapsed conformation.
One important caveat in interpreting the calculated distances from these results is that the experiments are accomplished with CaM labeled with relatively large fluorophores attached to the protein with a five-carbon linker. This can have a substantial impact when comparing the differences measured from FRET experiments from the theoretical distances obtained which consider the separation distance between the Cα of residues 34 and 110 (the sites of fluorophore attachment). Nevertheless, the results clearly show that CaM assumes a more collapsed conformation in the presence of crowding agents.
Details of the collapsed states of CaM in the absence and presence of a crowded environment
Motivated by the finding in Fig. S1 that the area of the basins consisting of non-native compact structures is broadened under a high level of crowding, we employ an asphericity parameter, Δ5, to better characterize structural differences among the compact structures. When Δ=0, the shape of an object resembles a sphere and when Δ=1 it resembles a rod. In addition, we also compare the structural changes in the N-lobe and the C-lobe separately in the presence of crowding by using the structural overlap functions, χN(or χC), of the N-lobe (or C-lobe). χN(or χc) measures the structural similarity between the N- (or C-) lobe of a CaM with the same lobe in the crystal structure of apoCaM. χN (or χC) varies between 0 and 1 and when it equals to 0 its structure is identical to the lobe of apoCaM.
Aided by a shape-sensitive parameter, we further compare the folding energy landscape of CaM from the perspective of the N-lobe and the C-lobe, respectively. At a temperature lower than Tf, we investigate the distribution of collapsed states in a crowder-free system (φc=0%) and in the presence of crowding (φc=40%). First, for the N-lobe, there is one dominant basin in the free energy landscape as a function of Δ and χN at φc=0% (circled in blue in Fig. 4(a)). The dominate structures taken from this basin characterized by 0.0 < χN< 0.1 and 0.2 < Δ < 0.4 are similar to a dumbbell-shaped apoCaM state and it is labeled the “Native-Dumbbell (ND)” state.
Figure 4.
Free energy diagram as a function of the structural overlap function of the N-lobe of CaM (χN) and the shape parameter (Δ) at T=0.96 Tf (a) in the bulk system and (b) in the presence of crowding agents at φc=40%. The color bar is scaled by kBT, where kB is the Boltzmann constant. The white area represents where the value of free energy exceeds 8.5 kBT. Typical ensemble structures pointing to circled basins are reconstructed to all-atomistic protein models using the SCAAL program7 The Ca2+-binding segments of CaM are: EF1 in blue, EF2 in cyan, EF3 in yellow and EF4 in red.
At φc=40% as shown in Fig. 4(b), the distribution of collapsed structures shifts to favor spherical conformations (Δ~0). The population of ND decreases and the populations of two other different ensemble structures that have spherical shapes emerge. First, the ensemble structures characterized by 0.0 < χN< 0.1 and 0.0 < Δ < 0.2 have a structurally native-like N-lobe and an unfolded C-lobe that collapsed over the N-lobe. It is named the “Native Collapsed” (NC) state and is shown in a red circle in Fig. 4(b). Second, the other ensemble structures characterized by 0.2 < χN < 0.4 and 0.0 < Δ < 0.2 are unfolded and collapsed in which both the N-lobe and C-lobe lost their native conformations. This state is called the “Completely Collapsed” (CC) state and it is shown in a green circle in Fig. 4(b). In the presence of high levels of crowding the distribution of compact CaM structures shift from an aspherical dumbbell to compact spherical states such as NC and CC. Such shape changes in the presence of crowding agreed with predictions from statistical-thermodynamics models61 and molecular simulations7; 62.
Interestingly, when we investigate the behavior of the C-lobe in the presence of crowding C-lobe is different from that of the N-lobe. We plot the folding energy landscape of CaM as a function of χcand Δ from the perspective of the C-lobe (Fig. 5). At φc=40% (Fig. 5(b)), there are only two dominant basins at a temperature lower than Tf. One of the two is ND, characterized by 0<χc<0.2 and 0<Δ<0.2. The other one is characterized by 0.3<χc<0.5 and 0<Δ<0.2 and is different from the crystal structure and contains a mixture of the NC and CC states. Unlike the N-lobe, the C-lobe is structurally unfolded in the collapsed structures. This suggested that the N-lobe of CaM is structurally stiffer than the C-lobe making it more resistant to collapse under a high level of crowding. This finding is in agreement with previous studies on the folding dynamics of the two lobes of CaM being asymmetric26. In the presence of crowding, the contrast of structural flexibility between the two lobes becomes more distinct.
Figure 5.
Free energy profiles as a function of the structural overlap function of the C-lobe of CaM (χC) and the shape parameter (Δ) at T=0.96 Tf (a) in the bulk system and (b) in the presence of crowding agents at φc=40%. The color bar is scaled by kBT, where kB is the Boltzmann constant. The white area represents where the value of free energy exceeds 8.5 kBT.
Structural plasticity in the CaM lobes
We further investigate the orientation of EF hands in each lobe associated with different collapsed structures. CaM has a pair of helix-coil-helix EF hands in each lobe that cooperatively bind Ca2+. We number these EF hands starting from the N-lobe where the first two, 1 and 2, lie within the N-lobe and the last two, 3 and 4, lie in the C-lobe. The EF hand 1 has two helices labeled (A/B) and EF hand 2 has the helices (C/D). Similarly, helices in EF 3 are (E/F) and those in EF 4 are (G/H) helices. In order to analyze the plasticity of CaM’s EF-hands of the three collapsed states, we measure the differences in the angles, θ, of the two helices that form the core of the EF hands in both N- and C-lobes (Fig. 6(a) and Fig. 7(a)).
Figure 6.
The distribution of the angles, ρ(cos(θ)), formed by adjacent helices in the individual EF hands of CaM. This distribution is shown for EF hands in the two lobes of CaM as well as three different collapsed states (ND, NC, and CC). (a) A schematic diagram shows the angle formed by adjacent helices in each EF hand. (b) ρ(cos(θ))of two helices (A/B) in EF 1 and (C/D) in EF2 in the N- lobe. (c) ρ(cos(θ)) of two helices (E/F) in EF 3 and (G/H) in EF4 in the C-lobe.
Figure 7.
The distribution of the angles, ρ(cos(θ)) formed by helices in different EF hands of CaM. This distribution is shown for EF hands in the two lobes of CaM as well as the three different collapsed states (ND, NC, and CC). (a) A schematic diagram shows the angle formed by helices of different EF hands within one lobe of CaM. (b) ρ(cos(θ)) of two helices (A/D) in the N-lobe and (E/H) in the C-lobe.
The probability distribution, ρ(cosθ), of the EF hands in different collapsed states and in each lobe is shown in Fig. 6(b). First, we focus on the ρ(cosθ) of helices in the N-lobe. ρ(cosθ) peaks at cosθ=0.92, and 0.89, for A/B, and C/D, respectively, in the ND state. However, in the NC state, this distribution does not change significantly. This indicates rigidity of EF hands in the N-lobe. Interestingly, in the CC state, the profile of ρ(cosθ) is flattened indicating a flexible EF hand that can sample any angle randomly.
Next, we focus on ρ(cosθ) of EF hands in the C-lobe. ρ(cosθ) peaks at cosθ=1, and 0.5 for E/F, and G/H, in the ND state, respectively, in Fig. 6(c). Unlike the N-lobe, the profiles of ρ(cosθ) are flat in both NC and CC states. This indicates that the structures of EF hands in the C-lobe are significantly more flexible than the ones in the N-lobe.
In addition, we measured the angles between helices A/D in the N-lobe, and E/H in the C-lobe as alterations in the orientation of these helices that underlie the promiscuity in protein target binding. The results are shown in Fig. 7, where ρ(cosθ) peaks at 0.3 and 0.5, for the A/D and E/H, in the ND state, respectively. The ρ(cosθ) of helices in A/D remains the same for the NC state as the ND state which then becomes flat in the CC state. However, ρ(cosθ) of helices in E/H in the C-lobe is quite different from that in the N-lobe. Its profile for E/H helices flattens dramatically in both the NC and CC states. It is suggested that the structures of target binding site (A/D) in the N-lobe is less flexible than that (E/H) in the C-lobe within a crowed environment.
III. Discussion
Despite the extensive characterization of CaM as a Ca2+ sensor, many questions remain open; particularly in the area of molecular recognition and conformational dynamics. The inherent flexibility of CaM along with the wealth of structural and kinetic studies makes it a unique case of study. NMR studies describe flexibility as the range of “motion” or poor RMSD 14 which is based on the dynamics of protein in solution while X-ray studies15 describe flexibility as large differences found between stable structural end-points when the protein is trapped in a crystalline state without knowledge of how stable these structures are in solution. Our description of flexibility agrees more with NMR in which the C-lobe can more easily sample a large number of conformations, but the extent of conformational changes may not necessarily be large.
CaM is responsible for translating changes in intracellular Ca2+ concentration to the activation of downstream targets. The four EF hands, two at each lobe, are responsible for sensing Ca2+, and convert this binding energy to changes in conformation that allows the activation of targets. In this report, we characterized the structural plasticity of CaM, in the presence of macromolecular crowding agents. Using a combined experimental and computational approach, we find that macromolecular crowding redistributes the population from an extended conformation of apoCaM to several dominant compact conformations each with different lobe-specific characteristics. Our thermodynamic analysis from molecular simulations qualitatively agree with ensemble studies of CaM using steady state fluorescence, where we observed an average decrease in the separation distance between the two lobes in the presence of crowding agents. The similarity of the N- to C-lobe separation distances of apoCaM in crowding agents with that of Ca2+/CaM in the absence of crowding agents suggested that crowding might be inducing conformations of CaM similar to the Ca2+/CaM bound state.
We further analyzed the plasticity of each EF hands within each lobe of CaM. Detailed analyses indicate that structural changes of EF-hands in the N-lobe in the ND, NC, and CC conformations are gradual in response to varying crowding levels. We conclude that the N-lobe can maintain its rigidity even at high concentrations of crowding agents, allowing the states of NC and CC to be populated. In contrast, the structure of the C-lobe is more susceptible to crowding levels resulting in enhanced flexibility of the EF-hands in the non-native states due to a loss of stabilizing tertiary contacts.
It is well known that upon Ca2+-binding, CaM exposes hydrophobic patches at the interface between the lobes and the linker that connects them and that these patches form the initial recognition sites for target binding. These hydrophobic patches are exposed by increases in the inter-helical angle between helix A and D (and B and C) in the N-lobe and E and H (and F and G) in the C-lobe63. Crowding was shown to randomize these inter-helical angles suggesting that partial exposure of these hydrophobic patches may lead to increased probability of target binding in the absence of Ca2+. Interestingly, in the presence of crowding, the inter helical angles A/D in the N-lobe were less flexible than those for E/H in the C-lobe, suggesting that crowding would more likely induce conformational transitions to favor target binding initiated in the C-lobe than the N-lobe.
A classical view of the mechanism of CaM is that upon Ca2+ binding a conformational transition in the EF hands is induced from a closed to open state acting as a switch mechanism12. With the results presented here, we expand this concept and propose that crowding changes the available free energy of the system allowing CaM to sample conformations in a controlled manner. Due to the intrinsic structural rigidity of the N-lobe, the impacts of crowding are more subtle permitting the N-lobe to function more like a rheostat. In contrast, the C-lobe which is inherently more flexible undergoes more dramatic, switch-like conformational transitions with an increase in free energy of the system due to macromolecular crowding. A model summarizing these main state transitions in the absence and presence of crowding is presented in Fig. 8. In this figure, the energy landscape over a hypothetical reaction coordinate is shown for the conformations found in the simulation. Also, the free energy is drawn as a reference to show the rheostat mechanism in the N-lobe and the switch like behavior of the C-lobe.
Figure 8.
Schematic representation of changes in the free energy profiles for the N- and C-lobes of CaM in the absence and presence of crowding agents. In buffer, the ND state is the dominant one in which the two lobes preserve their native contacts. The other less populated states (NC and CC) are more spherical than the ND state. However, they have different distribution of the native contacts. In both NC and CC states, the C-lobe has lost most of its native contacts while the N-lobe is more native like in the NC state and becomes less native like in the CC state. In the presences of crowding agents, an increase in free energy of the system increases the probability that transitions will occur between the different states. Because the N-lobe is less flexible, the relative proportion of molecules in each state remains similar. In contrast, the increased flexibility of the C-lobe causes a change in the distribution of the states with ND being less populated in favor of the NC and CC states as crowding increases the free energy. We propose that the less flexible nature of the N-lobe produces more subtle changes in its structure as the free energy of the system increases so that it behaves more like a rheostat. In contrast, as the free energy of the system increases the C-lobe behaves more like a switch as it transitions more abruptly from native to collapses states.
IV. Materials and Methods
Computational simulations
Coarse-grained model for proteins and crowding agents
A Sidechain-Cα (SCM) coarse-grained model is used to represent a CaM protein where each amino acid (except glycine) is modeled by two beads: a Cα bead and a side chain bead46; 51. Ficoll 70, the crowding agent, is modeled by rigid spheres with a radius of 55 Å5. The potential energy of the combined system consisting of CaM and Ficoll 70, is Ep+Ec+Epc. Epc is the interaction energy between a protein and crowding agents. Ep and Ec are the potential energies of a protein and crowding agents, respectively. Ep is Estructural+EHB+ENB where the structural energy, Estructural, is the sum of the bond-length potential, the side chain-backbone connectivity potential, the bond-angle potential, the dihedral potential, and the chiral interactions. The chiral energy accounts for an L-isoform preference of side chains. The backbone hydrogen bond (EHB) is represented by a direction-dependent nonbonded potential. The nonbonded interactions (NB) for side-chains, ENB, are represented by Lennard-Jones (LJ) potentials. Solvent-mediated interaction between pairs of side chains is based on the Betancourt-Thirumalai statistical potential52; 53. This statistical potential addresses sequence variations where the reference interaction, ε = 0.6kca / mol, is based on the Thr-Thr pairwise interaction. Pairwise interactions between the Cα and side chain beads, a protein and crowding agents (Epc), and crowding agents (Ec) themselves are repulsive5.
Simulation details
The thermodynamic properties of CaM in the bulk system and in crowded environments are studied by coarse-grained molecular simulations64 in which the Langevin dynamics equation of motion in a low friction region 65 is used. The simulations were done at constant volume and temperature (NVT) conditions with cubic periodic boundary conditions. The parameters and the methods for preparing the models used for a protein and crowding agents in our computer simulations can be found elsewhere7. Dynamics and ensemble structures of CaM in several volume fractions of crowding agents (φc) were studied (φc = 0%, 25%, and 40%). Such systems were simulated across a broad range of temperatures around the folding temperature of CaM, Tf. To enhance the sampling efficiency, the replica exchange method66; 67 that distributes the temperature of simulations from 0.75 Tf to 1.5 Tf over 20 replicas is implemented in which the probability of the exchanges is determined by Metropolis criterion. Each replica generates ≈45,000 conformations. Thermodynamic properties of CaM were analyzed by the weighted histogram analysis method68; 69.
Experimental preparations
FRET-pair
CaM 34C/110C was genetically engineered and labeled with Alexa488 as donor fluorophore and QSY9 as the acceptor quencher as described in ref70. The labeling ratio of the double-labeled CaM has a 1:1 stoichiometry between donor and acceptor. For each experiment, a stock of donor alone labeled CaM (CaMD) and the double labeled CaM (CaMDA) was brought to room temperature and the concentration of CaM was measured using the extinction coefficient of the donor fluorophore. Then 100 nM CaMD or CaMDA were mixed thoroughly with the crowders at different concentration in a base buffer consisting of 10 mM HEPES pH 7.4, with 1mM EGTA to remove any residual Ca2+. For saturated Ca2+ conditions, instead of EGTA, a 1 mM MgCl2 and 0.1 mM CaCl2 were used.
Steady State FRET
Fluorimetry measurements of CaMD and CaMDA were made in a temperature regulated PTI Quantamaster fluorimeter controlled by Felix 32 software. A typical experiment consisted of using a 470 nm excitation wavelength for CaMD, and the spectra from 480 nm to 600 nm was collected with 5 nm slit widths. Then the fluorescence spectra of CaMDA were measured at the same conditions. A numerical integration over the emission spectra, which accounts for the total fluorescence emission of the Alexa488, was performed to obtain FD and FDA, for CaMD and CaMDA respectively. The energy efficiency (E) was calculated using . To extract the separation distance rDA, we use the well-known relationship 71. For the Alexa488/QSY9 pair of dyes, Ro has a value in buffer of 64Å. A modest adjustment in Ro was made to compensate for changes in the index of refraction of the Ficoll solutions using the relationship where n is the index of refraction of buffer (1.33) and n' is the index of refraction of the Ficoll solution. The index of refraction of 20% and 40% Ficoll 70, measured with a refractometer, were 1.363 and 1.393, respectively.
Supplementary Material
Acknowledgement
MSC would like to thank TeraGrid and TLC2 for the computing resources and the UH GEAR program and the TcSUH seed grant from the University of Houston. MNW acknowledges the NIH/NINDS for supporting the experimental work (NS26086) and gratefully acknowledges an endowment from the William Wheless III Professorship. During a portion of this work, HS was supported by a training fellowship from the Keck Center Nanobiology Training Program of the Gulf Coast Consortia (NIH 2 R90 DK071054).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laurent TC, Ogston AG. The interaction between polysaccharides and other macromolecules. 4. the osmotic pressure of mixtures of serum albumin and hyaluronic acid. Biochem. J. 1963;89:249–253. doi: 10.1042/bj0890249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg OG. The influence of macromolecular crowding on thermodynamic activity: solubility and dimerization constants for spherical and dumbbell-shaped molecules in a hard-sphere mixture. Biopolymers. 1990;30:1027–1037. doi: 10.1002/bip.360301104. [DOI] [PubMed] [Google Scholar]
- 3.Minton AP. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: Macromolecular crowding and protein stability revisited. Biophys J. 2005;88:971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss H, Frisch H, Lebowitz JL. Statistical mechanics of rigid spheres. J. Chem. Phys. 1959;31:369–380. [Google Scholar]
- 5.Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates. Proc Natl Acad Sci U S A. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stagg L, Zhang S-Q, Cheung MS, Wittung-Stafshede P. Molecular crowding enhances native structure and stability of alpha/beta protein flavodoxin. Proc Natl Acad Sci U S A. 2007;104:18976–18981. doi: 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homouz D, Perham M, Samiotakis A, Cheung MS, Wittung-Stafshede P. Crowded, cell-like environment induces shape changes in aspherical protein. Proc Natl Acad Sci U S A. 2008;105:11754–11759. doi: 10.1073/pnas.0803672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, Tacheuchi T, Futaki S, Ito Y, Hiroaki H, Shirakawa M. High-resolution multidimensional NMR spectroscopy of proteins in human cells. Nature. 2009;458:106–110. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 9.Sakakibara D, Sasaki A, Ikeya T, Hamatsu J, Hanashima T, Mishima MMY, Hayashi N, Mikawa T, Walchli M, Smith BO, Shirakawa M, Guntert P, Ito Y. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 2009;458:102–106. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- 10.Kudlay A, Cheung MS, Thirumalai D. Crowding effects on the structural transitions in a flexible helical homopolymer. Phys Rev Lett. 2009;102:118101. doi: 10.1103/PhysRevLett.102.118101. [DOI] [PubMed] [Google Scholar]
- 11.Means AR, Dedman JR. Calmodolin - an introcellular calcium receptor. Nature. 1980;285:73–77. doi: 10.1038/285073a0. [DOI] [PubMed] [Google Scholar]
- 12.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends in Cell Biology. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 13.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu. rev. Biophys. Biomol. Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 14.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. A solution structure of calcium-free calmodulin. Nat. Struct. Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 15.Fallon JL, Ouiocho FA. A closed compact structure of native Ca(2+)-calmodulin. Structure. 2003;11:1303–1307. doi: 10.1016/j.str.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Ishida H, Nakashima K, Kumaki Y, Nakata M, Hikichi K, Yazawa M. The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property. Biochemistry. 2002;41:15536–15542. doi: 10.1021/bi020330r. [DOI] [PubMed] [Google Scholar]
- 17.Meador WE, Means AR, Quiocho FA. Modulation of Calmodulin Plasticity in Molecular Recognition on the Basis of X-Ray Structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 18.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J. Mol. Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 19.Barbato BI, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 20.Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interaction with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 21.Wriggers W, Mehler E, Pitici F, Weinstein H, Schulten K. Structure and dynamics of calmodulin in solution. Biophysical Journal. 1998;74:1622–1639. doi: 10.1016/S0006-3495(98)77876-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vigil DCGS, Trewhella J, Garcia AE. Functional dynamics of the hydrophobic cleft in the N-domain of calmodulin. Biophysical Journal. 2001;80:2082–2092. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, Jas GS, Kuczera K. Structure and dynamics of calcium-activated calmodulin in solution. Journal of Biomolecular Structure & Dynamics. 2001;19:247–271. doi: 10.1080/07391102.2001.10506736. [DOI] [PubMed] [Google Scholar]
- 24.Barton NP, Verman CS, Caves LS. Inherent flexibility of calmodulin domains: a normal-mode analysis study. J. Phys. Chem. B. 2002 [Google Scholar]
- 25.Chen Y-G, Hummer G. Slow conformational dynamics and unfolding of the calmodulin C-terminal domain. J Am Chem Soc. 2007;129:2414–2415. doi: 10.1021/ja067791a. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi S, Portman JJ. Inherent flexibility determines the transition mechanisms of the EF-hands of calmodulin. Proc Natl Acad Sci U S A. 2009;106:2104–2109. doi: 10.1073/pnas.0806872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Jas GS, Kuczera K. Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin. Biochimica et Biophysica Acta. 2004;1697:289–300. doi: 10.1016/j.bbapap.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Tan HW, Aa ZC, Chen GJ. Ligand-induced dimer formation of calmodulin. Journal of Molecular Recognition. 2008;21:267–274. doi: 10.1002/jmr.895. [DOI] [PubMed] [Google Scholar]
- 29.Ellis RJ. Macromolecular crowding: an important to neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 30.Ellis RJ, Minton AP. Join the crowd. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 31.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 32.Winzor DJ, Wills PR. Molecular crowding effects of linear polymers in protein solutions. Biophys. Chem. 2006;119:186–195. doi: 10.1016/j.bpc.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 33.van den Berg B, Ellis RJ, Dobson CM. Effects of macromolecular crowding on protein folding and aggregation. Embo J. 1999;18:6927–6933. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokuriki N, Kinjo M, Negi S, Hoshino M, Goto Y, Urabe I, Yomo T. Protein folding by the effects of macromolecular crowding. Protein Sci. 2004;13:125–133. doi: 10.1110/ps.03288104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ai X, Zhou Z, Bai Y, Choy W-Y. 15N NMR Spin Relaxation Dispersion Study of the Molecular Crowding Effects on Protein Folding under Native Conditions. J Am Chem Soc. 2006;128:3916–3917. doi: 10.1021/ja057832n. [DOI] [PubMed] [Google Scholar]
- 36.Du F, Zhou Z, Mo ZY, Shi JZ, Chen J, Liang Y. Mixed macromolecular crowding accelerates the refolding of rabbit muscle creatine kinase: Implications for protein folding in physiological environment. J. Mol. Biol. 2006;364:469–482. doi: 10.1016/j.jmb.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Ignatova Z, Krishnan B, Bombardier JP, Marcelino AMC, Hong J, Gierasch M. From the test tube to the cell: exploring the folding and aggregation of a beta-clam protein. Biopolymers. 2007;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasahara K, McPhie P, Minton AP. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J Mol Biol. 2003;326:1227–1237. doi: 10.1016/s0022-2836(02)01443-2. [DOI] [PubMed] [Google Scholar]
- 39.Qu Y, Bolen D. Efficacy of macromolecular crowding in forcing proteins to fold. Biophysical Chemistry. 2002;101-102:155–165. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 40.Spencer DS, Xu K, Logan TM, Zhou HX. Effects of pH, salt, and macromolecular crowding on the stability of FK506-binding protein: an integrated experimental and theoretical study. J Mol Biol. 2005;351:219–232. doi: 10.1016/j.jmb.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 41.Charlton LM, Barnes CO, Li C, Orans J, Young GB, Pielak GJ. Residue-level interrogation of macromolecular crowding effects on protein stability. J Am Chem Soc. 2008;130:6826–6830. doi: 10.1021/ja8005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perham M, Stagg L, Wittung-Stafshede P. Macromolecular crowding increases structural content of folded proteins. FEBS Letters. 2007;581:5065–5069. doi: 10.1016/j.febslet.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 43.Elcock AH. Atomic-level observation of macromolecular crowding effects. Proc Natl Acad Sci U S A. 2003;100:2340–2344. doi: 10.1073/pnas.0535055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Homouz D, Stagg L, Wittung-Stafshede P, Cheung MS. Macromolecular crowding modulates folding mechanism of α/β protein apoflavodoxin. Biophysical Journal. 2009;96:671–680. doi: 10.1016/j.bpj.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minh DDL, Chang CE, Trylska J, Tozzini V, McCammon JA. The Influence of Macromolecular Crowding on HIV-1 Protease Internal Dynamics. J Am Chem Soc. 2006;128:6006–6007. doi: 10.1021/ja060483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung MS, Thirumalai D. Crowding and confinement effects on structures of the transition state ensemble in proteins. J. Phys. Chem. B. 2007;111:8250–8257. doi: 10.1021/jp068201y. [DOI] [PubMed] [Google Scholar]
- 47.Friedel M, Sheeler DJ, Shea J-E. Effects of confinement and crowding on the thermodynamics and kinetics of folding of a minimalist beta-barrel protein. J. Chem. Phys. 2003;118:8106–8113. [Google Scholar]
- 48.Shaw MR, Thirumalai D. Free polymer in a colloidal solution. Physical Review A. 1991;44:R4797–R4800. doi: 10.1103/physreva.44.r4797. [DOI] [PubMed] [Google Scholar]
- 49.Zhou H-X. Effect of mixed macromolecular crowding agents on protein folding. Proteins. 2008;72:1109–1113. doi: 10.1002/prot.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung MS, Finke JM, Callahan B, Onuchic JN. Exploring the interplay of topology and secondary structural formation in the protein folding problem. J. Phys. Chem. B. 2003;107:11193–11200. [Google Scholar]
- 52.Betancourt MR, Thirumalai D. Pair potentials for protein folding: choice of reference states and sensitivity of predicted native states to variations in the interaction schemes. Protein Sci. 1999;8:361–369. doi: 10.1110/ps.8.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung MS, Thirumalai D. Nanopore-protein interactions dramatically alter stability and yield of the native state in restricted spaces. J Mol Biol. 2006;357:632–643. doi: 10.1016/j.jmb.2005.12.048. [DOI] [PubMed] [Google Scholar]
- 54.Zhang BW, Jasnow D, Zuckerman DM. Efficient and verified simulation of a path ensemble for conformational change in a united-residue model of calmodulin. Proc Natl Acad Sci U S A. 2007;104:18043–18048. doi: 10.1073/pnas.0706349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venturoli D, Rippe B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am. J. Physiol. Renal. Physiol. 2005;288:F605–F613. doi: 10.1152/ajprenal.00171.2004. [DOI] [PubMed] [Google Scholar]
- 56.Onuchic J, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Gruebele M. The fast protein folding problem. Ann. Rev. Phys. Chem. 1999;50:485–516. doi: 10.1146/annurev.physchem.50.1.485. [DOI] [PubMed] [Google Scholar]
- 58.Camacho CJ, Thirumalai D. Kinetics and thermodynamics of folding in model proteins. Proc Natl Acad Sci U S A. 1993;90:6369–6372. doi: 10.1073/pnas.90.13.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou HX. Loops, linkages, rings, catenanes, cages, and crowders: entropy based strategies for stabilizing proteins. Acc. Chem. Res. 2004;37:123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
- 60.Waxham MN, Tsai AL, Putkey JA. A mechanism for calmodulin (CaM) trapping by CaM-kinase II defined by a family of CaM-binding peptides. J. Biol. Chem. 1998;273:17579–17584. doi: 10.1074/jbc.273.28.17579. [DOI] [PubMed] [Google Scholar]
- 61.Minton AP. Excluded volume as a determinant of macromolecular structure and reactivity. Biopolymers. 1981;20:2093–2120. [Google Scholar]
- 62.Samiotakis A, Wittung-Stafshede P, Cheung MS. Folding, stability and shape of proteins in crowded environments: Experimental and computational approaches. International Journal of Molecular Sciences. 2009;10:572–588. doi: 10.3390/ijms10020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains. Nature Struct. Biol. 2001;8:990–997. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 64.Cheung MS, Chavez LL, Onuchic JN. The Energy Landscape For Protein Folding and Possible Connections to Function. Polymer. 2004;45:547–555. [Google Scholar]
- 65.Klimov DK, Thirumalai D. Viscosity dependence of the folding rates of proteins. Phys Rev Lett. 1997;79:317–320. [Google Scholar]
- 66.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics methods for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 67.Sanbonmatsu KY, Garcia AE. Structure of Met-Enkephalin in explicit aqueous solution using replica exchange molecular dynamics. Proteins. 2002;46:225–234. doi: 10.1002/prot.1167. [DOI] [PubMed] [Google Scholar]
- 68.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules I. The method. Journal of Computational Chemistry. 1992;13:1011–1021. [Google Scholar]
- 69.Chodera JD, Swope WC, Pitera JW, Seok C, Dill KA. Use of the weighted histogram analysis method for the analysis of simulated and parallel tempering simulations. J. Chem. Theory Comput. 2007;3:26–41. doi: 10.1021/ct0502864. [DOI] [PubMed] [Google Scholar]
- 70.Allen MW, Urbauer RJ, Zaidi A, Williams TD, Urbauer JL, Johnson CK. Fluorescence labeling, purification, and immobilization of a double cysteine mutant calmodulin fusion protein for single-molecule experiments. Anal. Biochem. 2004;325:273–284. doi: 10.1016/j.ab.2003.10.045. [DOI] [PubMed] [Google Scholar]
- 71.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.