Abstract
Neural crest cells (NCCs) are a unique population of multipotent cells that migrate along defined pathways throughout the embryo and give rise to many diverse cell types including pigment cells, craniofacial cartilage and the peripheral nervous system (PNS). Aberrant migration of NCCs results in a wide variety of congenital birth defects including craniofacial abnormalities. The chemokine Sdf1 and its receptors, Cxcr4 and Cxcr7, have been identified as key components in the regulation of cell migration in a variety of tissues. Here we describe a novel role for the zebrafish chemokine receptor Cxcr4a in the development and migration of cranial NCCs (CNCCs). We find that loss of Cxcr4a, but not Cxcr7b results in aberrant CNCC migration, defects in the neurocranium, as well as cranial ganglia dismorphogenesis. Moreover, overexpression of either Sdf1b or Cxcr4a causes aberrant CNCC migration and results in ectopic craniofacial cartilages. We propose a model in which Sdf1b signaling from the pharyngeal arch endoderm and optic stalk to Cxcr4a expressing CNCCs is important for both the proper condensation of the CNCCs into pharyngeal arches and the subsequent patterning and morphogenesis of the neural crest derived tissues.
Keywords: chemokine, cxcr4, sdf1, cxcr7, neural crest, migration, neurocranium, endoderm
Introduction
The neural crest (NC) is an evolutionarily conserved cell population among vertebrates that has long fascinated researchers due to the ability of neural crest cells (NCCs) to migrate and differentiate into a number of different cell types (LaBonne and Bronner-Fraser, 1998) (Ayer-Le Lievre and Le Douarin, 1982). NCCs are induced at the border of the neural plate and the non-neural ectoderm, delaminate from the neural tube and migrate throughout the body, giving rise to various cell types including pigment cells, craniofacial cartilage and the peripheral nervous system (PNS) (Ayer-Le Lievre and Le Douarin, 1982) (Brugmann et al., 2006) (Huang and Saint-Jeannet, 2004) (Raible et al., 1992) (Kontges and Lumsden, 1996) (Trainor, 2005) (Trainor and Krumlauf, 2000) (Schilling and Kimmel, 1994) (Serbedzija et al., 1994) (Serbedzija et al., 1990). Cranial neural crest cells (CNCCs) migrate in three streams, with the anteriormost CNCCs emigrating first, followed by the emigration of more caudally located CNCCs. The segmental fate of the CNCCs is dictated through the premigratory position of the CNCCs along the antero-posterior axis of the embryo. Once the CNCCs reach their destination in the arches, signaling via tissue interactions between endoderm, mesoderm, ectoderm and CNCCs directs the final cell fate (Clouthier and Schilling, 2004).
The peripheral nervous system in the anterior region of the embryo consists of a series of cranial ganglia and glia that are derived from both CNCCs and ectodermal placodes. While the precise contributions of the placodes and CNCCs to many of the zebrafish cranial ganglia is not known, the trigeminal sensory ganglia are derived from both CNCCs and placodal cells (Holzschuh et al., 2005; Knaut et al., 2005; Schilling and Kimmel, 1994). Trigeminal neurons extend axonal projections throughout the head and are important for detecting chemical, mechanical and thermal stimuli (Knaut et al., 2005).
In the zebrafish embryo, CNCCs of the pharyngeal arches give rise to the ventral pharyngeal skeletal elements. Specifically, pharyngeal arch one gives rise to the Meckel’s and palatoquadrate cartilages, arch two forms the basihyal, ceratohyal and hyosymplectic cartilages and arches three through seven give rise to the ceratobranchial cartilages (Schilling et al., 1996) (Piotrowski et al., 1996). The dorsal craniofacial cartilages make up the neurocranium or “braincase”. The anterior neurocranium is composed of two rod-like structures, termed trabeculae, which fuse anteriorly and connect to the ethmoid plate. The anterior neurocranium is derived from the anteriormost CNCCs that migrate out from the dorsal neural tube and reach the dorsal anterior aspect of the head following a path along the optic stalks, medial to the eyes (Wada et al., 2005) (Eberhart et al., 2006; Langenberg et al., 2008).
Several recent studies have implicated multiple key signaling pathways in the development of craniofacial cartilages and ganglia. Previous studies have established that defects in Shh signaling in zebrafish cause aberrant neurocranium and viscerocranial development (Wada et al., 2005) (Eberhart et al., 2006). Additionally, Fgf signaling from the pharyngeal endoderm is important for normal arch and subsequent cartilage development (Crump et al., 2004; Piotrowski and Nusslein-Volhard, 2000). Finally, Retinoic Acid (RA) signaling has also been shown to be important during craniofacial development, specifically for endodermal pouch morphogenesis (Kopinke et al., 2006). Thus, complex signaling interactions are required for proper development of the craniofacial skeleton. The zebrafish NC provides an outstanding model for the study of migration of diverse cell types and their subsequent differentiation. Yet while many studies have uncovered various molecular mechanisms for NCC differentiation and patterning, little is known about the signaling effectors that regulate migration of CNCCs (Yelick and Schilling, 2002).
Chemokines are small, secreted chemoattractants that have been well studied for their roles in regulating cell migration during embryogenesis, immune response and cancer (Kucia et al., 2004). The chemokine, Stromal cell derived factor (Sdf1), which preferably binds to the receptors Cxcr4 and Cxcr7 (Balabanian et al., 2005) (Bleul et al., 1996) (Boldajipour et al., 2008; Horuk, 2001), has been reported to regulate cell trafficking of hematopoetic stem cells (Horuk, 1998) and to promote migration of germ cells (Doitsidou et al., 2002); (Thorpe et al., 2004); (Dumstrei et al., 2004; Knaut et al., 2003; Molyneaux et al., 2003) (Sasado et al., 2008). Recently, Boldajipour and colleagues (2008) have shown that during germ cell migration, Sdf1 signals through Cxcr4, while Cxcr7 instead functions as a mock receptor for Sdf1. Cxcr7 thus functions in ligand sequestration and is thought to play a role in the generation of an Sdf1 gradient that directs germ cell migration into the presumptive gonad (Boldajipour et al., 2008).
cxcr4 has also been shown to be expressed in migrating NCCs in the mouse embryo. sdf1, conversely, is expressed within the migratory path of the NCCs and has been shown in mouse to signal through Cxcr4 to mediate positioning of dorsal root ganglia (DRG) (Belmadani et al., 2005). In addition, expression analysis of sdf1 in the chick embryo reveals an sdf1 expression domain within the branchial arch region (Rehimi et al., 2008).
Two cxcr4 genes, cxcr4a and cxcr4b, have been isolated in zebrafish and are closely related to mammalian cxcr4. Similar to mouse, zebrafish cxcr4a is expressed within NCCs (Chong et al., 2001). Although a functional analysis of the cxcr4 genes in NCC migration has not been reported for zebrafish, cxcr4b has been shown to be important for proper positioning of the partially NC-derived trigeminal sensory ganglia (Knaut et al., 2003). Moreover, Sdf1a signaling has been implicated in melanophore patterning of the zebrafish embryo, suggesting a link between Sdf1-Cxcr4 signaling and NC migration (Svetic et al., 2007). Here we describe a new role for Cxcr4a and Sdf1b, but not Cxcr7b in CNCC migration and patterning during zebrafish craniofacial development.
Materials and Methods
Animals
The zebrafish were maintained according to Westerfield (1993) and staged by hours post fertilization and morphology according to Kimmel (1995). ntlb195 and flhn1 mutant lines were obtained from the Zebrafish International Resource Center.
Embryo manipulation and analysis
Whole-mount in situ hybridization was adapted from Thisse and Thisse (1998) and Brent and colleagues (2003) (Brent et al., 2003). Double fluorescent in situ hybridization was performed as described by Pineda (2006). Immunohistochemistry was performed as described (Ungos et al., 2003). anti-Zn8 primary antibody (ZIRC) was used at a 1:25 dilution. Anti-HuC/D (Molecular Probes) was used at a 1:1000 dilution. Alcian Blue staining was performed as described (Schilling et al., 1996).
For live imaging, 12 hpf embryos were mounted in 0.7% low melt agarose in fish water and oriented in a dorso-lateral angle. CNCC migration was followed using a Zeiss LSM 510 confocal microscope equipped with a heated stage set to 28°C. Z stack images were taken once every 5 minutes. Double in situ hybridizations were visualized using Zeiss LSM 510 confocal microscope. Quantification of expression overlap was preformed using Zeiss Meta software. The colocalizaion coefficients were calculated based on relative number of pixels that overlap, expressed over total number of pixels for a value of 0 (not colocalized) to 1 (colocalized). cxcr4a expression with neural crest cell markers (barx1, dlx2a, hand2, crestin), and both sdf1b and cxcr7b with the endodermal marker nkx2.3 have 0.9–1 colocalization coefficients, showing regions of pixel overlap.
Sections were completed following embedding of embryos in either 1.5 % agar in 5% sucrose or in OCT and were sectioned using a crysotat for a thickness between 9–12 µm.
Antisense morpholino oligonucleotide injections
Antisense Morpholino Oligonucleotide (MO) for cxcr4a, cxcr7b and sdf1b have been previously described: cxcr4a MO1 5’-ATAAGCCATCTCTAAAAGACTTCTC-3’; cxcr4a mismatch MO 5’ATAAACCATATCTAAGAGACGTCT-3’; cxcr4a MO3 5’GACTTCTCCCGTTCCTTCAGTCTCC-3’ (Chong et al., 2007);sdf1b 5’-CGCTACTACTTTGCTATCCATGCCA-3'(Knaut et al., 2005); cxcr7b 5’ TCATTCACGTTCACACTCATCTTGG-3’ (Dambly-Chaudiere et al., 2007). Universal Control MO sequence: 5’-CCTCTTACCTCAGTTACAATTTATA-3’. The oligonucleotides were dissolved in distilled water. cxcr4a MO3 or cxcr4a MO1 were injected into 1– to 4-cell-stage embryos together with rhodamine dextran (Molecular Probes). Universal control morpholino and cxcr4a mismatch morpholinos did not result in any craniofacial phenotypes. cxcr4a MO1 and cxcr4a MO3 gave similar craniofacial phenotypes.
DNA constructs and RNA overexpression
For overexpression, the cxcr4a or sdf1b ORF was cloned into the pCS2 vector. RNA was prepared using the mMessage mMachine capped RNA transcription kit (Ambion). Capped RNA was injected into 1-cell-stage embryos together with rhodamine dextran (Molecular Probes).
Results
The chemokine receptors cxcr4a and cxcr7b are expressed within the pharyngeal arches
Previous work has shown that the chemokine receptor cxcr4 is expressed in the trunk NC in mouse embryos and is important for proper DRG positioning (Belmadani et al., 2005). We asked whether (1) cxcr4 has a conserved expression pattern within the NC among vertebrates and (2) if cxcr4 is required for NCC migration in the zebrafish embryo. We first analyzed the expression patterns of both cxcr4a and cxcr4b during zebrafish embryonic development. At 14–17 hours post fertilization (hpf), when CNCCs are migrating, cxcr4b mRNA is not expressed in the NC (data not shown; (Chong et al., 2001), whereas cxcr4a is expressed within the cranial NCCs but not in trunk NCCs of the embryo (Figure 1 A,B) (Chong et al., 2007); (Chong et al., 2001). At 17 hpf, cxcr4a is also expressed in the region of the optic stalk (Figure 1B; arrow, inset shows higher magnification view) and by 24 hpf, cxcr4a is expressed both in the optic stalk region (Figure 1C, arrow) and throughout the pharyngeal arches. We do however note that cxcr4a is excluded from the central region of arch 1, which may correspond to the mesodermal core (Figure 1C). Although we also observe this core upon sectioning, we cannot completely rule out the possibility that cxcr4a is expressed at low levels in the mesodermal core. At 25 hpf, as the pharyngeal arches begin to condense into distinct structures, cxcr4a expression begins to form discrete expression domains that correspond to the pharyngeal arches (Figure 1 D). By 36 hpf, cxcr4a expression is prominent in the optic stalk (Chalasani et al., 2007; Li et al., 2005) and at a lower level throughout the arches (Figure 1E).
Figure 1. cxcr4a, cxcr7b, and sdf1b expression in the zebrafish embryo.
Dorsal views (A,B,C,F,G,H,K,L,M), lateral views (D,E,I,J,N,O) and anterior to the left. (A–E) cxcr4a expression. cxcr4a is observed in NCCs migrating to the arches (A). At 17 hpf (B) and 24 hpf (C) is expressed in the optic stalk (B,C; arrows) and pharyngeal arch region but is excluded from the central arch, likely corresponding to the mesodermal core. By 25 hpf, cxcr4a expression condenses into discreet arches (D) At 36 hpf, cxcr4a is expressed in the optic stalk and throughout the arches (E).
(F–J) cxcr7b expression. From 14–17hpf, cxcr7b is expressed in the midbrain, otic placode and in rhombomeres 3, 5 and 6 and, at 17 hpf, in the optic stalk (F,G; arrow in G to optic stalk). By 24 hpf, cxcr7b is expressed in the ventral region of the posterior pharyngeal arches (H, arrow). From 28–48 hpf, cxcr7b expression is seen throughout the pharyngeal arches (I–J).
(K–O) sdf1b expression. At 14–17hpf, sdf1b is expressed in the region of the migrating CNCCs, within the optic stalk and dorsal rim of eye (K,L; arrows in L and M). By 24 hpf (M) sdf1b is expressed in the typical scalloped pattern of the pharyngeal endoderm. At 25hpf, sdf1b is expressed in endodermal pouches 1–3 (N) and pouches 1 and 2 at 36 hpf (O).
Recent reports have shown that the chemokine sdf1 binds both cxcr4 and cxcr7 chemokine receptors, although cxcr7 functions as a mock receptor (Balabanian et al., 2005) (Boldajipour et al., 2008; Horuk, 2001). Moreover, cxcr7 and cxcr4 genes function together to guide germ cells to the presumptive gonad (Sasado et al., 2008) and to guide migration of the lateral line during zebrafish development (Dambly-Chaudiere et al., 2007) (Valentin et al., 2007). We therefore analyzed the expression pattern of cxcr7b throughout the development of the zebrafish embryo to ascertain whether or not cxcr4a and cxcr7b are similarly expressed in the pharyngeal arches. From 14–17 hpf, cxcr7b is expressed in the midbrain, the region surrounding the otic placode, optic stalk and in rhombomeres 3, 5 and 6 (Figure 1 F,G; arrow) but is not expressed in the CNCCs at these early stages. By 24 hpf, cxcr7b is expressed in the posterior arch region, ventral to the otic vesicle (Figure 1 H; arrow) and from 28–48 hpf is expressed throughout the pharyngeal arches (Thisse, 2001) (Figure 1 I,J). Taken together, the results of the expression analysis show that both cxcr4a and cxcr7b, but not cxcr4b, are expressed in the pharyngeal arch region. However, the expression patterns of these chemokine receptors within the pharyngeal region are distinct from one another suggesting they may function within different tissues.
The chemokine Sdf1b is expressed in the pharyngeal endoderm
We next examined the expression patterns of the Cxcr4 receptor ligands sdf1a and sdf1b to determine if they might function with cxcr4a in CNCC development. We did not detect sdf1a in the CNCC (data not shown;(Thisse, 2001), but found sdf1b expressed within the domain of CNCC migration from 14–17 hpf, as has been previously described (Chong et al., 2007) (Figure 1 K,L). At 17hpf, sdf1b is also expressed within the optic stalk region and along the dorsal rim of the eye (Figure 1 L, arrows). At 24 hpf, sdf1b is expressed in the optic stalk (Figure 1 M, arrow) and in a scalloped pattern that is typical of the endoderm of the pharyngeal arches (Figure 1 M). As pharyngeal arch development proceeds, the pharyngeal endoderm migrates laterally to form pouches that interdigitate each of the pharyngeal arches (Crump et al., 2004). Between 25–36 hpf, sdf1b is seen in pharyngeal endodermal pouches 1, 2 and faintly in pouch 3 (Figure 1 N,O). Previous studies have also shown that sdf1b is expressed posterior and dorsal to the optic chiasm at 48 hpf and functions in retinal axon guidance (Chalasani et al., 2007; Li et al., 2005). We note, however, that expression of sdf1b is already evident in the region of the optic stalk by 17 hpf (Figure 1L, arrows). Importantly, it has been reported that anterior CNCCs travel along the dorsal optic vesicle to populate the region that will give rise to the neural crest derived anterior neurocranium (Wada et al., 2005) (Eberhart et al., 2006; Langenberg et al., 2008).
Signaling between various tissue types including ectoderm, CNCCs and endoderm is integral to proper pharyngeal arch development (Graham and Smith, 2001) (Knight et al., 2005) (Piotrowski and Nusslein-Volhard, 2000). In order to rule out the possibility, that sdf1b and cxcr4a are also expressed within the ectoderm, we prepared transverse and sagital cryosections through the pharyngeal arch region of embryos 25 hpf that were labeled by in situ hybridization for sdf1b and cxcr4a mRNA. From this analysis, we find that neither sdf1b nor cxcr4a are expressed in the ectoderm within the pharyngeal arch region, although we cannot rule out the possibility that there is expression that is not detectable by in situ hybridization (Supplemental Figure S1).
cxcr4a is expressed in the CNCC of the pharyngeal arches
We next performed additional expression analyses to determine which tissues cxcr4a, sdf1b and cxcr7b are expressed in during pharyngeal arch morphogenesis. To determine whether cxcr4a, cxcr7b or sdf1b are expressed in the NC of the pharyngeal arches, we performed double fluorescent in situ hybridization (Pineda et al., 2006) using the CNCC marker dlx2a, which is expressed throughout the pharyngeal arch NC (Kimmel and Eberhart, 2008). cxcr4a has been reported to be expressed in CNCCs in zebrafish during CNCC migration (Chong et al., 2001). We find that cxcr4a and the neural crest marker dlx2a partially overlap in the anterior arches at 25 hpf after CNCC migration has ceased (Figure 2 A–C, arrow). We also find that sdf1b and cxcr7b are not co-expressed with dlx2a in the anterior arches. cxcr7b is, however, coexpressed in a small subset of dlx2a positive cells within the posterior arches (Supplemental Figure S2 J–L; data not shown). Moreover, cxcr7b is expressed in cells that surround and interdigitate each of the dlx2a positive pharyngeal arches at 28hpf (Supplemental Figure S2 J–L); this pattern is similar to the expression of markers known to be expressed in the pharyngeal endoderm.
Figure 2. Expression of cxcr4a, cxcr7b and sdf1b in the developing pharyngeal arches.
Lateral views, anterior to the left. Single channel (A, B, D, E, G, H, J, K, M, N, P, Q, S, T) and merged (C, F, I, L, O, R, U) images of confocal micrographs of double fluorescent in situ hybridization of cxcr4a, cxcr7, and sdf1b along with tissue specific marker expression in the pharyngeal arch region. Neural crest markers are indicated in green with cxcr4a (red) in A–O, and endodermal marker nkx2.3 (red) with sdf1b or cxcr7b (green) in P–U.
(A–C) CNCC marker dlx2a expression (green) overlaps cxcr4a (red) in a subset of CNCCs (yellow, arrows) at 25hpf. (D–F) barx1 is coexpressed in a subset of cxcr4a cells at 25hpf. (G–I) At 28hpf, hand2 is expressed in the ventral most domain of arch CNCCs and partially overlaps with cxcr4a in this domain (yellow, arrows). (J–L) cxcr4a (red) is mostly excluded from sox10 nonectomesenchymal expression at 19hpf (green). (M–O) The pan CNCC marker crestin is expressed in a broad domain in the pharyngeal arch region, where cxcr4a is also expressed. cxcr4a is excluded from the dorsal crestin domain that corresponds to nonectomesenchymal NCCs. (P–R) sdf1b (green) and the endodermal marker nkx2.3 (red) overlap in pouch 2 (arrows) at 28 hpf. (S–U) cxcr7b (green) is coexpressed with nkx2.3 (red) throughout pharyngeal arch endoderm at 31hpf (yellow; arrowheads).
We next examined whether cxcr4a colocalizes with barx1, which is expressed in the pharyngeal arch NC (Sperber and Dawid, 2008). We find that at 25hpf barx1 is coexpressed in a subset of NCCs with cxcr4a in the anterior pharyngeal arches. This colocalization is restricted to the ventral and medial portion of the arches, but not to the dorsal region of the arches where barx1 expression is absent. We also note that cxcr4a is expressed in a region dorsal to the arches between arches 2 and 3, which does not correspond to the pharyngeal arch crest (Figure 2 D–F). In order to confirm the exact boundaries of barx1 expression within the pharyngeal arch crest, we examined the colocalization of barx1 and dlx2a at 25hpf. Consistent with our findings regarding the colocaliztion of barx1 and cxcr4a, we find that in the anterior arches, barx1 expression is excluded from the dorsal domain of dlx2a but is coexpressed in the medial and ventral domain of dlx2a expression (Supplemental Figure S2 M–O). hand2 is expressed in CNCCs that reside in the ventral-most region of the pharyngeal arches (Miller et al., 2003). Consistent with the expression results with barx1, we find that cxcr4a and hand2 partially overlap in the ventral-most CNCCs of the anterior pharyngeal arches at 28hpf (Figure 2 G–I; arrows).
In order to ascertain whether cxcr4a is also expressed in the nonectomesenchymal CNCCs that give rise to the cranial ganglia and glia, we examined whether or not cxcr4a is coexpressed with the nonectomesenchymal NC marker sox10 during pharyngeal development (Blentic et al., 2008). We find that at 19hpf cxcr4a is excluded from the sox10 expression domain in the anterior stream but does colocalize with a small subset of sox10 expressing cells midway between the eye and otic vesicle (Figure 2 J–L). We next examined whether cxcr4a colocalizes with the pan-NC marker crestin. At 28 hpf, cxcr4a and crestin are partially colocalized throughout the anterior pharyngeal arch crest, but cxcr4a is excluded from the nonectomesenchymal NC, located dorsal to the anterior arches (Figure 2 M–O). Taken together, these results show that cxcr4a is expressed throughout CNCC migration and this expression persists within the CNCCs of the anterior pharyngeal arches post-migration, during pharyngeal arch condensation and patterning.
sdf1b and cxcr7b are expressed in the pharyngeal endoderm
In order to determine whether sdf1b and cxcr7b are expressed in the endoderm, we performed double fluorescent in situ hybridization using the endodermal marker nkx2.3. We find that sdf1b is co-expressed with nkx2.3 in pharyngeal pouch 2 (Figure 2 P–R; arrows). Moreover, while nkx2.3 is only weakly expressed in pouch 1, sdf1b is strongly expressed in the first pouch. Additionally, cxcr7b is partially coexpressed with nkx2.3 in the pharyngeal arch endoderm (Figure 2 S–U; arrowheads). These results indicate that cxcr7b and sdf1b are expressed in the pharyngeal endoderm.
cxcr4a, cxcr7b and sdf1b expression within the pharyngeal arches
We next sought to determine whether the expression of these chemokine receptors and sdf1b might overlap using double fluorescent in situ hybridization analyses. sdf1b and cxcr4a are expressed in a complementary manner at 25 hpf, with only a subset of coexpressing cells at the border of the endoderm and arch crest (Supplementary Figure S2 A–C). Conversely, we find that cxcr7b and sdf1b are coexpressed in both the medial endoderm and within the first 3 pharyngeal pouches at 28hpf (Supplementary Figure S2 D–F). At 28 hpf, cxcr4a and cxcr7b are not coexpressed within the endoderm, but do show coexpression in a small subset of cells within the posterior arches (Supplementary Figure S2 G–I).
In summary, these results indicate that cxcr4a is expressed predominantly in the anterior pharyngeal arch CNCCs, while sdf1b and cxcr7b are expressed predominantly in the endoderm during pharyngeal arch development. This is consistent with previous studies that have shown that the chemokines sdf1a and/or sdf1b are often expressed in adjacent and complementary tissues to cxcr4a or cxcr4b expressing domains (Li et al., 2005) (Schwarting et al., 2006) (Miyasaka et al., 2007) (Chalasani et al., 2007).
Loss of Cxcr4a signaling results in neurocranium defects
The expression of cxcr4a in the arch neural crest during pharyngeal arch development and within the optic stalk suggests that cxcr4a might function in the development of the ventral and dorsal craniofacial cartilages. In order to investigate the function of Cxcr4a in craniofacial development, we used Morpholino-mediated depletion of the cxcr4a gene product (Chong et al., 2007). We utilized 2 Morpholinos designed to knockdown cxcr4a, a control mismatch Morpholino (Chong et al., 2007) and a universal control Morpholino to verify specificity of the cxcr4a Morpholinos. We find that both cxcr4a morpholinos result in similar craniofacial phenotypes (described below), whereas, even high doses (up to 25ng) of the control mismatch and universal control Morpholino do not result in any craniofacial phenotypes. Moreover, when cxcr4a mRNA is coinjected with either cxcr4a Morpholino, the craniofacial phenotype is partially rescued (Table 1). Taken together, these results indicate that the cxcr4a Morpholinos are specific for cxcr4a mRNA.
Table 1.
Quantification of results from migration, alcian blue and in situ hybridization analyses in cxcr4a morphant embryos.
| Injection | Cartilage Phenotype | ||
|---|---|---|---|
| Ethmoid Plate Reduced |
Hyposym- pletic Defects |
Ectopic | |
|
cxcr4a Morpholino (MO) (ng/nl) |
26/65 (40%) |
||
|
cxcr4a MO (as control for rescue) |
21/52 (40%) |
||
|
cxcr4a/cxcr7b* Morpholino (ng/nl) |
37/64 (54%) |
||
|
sdf1b Morpholino (ng/nl) |
30/53 (56%) |
||
|
cxcr4a mRNA overexpression |
16/62 (26%) |
22/62 (35.5%) |
13/62 (21%) |
|
cxcr4a Morpholino+cxcr4a mRNA(200pg) rescue |
11/42 (26%) |
6/42 (14%) |
|
cxcr7b morphants alone have no craniofacial phenotype
At 6.5 dpf, cxcr4a morphants have reduced forebrains as compared to wildtype and control Morpholino injected embryos, resulting in a “dolphin-like” appearance (Supplementary Figure S3 G,H). We performed alcian blue staining to visualize the cartilage of these morphant larvae. While disrupting Cxcr4a signaling does not result in defects in the ventral pharyngeal skeleton, we find that the ethmoid plate is severely reduced or absent and that the trabeculae of the neurocranium are fused and thickened (Figure 3 A,B). This reduction in the neurocranium accounts for the “dolphin-like” appearance of cxcr4a morphants.
Figure 3. cxcr4a morphants have neurocranium defects.

Dorsal views and anterior to the top (A–C) and lateral views, anterior to the left (D, E). (A) wild type (WT) neurocranium at 6.5 dpf. (B) cxcr4a morphants lack an ethmoid plate (ep) and have fused trabeculae (t). (C) sdf1b morphants phenocopy cxcr4a morphants, and have more severe anterior basicapsular commissure defects (arrow). (D, E) Wildtype (D) and cxcr4a morphant embryo (E) labeled with anti-HuC 52 hpf to indicate the cranial ganglia. Morphant embryos have a reduction in cranial ganglia IX and X (arrow). bc, basicapsular commissure; bp, basil plate; oa, occipital arch; ep, ethmoid plate; t, trabeculae.
If Sdf1b and Cxcr7b function with Cxcr4a in its role in craniofacial development, then knockdown of sdf1b and cxcr7b should result in a similar phenotype to cxcr4a morphants. Indeed we find that sdf1b morphants phenocopy the craniofacial defects seen in cxcr4a morphants, and have additional defects in anterior basicapsular commisure formation (Figure 3 C, arrow). Knock down of cxcr7b however, does not result in craniofacial defects (data not shown). These results suggest that Sdf1b and Cxcr4a function together in development of the neurocranium.
Although both cxcr7b and cxcr4a are expressed in the pharyngeal arches, knockdown of either gene surprisingly does not result in ventral craniofacial cartilage defects. To rule out the possibility that these genes might function redundantly with one another during morphogenesis of the pharyngeal skeleton, we performed double knockdown of cxcr7b and cxcr4a and analyzed the resulting cartilage phenotype. cxcr4a and cxcr7b double morphants do not have a phenotype within the pharyngeal skeleton, suggesting they do not function redundantly with one another during pharyngeal arch morphogenesis (not shown).
In order to ascertain whether cxcr4b might also play a role in craniofacial development, we performed alcian blue staining on the cxcr4b mutant, odysseus (ody) at 5 dpf (Knaut et al., 2003). We find that ody mutants have normal craniofacial patterning, suggesting that cxcr4b does not play a role in craniofacial development (data not shown). This observation is supported by absence of cxcr4b expression within CNCCs and pharyngeal arches (Chong et al., 2001).
Two recent reports by Nair and Schilling (2008) and Mizoguchi et al (2008) show that cxcr4a is important for endodermal cell migration during zebrafish gastrulation. The studies show that impaired Cxcr4a signaling leads to aberrant endodermal cell migration during gastrulation. Moreover, while Nair and Schilling report duplication of endodermal organs in morphants, Mizoguchi and colleagues report loss or reduction of these endodermal organs (Mizoguchi et al., 2008; Nair and Schilling, 2008). In order to rule out the possibility that endodermal duplications as reported by Nair and Schilling (2008) for cxcr4a morphant embryos can cause craniofacial defects similar to sdf1b/cxcr4a morphants, we analyzed the craniofacial skeletons of notail (ntl) and floating head (flh) mutant larvae, which also display endodermal duplications (Chen et al., 2001). We find that neither ntl nor flh mutant larvae have defects in the craniofacial skeleton that are similar to sdf1b or cxcr4a morphants (Supplementary Figure 9). Both mutants have overall smaller heads and thus the skeletal elements are reduced in size. Additionally, both mutant lines show loss of the more posterior ceratobranchial cartilages. The dorsal skeleton, however, including the ethmoid plate and tribecula appear normal (Supplementary Figure S9). Thus, we find that neither ntl nor flh mutant larvae phenocopy the craniofacial defect observed in the sdf1b or cxcr4a morphants, suggesting that the craniofacial phenotypes associated with sdf1b/cxcr4a knockdown are not a consequence of early endodermal migration defects.
cxcr4a morphants have reduced cranial ganglia
Trigeminal sensory neurons have been shown to express cxcr4b during development (Knaut et al., 2005). Moreover, assembly of trigeminal neurons into a single ganglion cluster is mediated through signaling by Sdf1a and Sdf1b through Cxcr4b. In the absence of sdf1a, sdf1b or cxcr4b, the trigeminal neurons form two ganglion clusters instead of a single bilateral cluster (Knaut et al., 2005).
We reasoned that since cxcr4a is expressed in the CNCCs and is important for CNCC migration, cranial ganglia might also be affected in the absence of cxcr4a signaling. We therefore analyzed the cranial ganglia in morphants at 52 hpf using antibodies against HuC/D, which is expressed in all differentiated neurons. At 52 hpf, we find that cranial ganglia IX and X are often reduced or missing in cxcr4a morphants (Figure 3 D,E; arrow). Conversely, cxcr7b morphants do not have defects in posterior cranial ganglia development (data not shown). These results suggest that cxcr4a is important for the development of cranial ganglia IX and X.
cxcr4a morphants have disorganized pharyngeal arches
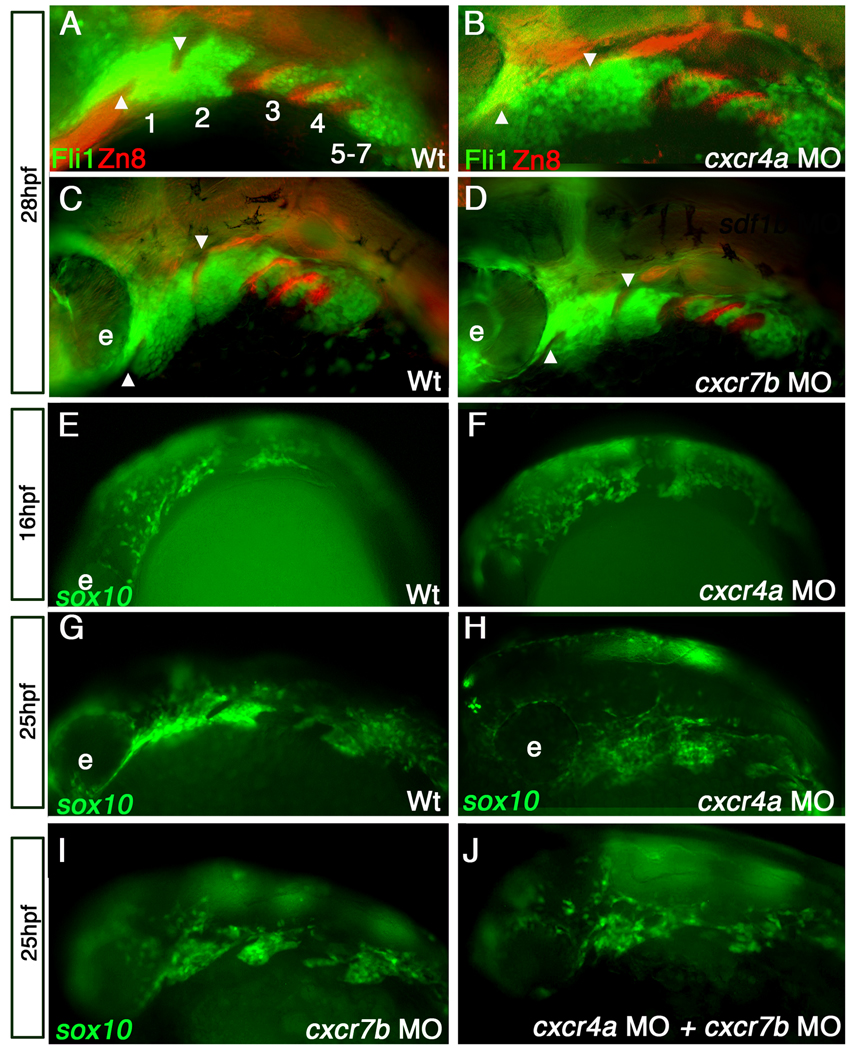
cxcr4a is expressed in the CNCCs during migration, while sdf1b is expressed in the migratory pathway of CNCCs (Chong et al., 2001) (Chong et al., 2007) from 12–20 hpf. Moreover, after CNCC migration is complete (between 20–21 hpf), cxcr4a and sdf1b are expressed within the pharyngeal arches. We therefore asked whether cxcr4a morphants display normal arch morphology by examining pharyngeal arches of cxcr4a morphants at 28 hpf in a tg{fli1::eGFP} background. tg{fli1::eGFP} is expressed in postmigratory CNCCs and the developing vasculature of the embryo (Lawson and Weinstein, 2002). While cxcr4a morphants have overall normal forming arches, they do however have loose and disorganized anterior arch structure as compared to the condensed arches of control siblings. In morphant embryos, the stomodeum is not well defined and CNCCs appear within the region of pharyngeal pouch 1, which normally creates a separation between arches 1 and 2 (Figure 4 compare A,C with B arrowheads mark position of endodermal pouch 1 and stomodeum; n= 24/42 or 57% affected). Conversely, we find that cxcr7b morphants have properly condensed arches at 28hpf (Figure 4 D). This suggests that Cxcr4a, but not Cxcr7b, is involved in NCC compaction into the pharyngeal arches.
Figure 4. cxcr4a morphants have aberrant CNCC migration and arch condensation.
Lateral views, anterior to the left. (A–D) tg{fli1::eGFP} (green) embryo marking postmigratory CNCCs double labeled with the endoderm marker Zn-8 (red). (E–J) tg{sox10::eGFP}(green) labeling neural crest migration and condensation into the pharyngeal arch region. (A, B) cxcr4a morphants (B) show loose arch structure, as compared to wild type embryos (A) Arrowheads mark endodermal pouch1 and stomodeum in A–D as a reference for compaction. In addition, endodermal specification and morphogenesis is unaffected as seen by Zn8 expression (red). (C, D) cxcr7b MO injected embryos (D) have normal CNCC (green) compaction in the pharyngeal arch and endodermal expression of Zn8 (red) as compared to wildtype at 28hpf (C).
(E, G) Wildtype tg{sox10::eGFP} embryos display normal onset of CNCC migration from the neural tube toward the ventral arch region, migrate normally mid-migration at 16 hpf (E) and are properly organized post-migration at 25 hpf (G). Early to mid CNCC migration is unaffected in cxcr4a morphants at 16hpf, as CNCCs are migrating ventrally from the neural tube (F) but show loose organization of CNCC in the anterior arches at 25 hpf (H). cxcr7b morphant embryos have normal arch morphology (I) and double cxcr4a and cxcr7b morphants (J) show similar disorganized arch phenotype as cxcr4a single morphants.
As signaling between tissues is important in pharyngeal arch development, we next asked whether endodermal pouch formation is normal in cxcr7b, cxcr4a and sdf1b morphants using the endoderm marker Zn8. We find that cxcr7b, cxcr4a and sdf1b morphants are capable of forming endodermal pouches (Figure 4 A–D red; data not shown). Thus although sdf1b and cxcr7b are expressed in the pharyngeal endoderm, they do not play a role in endoderm specification or morphogenesis of the endodermal pouches.
Since cxcr4a morphants display disorganized cells within the arches and cxcr4a is expressed in the CNCCs during migration, we asked whether CNCC migration is affected in morphants using a tg{sox10::GFP} reporter, which is expressed in migrating NCCs (Carney et al., 2006). At 16 hpf, CNCCs in cxcr4a morphant embryos are capable of initiating migration and NCCs are seen migrating ventrally from the dorsal neural tube into the pharyngeal arch region (Figure 4 E,F). At 25 hpf, however, morphant CNCCs appear loosely organized and more spread out in the pharyngeal arches, as compared to control siblings, in which CNCCs have begun to condense into clearly distinguishable arches (Figure 4 G,H; n = 25/37 or 68% affected).
We next asked whether cxcr7b knockdown also results in disorganized pharyngeal arches using a tg{sox10::GFP} reporter. We find that cxcr7b morphants have normal arch morphology at 25 hpf (Figure 4 I). Since cxcr7b expression within the arches does not commence until well after the NCCs have reached the arches, it is not surprising that cxcr7b is not necessary for establishing the general structure of the pharyngeal arches. Moreover, knocking down both cxcr4a and cxcr7b simultaneously results in a phenotype similar to cxcr4a single morphants (Figure 4 J). These results suggest that cxcr7b is dispensable for pharyngeal arch development, while cxcr4a is important for the formation of discrete pharyngeal arches.
cxcr4a morphants have CNCC migration defects
We next followed migration of CNCCs using time lapse confocal microscopy in wild type embryos and cxcr4a morphants using a tg{sox10::GFP} reporter line. We followed CNCCs from the onset of migration until after the CNCCs have condensed into the arches. We find that although cxcr4a morphant CNCCs are capable of initiating migration from the neural tube, CNCCs migrate to ectopic sites in the embryo (Figure 5 E–J; Supplementary movie 1 and Supplementary movie 2). For example, we find that while in wild type control embryos, CNCCs avoid the eye during the early stages of CNCC migration (Figure 5 A–D)(Langenberg et al., 2008), morphant CNCCs precociously migrate over the eye (Figure 5 F–I). In addition, after CNCCs have ceased migrating in control embryos, we observe CNCCs in morphant embryos continuing to migrate out over the yolk of the embryo (Figure 5 E,J; arrows in J; Supplementary movie 1 and Supplementary movie 2).
Figure 5. Time lapse imaging of aberrant CNCC migration in cxcr4a morphant embryos.

Lateral views, anterior to left. Stills taken from live time lapse imaging from 14–23 hpf. WT CNCC migration from 14 to 23hpf (A–D). WT CNCCs avoid the eye during early CNCC migration (B, arrow). cxcr4a morphant CNCCs do not avoid the eye during early phases of migration (F–I; B, arrow). (E,J) Brightfield and fluorescent (E) and fluorescence only (J) images of cxcr4a morphant CNCCs migrating ectopically over the yolk. e, eye. Numbers denote arches.
We conclude that cxcr4a is important for normal CNCC migration into the pharyngeal arches. We do however find that morphants display only a mild migration phenotype as most CNCCs reach the pharyngeal arches. This observation is supported by alcian blue analysis of morphants, which reveals normal ventral craniofacial cartilages in morphant embryos. Moreover, analysis of the pharyngeal arch markers endothelin-1, hand2 and sox9a, shows that while morphant embryos initially have a delay or reduced expression of these genes, they recover shortly thereafter and have normal arch expression (Supplementary Figure S3). Conversely, the anterior migration of CNCCs to the presumptive neurocranium may be particularly sensitive to aberrant migration and cxcr4a/sdf1b levels, resulting in defects within the neurocranium.
cxcr4a and sdf1b overexpression result in CNCC migration defects and ectopic craniofacial cartilages
Loss of cxcr4a or sdf1b via morpholino mediated knockdown results in phenotypes in dorsal craniofacial cartilages, but not in the ventral pharyngeal cartilages. cxcr4a and sdf1b are however expressed in the ventral pharyngeal arches throughout development. We thus asked whether overexpression of either sdf1b or cxcr4a throughout the embryo would result in phenotypes associated with both viscerocranium and neurocranium development. Although informative, whole embryo overexpression does have some caveats. While we coinject with a caged dextran and screen the embryos for expression, there remains the possibility that the mRNA and consequently protein, is in fact mosaic. However, we feel that there is sufficient RNA to remain during CNCC migration and early branchial arch patterning, during the time of sdf1b and cxcr4a function.
Overexpressing cxcr4a or sdf1b mRNA results in ectopic cartilages, abnormal hyosymplectic elements, unilateral loss of anterior cartilages (Supplementary Figure S4, arrowheads, shows cxcr4a overexpression), and fusions of ventral pharyngeal cartilages (Supplementary Figure S4; Supplementary Figure S5). Embryos injected with either cxcr4a or sdf1b mRNA have small ectopic cartilages located near the Meckel’s and ceratohyal cartilages (Supplementary Figure S4 A,B,D for cxcr4a; Supplementary Figure S5 for sdf1b). Higher doses of cxcr4a mRNA (250 pg) cause fusions of the Meckel’s and ceratohyal cartilages and less frequently, fusions of the posterior ceratobranchial elements (Supplementary Figure S4 H; arrows). Similarly, sdf1b overexpression results in fusions of pharyngeal arch 1 and arch 2 derived skeletal elements (Supplementary Figure S5 C). In addition, the hyosymplectic cartilage is often misshapen or severely reduced in embryos ectopically expressing either sdf1b or cxcr4a mRNA (Supplementary Figure S4 H; Supplementary Figure S5 C). Taken together, these results suggest that both sdf1b and cxcr4a play important roles during arch development. Conversely, injection of up to 350 pg of gfp mRNA does not result in any craniofacial defects or ectopic cartilages (Supplementary Figure S4 E,G).
Overexpression of sdf1b or cxcr4a also results in defects within the neurocranium. Lower doses of cxcr4a mRNA (100 pg) or sdf1b mRNA cause reduction of the trabeculae and a cleft within the ethmoid plate (Supplementary Figure S4 F; Supplementary Figure S5 D). Moreover, ectopic cartilages are often present near the ethmoid plate (Supplementary Figure S4 F; Supplementary Figure S5 D). This phenotype is the opposite of cxcr4a morphant larvae, where the ethmoid plate is absent/reduced and the trabeculae are fused and thickened. At higher doses of sdf1b or cxcr4a mRNA, however, we find that the ethmoid plate phenotype resembles that of cxcr4a and sdf1b morphant larvae (data not shown). This suggests that ethmoid plate development is particularly sensitive to the levels of sdf1b and cxcr4a expression. Similar results have been reported where the overexpression of sdf1b mimics the sdf1b morphant phenotype in trigeminal ganglion development, as a localized source of Sdf1b is needed for directing migration of cell types (Knaut et al., 2005). In the absence of Sdf1b or in the overexpression of Sdf1b, migrating cells lack a directional cue and are unable to reach their destination.
In order to determine whether these skeletal defects due to ectopic sdf1b or cxcr4a expression occur early, during migration into the pharyngeal arches, or whether the effects are due to later postmigratory patterning defects, we analyzed the effects of sdf1b and cxcr4a overexpression on the CNCCs of the pharyngeal arches in tg{sox10::GFP} embryos. We find that ectopic sdf1b or cxcr4a mRNA results in disorganized arches, with anterior arches often fused (Supplementary Figure S6 C) or reduced (Supplementary Figure S6 B). Posterior arch formation is typically unaltered in embryos overexpressing either sdf1b or cxcr4a. Significantly, overexpression of either sdf1b or cxcr4a results in ectopic clusters of CNCCs along the yolk and anteriorly within the head (Supplementary Figure S6 A–D). The presence of ectopic CNCCs throughout the embryo suggests that sdf1b and cxcr4a overexpression results in CNCC defects during the early stages of pharyngeal arch development that correspond to migration of CNCCs into the arches and the formation of discrete arches.
Ap2a lies upstream of cxcr4a during CNCC development
tfap2a has been shown to be important for neural crest development, survival and for craniofacial cartilage development (Knight et al., 2004; Knight et al., 2005; Knight et al., 2003). We therefore asked whether cxcr4a and sdf1b expression might be affected in the tfap2a mutant lockjaw (low). At 25 hpf, sdf1b and cxcr7b expression is normal in low mutant embryos (data not shown) suggesting that endoderm formation is unaffected in low mutant embryos. We do however find a reduction of cxcr4a expression in the pharyngeal region of low embryos at 25 hpf (Supplementary Figure S7). This result is consistent with previous observations that CNCC markers such as dlx2 (Knight et al., 2003) and fli1 (Knight et al., 2005) are reduced or absent in low mutant embryos.
The bHLH transcription factor hand2, a target of endothelin signaling, is expressed in ventral CNCCs during pharyngeal arch development and is required for ventral skeletal cartilage development (Miller et al., 2003) (Ruest et al., 2003). We next asked whether hand2 might regulate cxcr4a, cxcr7b and sdf1b expression. We find that at 25 hpf, hanS6 mutant embryos display normal sdf1b, cxcr7b and cxcr4a expression, suggesting hand2 does not regulate cxcr7b, cxcr4a or sdf1b (data not shown). This data is consistent with the observation that early ventral pharyngeal arch markers are unchanged in hand2 mutant embryos (Miller et al., 2003).
Retinoic acid signaling regulates cxcr7b, cxcr4a and sdf1b expression
In order to ascertain whether RA signaling might regulate sdf1b, cxcr4a and cxcr7b expression, we treated embryos with both RA (100 µM) and the RA synthesis inhibitor DEAB (50 µM) between 21–25 hpf and then examined the expression of cxcr4a, sdf1b and cxcr7b. Embryos treated with RA have a reduction in the expression of cxcr4a and cxcr7b (Figure 6 E, G as compared to A, C; arrowheads demarcate same position in A, E, I as reference and arrows in C, G, K point to the otic vesicle) and a very slight reduction of sdf1b in pouch 2 (Figure 6 F, compared to B). We do not, however, find a change in optic stalk expression of sdf1b or cxcr4a in response to RA treatment (Figure 6 E, F). The modest decrease in sdf1b in response to RA treatment is consistent with previous reports that pharyngeal pouches 1 and 2 are insensitive to RA treatment (Kopinke et al., 2006). Treating embryos with RA from 24–28 hpf also reduces cxcr7b expression in the region of the otic vesicle and pharyngeal endoderm but not in the forebrain (Figure 6 D, H).
Figure 6. Retinoic Acid signaling is upstream of cxcr4a and sdf1b.
Lateral views, anterior to the left of 25hpf (A–C, E–G, I–K) and 28hpf embryos (D,H,L). (A–D) Control embryos treated with DMSO between 21 and 25 hpf show normal expression of cxcr4a (A), sdf1b (B), and from 21–25hpf and 24–28 hpf showing normal cxcr7b expression (C, D respectively). Embryos treated with 100µM RA from 21–25 hpf show reduced expression of cxcr4a (E as compared to A; arrowheads), a mild reduction in pouch 2 of sdf1b (F) and mild reduction of cxcr7b (G) below the otic vesicle (arrows point to otic vesicle region in C, G, K). Treatment from 24–28hpf (H) causes a reduction of cxcr7b expression in the endoderm but not the forebrain as compared to wildtype embryos. Embryos treated with the 50µM RA synthesis inhibitor DEAB show an expansion of cxcr4a (I) and sdf1b (J) expression posterior to the otic vesicle (ov) as compared to DMSO treated embryos in (A,B). Embryos treated with the Fgf inhibitor SU5402 from 21–25hpf (K) and 24–28 hpf (L) have reduced expression of cxcr7b in the otic region (K,L) and within the pharyngeal endoderm (L) as compared to DMSO treated controls (C,D).
Embryos treated with DEAB show no effect on cxcr7b expression or sdf1b and cxcr4a expression within the optic stalk (Figure 6 I, J). DEAB treated embryos do, however, have an expansion of both cxcr4a and sdf1b throughout the pharyngeal arch region (Figure 6 I,J). Moreover, expanded sdf1b expression does not take on the normal shape of the pharyngeal endoderm. These results are consistent with previous studies on the effects of DEAB treatment on endodermal pouch morphogenesis, which causes enlargement of endodermal pouches and disrupts posterior pouch formation (Kopinke et al., 2006). Results from our expression analysis in response to RA and DEAB drug treatments suggest that RA signaling lies upstream of cxcr4a, sdf1b and cxcr7b during pharyngeal arch development.
Fgf signaling regulates cxcr7b but not cxcr4a and sdf1b expression
Previous studies have established that Fgf signaling from the pharyngeal endoderm is important for normal arch development (Crump et al., 2004; Piotrowski and Nusslein-Volhard, 2000). We therefore asked whether pharmacological disruption of Fgf signaling would disrupt sdf1, cxcr7b and cxcr4 expression. Treating embryos between 21–25 hpf with the Fgf inhibitor SU5402 does not affect sdf1b or cxcr4a expression, suggesting that Fgf signaling does not regulate sdf1b and cxcr4a expression (not shown). We do however find that treating embryos with SU5402 between 21–25 hpf does result in decreased expression of cxcr7b in the region of the otic placode (Figure 6 C, K). As cxcr7b expression commences throughout the pharyngeal region after 28 hpf, we treated embryos with SU5402 from 24–28 hpf and examined expression of cxcr7b at 28 hpf. SU5402 treatment between 24–28 hpf results in reduced cxcr7b expression in the pharyngeal endoderm (Figure 6 D, L). To ensure that our results were a direct result of Fgf signaling on cxcr7b expression and not due to defects in endoderm morphogenesis caused by SU5402 treatment, we treated tg{fli1::eGFP} embryos from 24–28 hpf with SU5402 and visualized the endoderm using the Zn8 antibody. We find that SU5402 treatment at this time period does not result in loss of endodermal pouches. Instead, pouches are slightly misshapen and are not as elongated as DMSO treated control embryos (Supplementary Figure S8). We conclude that this mild pouch phenotype cannot completely account for the reduction of cxcr7b expression. In sum, reduced cxcr7b expression is likely due to transcriptional regulation of cxcr7b via Fgf signaling.
Hedgehog signaling regulates expression of sdf1b within the optic stalk
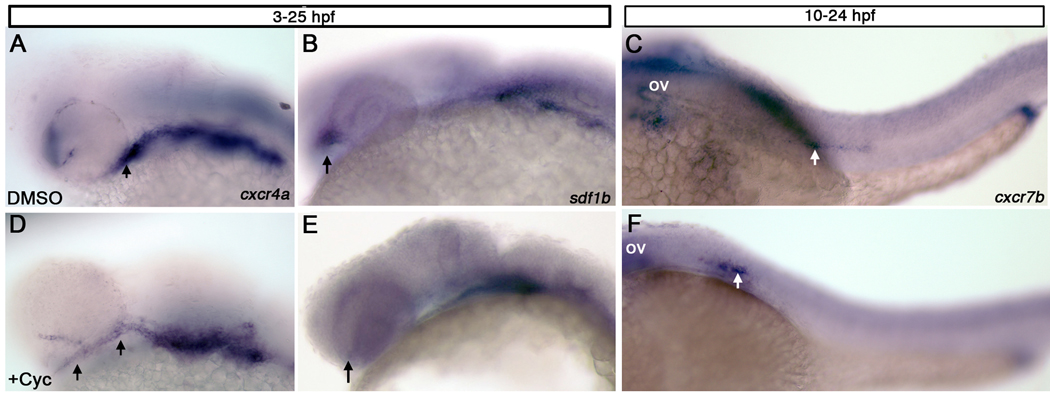
Sonic hedgehog (Shh) signaling has been shown to be important in the development of the anterior neurocranium and viscerocranial cartilages (Eberhart et al., 2006; Wada et al., 2005). In fact, shh mutants and embryos treated with the Hedgehog (Hh) inhibitor cyclopamine have fusion of trabeculae and loss of the ethmoid plate (Wada et al., 2005), similar to cxcr4a and sdf1b morphants (this study). We treated embryos with cyclopamine throughout development (3 hpf–25 hpf) and examined the expression of cxcr4a, cxcr7b and sdf1b. We find that cyclopamine does not disrupt expression of cxcr7b or sdf1b within the pharyngeal arches (Figure 7 B,E; data not shown). However, cyclopamine treatment does result in reduced cxcr4a expression in the first pharyngeal arch and in mispositioning of the optic stalk (Figure 7 A,D; arrows). Interestingly, cyclopamine treatment also results in loss of sdf1b expression in the optic stalk, suggesting that Hh signaling lies upstream of sdf1b in the development of the anterior neurocranium (Figure 7 B,E). Thus a possible mechanism of Shh action may in part be mediated by sdf1b signaling.
Figure 7. Hedgehog signaling regulates expression of sdf1b within the optic stalk.
Lateral views, anterior to the left observed at 25hpf (A, B, D, E) and 28hpf (C, F). (A, B) Embryos treated with DMSO or cyclopamine (D, E) from 3 hpf–25hpf show mispositioned cxcr4a expression within the optic stalk and less expression in the first arch of cyclopamine treated embryos (D; arrows), as compared to DMSO treated control embryos (A). sdf1b expression in the optic stalk is lost in cyclopamine treated animals (E; arrow) in comparison to DMSO treated controls (B; arrows point to optic stalk in B,E). (C, F) Embryos treated with cyclopamine from 10–24 hpf and observed at 28 hpf with cxcr7b expression show lateral line migration defects. The lateral line primordium in cyclopamine treated embryos (F) does not migrate posteriorly as compared to DMSO treated embryos (C; white arrows). ov, otic vesicle.
In addition, embryos treated with cyclopmamine from 10–24 hpf and subsequently fixed at hpf show lateral line migration defects. Specifically, we find that while cxcr7b is still expressed in the lateral line, migration of the lateral line is inhibited (Figure 7 C, F). In control embryos, the lateral line has reached the anterior portion of the yolk sac extension by 28hpf, while in cyclopamine treated animals, the lateral line lies more anteriorly midway between the otic vesicle and the anterior portion of the yolk sac extension.
Discussion
The results presented in this study highlight a previously undescribed role for chemokine signaling through Sdf1b/Cxcr4a in CNCC migration and in the patterning of CNCC derivatives. Knockdown of Cxcr4a results in aberrant migration of CNCCs to ectopic destinations and in the failure of CNCCs to fully condense within the pharyngeal arches. We hypothesize that the aberrant condensation of CNCCs is due to a role for cxcr4a/sdf1b in the final steps of CNCC migration. Overexpression of sdf1b and cxcr4a results in defects in CNCC migration, pharyngeal arch morphogenesis and patterning of craniofacial cartilage elements. Moreover, both knockdown and overexpression of sdf1b and cxcr4a leads to migration and craniofacial defects, suggesting that a localized source and strict balance of Sdf1b signaling is required for CNCC migration. Thus, Sdf1b signaling through Cxcr4a functions in the guidance of CNCCs during migration (Knaut et al., 2005); this study). The fact that the CNCCs can reach their final destination suggests that Cxcr4a/Sdf1b signaling is required for directed migration and condensation but not for overall targeting. This hypothesis is consistent with a conserved role for cxcr4a/sdf1b in directing the migration of a variety of cell types, including mouse DRG and germ cells (Doitsidou et al., 2002); (Thorpe et al., 2004); (Dumstrei et al., 2004);(Knaut et al., 2003); (Molyneaux et al., 2003) (Belmadani et al., 2005; Sasado et al., 2008).
Interestingly, cxcr4a and sdf1b expression persists in the pharyngeal arches throughout craniofacial development, after CNCCs have ceased migrating. Additionally, we find that posterior cranial ganglia IX and X are affected in cxcr4a morphant embryos suggesting a role for Cxcr4a in patterning the cranial ganglia. While the exact contributions of the CNCCs to the zebrafish cranial ganglia are not known, in chick, the CNCC give rise to both the glossopharyngeal (IX) and vagal (X) cranial ganglia (reviewed by (Barlow, 2002). Additionally, craniofacial cartilage phenotypes associated with the overexpression of cxcr4a suggest that in addition to being important for migration, cxcr4a signaling may also be important for patterning of the craniofacial skeleton. Moreover, while Cxcr7 has been shown to be important in migration of germ cells by helping to establish an Sdf1b gradient (Boldajipour et al., 2008), we find that Cxcr7b does not function with Sdf1/Cxcr4 in CNCC migration and subsequent development of the craniofacial skeleton and ganglia.
A role for Sdf1b/Cxcr4a in CNCC migration
The ability of NCCs to migrate throughout the embryo to reach their final destinations is imperative for the development of multiple structures including craniofacial cartilage and the peripheral nervous system. In the cranial region, CNCC migration is thought to occur by relying on cell-cell communication with each other, as well as with the environment through which they migrate (Kulesa and Fraser, 2000). While work has elucidated the roles of various signaling molecules in the patterning of these NC derivatives, the molecular mechanisms involved in directing NCC migration remain unclear (Clouthier and Schilling, 2004; Yelick and Schilling, 2002). Ephrins and Ephrin receptors have, however been implicated in regulating CNCC migration and are thought to function by altering cell adhesion, likely through interactions with integrins. In the chick, Ephrin-B1 inhibits NCC migration in the caudal somite and is required for segmental organization of migration (Krull, 1998) (Krull et al., 1997). In the mouse embryo, Ephrin-B2 is important for CNCC migration into branchial arch 2, while Ephrin-B1 is required for multiple steps during CNCC migration and mutant mice exhibit a cleft palate (reviewed by (Davy and Soriano, 2005). The Neuropilin family has also been shown to be required for NC migration. Knockdown of Neuropilin-1 in chick results in a reduction in the number of CNCCs that reach the branchial arches (McLennan and Kulesa, 2007). Previous studies have also shown that signaling through Neuropilin-2 by Semaphorin 3F is important for proper CNCC migration and condensation of the trigeminal sensory ganglia. Interestingly, while CNCC migration is affected in these mutants, no skeletal defects are apparent (Gammill et al., 2006). sdf1b and cxcr4a morphants also show aberrant CNCC migration, similar to Neuropilin2/Semaphorin3F mutants, but they do not exhibit defects in the viscerocranium, suggesting that most CNCCs are able to reach the arches and become normally patterned. Thus, it is likely that multiple redundant mechanisms are employed to ensure proper craniofacial development.
The results from this study identify a new signaling mechanism that is important for CNCC migration and craniofacial skeleton morphogenesis. We find that Cxcr4a/Sdf1b signaling is important for the migration of CNCCs to the pharyngeal arches and around the eye. cxcr4a morphants show aberrant migration of CNCCs across the eye and along the yolk, as well as defects in the condensation of CNCCs into pharyngeal arches. Moreover, overexpression of cxcr4a and sdf1b results in ectopic migration of CNCCs throughout the anterior embryo.
Recent studies have shown that proper migration of CNCCs around the eye is important for development of the neurocranium (Eberhart et al., 2008) (Langenberg et al., 2008). Eberhart and colleagues (2008) recently described a role for the microRNA Mirn140 in modulating the attraction of CNCCs via platelet derived growth factor (pdgfaa) signaling during zebrafish neurocranium development. In embryos lacking Mirn140, CNCCs accumulate around the optic stalk near the attractive Pdgfaa signal and fail to migrate further to the oral ectoderm, resulting in loss of the ethmoid plate and in palatal clefting (Eberhart et al., 2008).
Another study by Langenberg and colleagues (2008) demonstrated that the eye is important for organizing CNCC migration within the zebrafish embryo. In wild type embryos, CNCCs migrate anteriorly around the eye and are not detected on the eye surface until 23–24 hpf. Soon after, however, CNCCs will cover the surface of the eye. In chokh(rx3) mutant embryos that fail to develop eyes, CNCCs do not migrate anteriorly and instead remain disorganized in a region posterior to the normal eye location. Analysis of craniofacial cartilages in chokh(rx3) mutant larvae shows loss of the ethmoid plate and fusion of the trabeculae. chokh(rx3) mutant larvae do, however, have normal viscerocranium development (Langenberg et al., 2008).
Our results implicate Sdf1b/Cxcr4a signaling in organizing the migration of CNCCs around the eye to the presumptive anterior neurocranium. Loss of cxcr4a or overexpression of sdf1b results in precocious migration over the surface of the eye, as well as loosely organized pharyngeal arches. Other studies have indicated that Sdf1b signals from the optic stalk to cxcr4b expressing retinal ganglion cells and functions in retinal axon guidance (Chalasani et al., 2007; Li et al., 2005). Thus Sdf1b signaling is important for organizing multiple migratory events around the eye during zebrafish embryonic development.
A role for Cxcr4a/Sdf1b signaling in patterning the craniofacial elements
After neural crest cells have migrated into the branchial arches or anterior optic stalk region, communication between various cell types and tissue layers is paramount to normal patterning of CNCCs (reviewed in (Clouthier and Schilling, 2004; Graham and Smith, 2001). Previous reports indicate that the proper formation of endodermal pouches is necessary for craniofacial patterning (Crump et al., 2004; Piotrowski and Nusslein-Volhard, 2000). Moreover, studies regarding the role of the ectoderm in craniofacial development have uncovered the importance of signaling between the ectoderm and CNCCs during craniofacial patterning (Knight et al., 2005) (Eberhart et al., 2006). Our analyses of gene expression for sdf1b suggest that Sdf1b is secreted from the optic stalk and from the anterior endodermal pharyngeal pouches. This observation, coupled with previous studies that have shown that Sdf1 preferentially binds the Cxcr4 receptor (Horuk, 2001), make it likely that Sdf1b signals from the optic stalk and endoderm to Cxcr4a expressing NCCs to direct migration, craniofacial patterning and morphogenesis. Our hypothesis is supported by the observation that sdf1b morphant embryos phenocopy the skeletal defects observed in cxcr4a morphants. Our observations further support the idea of crosstalk between tissues, specifically between Sdf1b in the optic stalk and endoderm to Cxcr4a expressing CNCCs during craniofacial development and identify new players in communication between these tissues.
Studies have shown that Fgf and RA signaling are important for patterning the endoderm during craniofacial development. Specifically, RA and Fgf signaling are dispensable for endoderm specification but have been implicated in regulating endodermal pouch morphogenesis (Crump et al., 2004) (Kopinke et al., 2006). We find that RA signaling is important for the regulation of sdf1b and cxcr4a expression within the pharyngeal arches but not within the optic stalk during zebrafish development. Conversely, we find that Fgf signaling does not regulate sdf1b/cxcr4a expression.
Analysis of cxcr4a/sdf1b morphant craniofacial phenotypes with Alcian Blue staining revealed defects in the neurocranium, including fusion of the tribeculae and loss of the ethmoid plate. These defects phenocopy loss of Hh signaling via the genetic mutant sonic you and through pharmacological treatments using the Hh inhibitor cyclopamine (Wada et al., 2005) (Eberhart et al., 2006). Hh signaling has been shown to act multiple times during development to initially direct the condensation of CNCCs onto the roof of the stomodeum and later to pattern the neurocranium (Eberhart et al., 2006). The signaling components functioning downstream of Hh in neurocranium development have not, however, been uncovered. We find that sdf1b expression within the optic stalk is regulated by Hh signaling. Inhibition of Hh signaling via cyclopmaine results in loss of sdf1b expression. Taken together, we suggest that sdf1b might function downstream of Hh signaling in the development of the anterior neurocranium.
In conclusion, the developmental program that functions to pattern the craniofacial cartilages is a complex process that involves various signaling molecules and communication between various cell types. Here we show that zebrafish chemokine signaling plays a novel role in craniofacial development. Specifically, our data suggests that Sdf1b signaling through the chemokine receptor Cxcr4a is necessary for the condensation of CNCCs into discreet arch structures and in the development of the craniofacial skeleton.
Supplementary Material
CNCC migration beginning at 14 hpf in a WT tg{sox10::eGFP}embryo.
Lateral views, anterior to the left. (A,B) tg{fli1::eGFP} (green) embryo marking postmigratory CNCCs double labeled with the endoderm marker Zn-8 (red). (A) Control embryo treated from 24–28 hpf with DMSO shows normal arch and pharyngeal pouch development. (B) Embryo treated with SU5402 from 24–28 hpf shows reduced arches 3 and 4, as well as broad and less elongated posterior pharyngeal pouches as compared to DMSO treated control sibling.
Whole mount (A–C, E–G) and dissected (D,H) alcian blue stained cartilage at 4.5dpf, anterior to the top. Ventral cartilages of a wildtype or heterozygous larva (A). floating head (flt) (B) and notail (ntl) mutant larvae (F) show normal anterior ventral cartilage development, but are missing posterior ceratobranchials. floating head (C) and notail (G) mutant larvae form an ethmoid plate and trabeculae, but are missing the posterior neurocranium as compared to control neurocranium (E). Dissected neurocranium of floating head (D) and notail mutant larvae (H) showing a normal ethmoid plate. wt/het, wildtype or heterozygous; m, Meckel’s cartilage; ch, ceratohyal; cb1, 2, 3, ceratobranchial 1, 2, 3; ep, ethmoid plate; tr, tribeculae
(A) Sagittal section of 25 hpf wild type embryo stained for cxcr4a mRNA expression, arrow points to expression throughout the mesenchyme and arrowheads shows exclusion from the ectoderm. cxcr4a is also significantly reduced in the mesodermal core (B) Transverse section of 25 hpf wild type embryo stained for sdf1b expression. Expression is absent from the ectoderm (arrowhead) but present in the endoderm (arrow). Nt, neural tube; m, mesoderm
Lateral views, anterior to the left. Confocal micrographs of double fluorescent in situ hybridization of cxcr4a, cxcr7, sdf1b, dlx2a and barx1 in the pharyngeal arch region. (A–C) cxcr4a (red) is largely excluded from sdf1b (green) in the arches, but is slightly coexpressed with sdf1b at the endodermal border (D–F) cxcr7b (red) is coexpressed with sdf1b (green) throughout the medial endoderm and anterior pharyngeal pouches at 28hpf (arrowheads). (G–I) cxcr4a (green) is mostly excluded from cxcr7b (red) expressing cells, but does show some coexpression in the posterior pharyngeal region. (J–L) cxcr7b (red) is excluded from the dlx2a expressing CNCCs in the anterior arches but does show partial overlap with dlx2a expressing cells of the posterior pharyngeal arches. (M–O) barx1 is coexpressed with dlx2a in the ventral and medial portion of the pharyngeal arches but is reduced or absent in the dorsal arch (arrows).
CNCC migration beginning at 14hpf in a cxcr4a morphant embryo shows CNCC aberrantly migrating over the yolk at the end of migration.
Lateral views, anterior to the left. Expression of hand2, edn-1 and sox9a in wildtype (A, C, E) and cxcr4a morphant embryos (B, D, F) shows that expression is reduced but not absent after cxcr4a knockdown. (G) Wildtype larva at 6.5 dpf. (H) cxcr4a morphant larva at 6.5 dpf has a reduced forebrain and “dolphin” like phenotype (arrow and arrowhead).
Whole mount (A–D) and dissected (E–H) alcian blue stained cartilage at 6.5dpf, anterior to the top. Wildtype (A) and gfp mRNA injected controls (E,G) and examples of cxcr4a mRNA injected embryos (B–D,F,H). Injection of 100pg cxcr4a mRNA results in small ectopic cartilages often located near the Meckel’s and ceratohyal cartilages (B, D; arrow). We also often observe unilateral loss of Meckel’s cartilages and abnormal palatoquadrate elements (C; arrowheads). Injection of 250pg cxcr4a mRNA results in abnormal hyosymplectic elements (hs), fusions of the Meckel’s and ceratohyal cartilages, as well as fusions of the posterior ceratobranchial elements (H, arrow) as compared to control embryos injected with gfp mRNA (G). Injection of 100pg of cxcr4a mRNA causes ectopic cartilages and neurocranium defects with a reduction of the trabeculae (t) and a cleft within the ethmoid plate (ep; F; arrow) as compared to control embryos injected with gfp mRNA (E; arrow at same location). bc, basicapsular commissure; cb1–5, ceratobranchial cartilage 1–5; ch, ceratohyal; hs, hyosymplectic; pq, palatoquadrate; ep, ethmoid plate; t, trabeculae.
Dissected alcian blue stained cartilage at 6.5dpf, anterior to the top. Wildtype control larva (A, B) and sdf1b mRNA injected larvae (C, D). Injection of 250pg sdf1b mRNA results in small ectopic cartilages (C asterisk, arrow, D), abnormal hyosymplectic elements (C, arrow), fusions of the Meckel’s and palatoquadrate cartilages (C, arrowhead) as compared to control larvae (A). sdf1b mRNA overexpression causes ectopic cartilages and neurocranium defects with a reduction of the trabeculae, a clefted palate and loss of the ethmoid plate (D), as compared to control larvae (B).
Lateral views, anterior to the left of 28 hpf embryos (A,B). (A) WT tg{sox10::eGFP} embryos show normal pharyngeal arch patterning, while embryos injected with cxcr4a mRNA (B) show pharyngeal arch defects, with much of arch 1 missing and ectopic CNCCs ventro-lateral to the arch region along the yolk (arrows). Embryos injected with sdf1b mRNA (C) show similar arch defects to cxcr4a mRNA overexpression with fusion of arch 1 and 2. Lower (10x) magnification of embryo injected with sdf1b mRNA showing ectopic CNCCs along the yolk ventral to the eye and pharyngeal arches (D, arrows). Inset shows 20x magnification of ectopic CNCCs along yolk. eye is outlined with white line in panels A–C.
(A) Wildtype embryo stained for cxcr4a mRNA expression 25hpf. (B) lockjaw mutant embryos have reduced cxcr4a expression within the pharyngeal arches at 25hpf.
Acknowledgements
We thank L. Hernandez, C. Johnson and D. Killian for discussion and support; M. Singleton for excellent fish care; A. Schier for generously supplying us with the ody line; A. Schier, T. Schilling, T. Piotrowski, S. Sperber, I. Dawid and D. Stock for constructs; and M. Wright and C. C. Rossi for double Fluorescent in situ protocol; L. Niswander and C. Pyrgaki for use of confocal microscope and advice for live imaging. D. Clouthier for critical reading of this manuscript. We gratefully acknowledge the support of NIH P30 NS048154 Neuroscience Zebrafish and Imaging Cores, HD050698 and DE017699 to K.B.A, NIH F32 HD056779 to E.O.K. and F32 DE018594 to D.A.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayer-Le Lievre CS, Le Douarin NM. The early development of cranial sensory ganglia and the potentialities of their component cells studied in quail-chick chimeras. Dev Biol. 1982;94:291–310. doi: 10.1016/0012-1606(82)90349-9. [DOI] [PubMed] [Google Scholar]
- Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- Barlow LA. Cranial Nerve Development: Placodal Neurons Ride the Crest. Current Biology. 2002;12:R171–R173. doi: 10.1016/s0960-9822(02)00734-0. [DOI] [PubMed] [Google Scholar]
- Belmadani A, Tran PB, Ren D, Assimacopoulos S, Grove EA, Miller RJ. The chemokine stromal cell-derived factor-1 regulates the migration of sensory neuron progenitors. J Neurosci. 2005;25:3995–4003. doi: 10.1523/JNEUROSCI.4631-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blentic A, Tandon P, Payton S, Walshe J, Carney T, Kelsh RN, Mason I, Graham A. The emergence of ectomesenchyme. Dev Dyn. 2008;237:592–601. doi: 10.1002/dvdy.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Tapadia MD, Helms JA. The molecular origins of species-specific facial pattern. Curr Top Dev Biol. 2006;73:1–42. doi: 10.1016/S0070-2153(05)73001-5. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Sabol A, Xu H, Gyda MA, Rasband K, Granato M, Chien CB, Raper JA. Stromal cell-derived factor-1 antagonizes slit/robo signaling in vivo. J Neurosci. 2007;27:973–980. doi: 10.1523/JNEUROSCI.4132-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JN, van Bebber F, Goldstein AM, Serluca FC, Jackson D, Childs S, Serbedzija G, Warren KS, Mably JD, Lindahl P, Mayer A, Haffter P, Fishman MC. Genetic steps to organ laterality in zebrafish. Comp Funct Genomics. 2001;2:60–68. doi: 10.1002/cfg.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SW, Emelyanov A, Gong Z, Korzh V. Expression pattern of two zebrafish genes, cxcr4a and cxcr4b. Mech Dev. 2001;109:347–354. doi: 10.1016/s0925-4773(01)00520-2. [DOI] [PubMed] [Google Scholar]
- Chong SW, Nguyet LM, Jiang YJ, Korzh V. The chemokine Sdf-1 and its receptor Cxcr4 are required for formation of muscle in zebrafish. BMC Dev Biol. 2007;7:54. doi: 10.1186/1471-213X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Schilling TF. Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res C Embryo Today. 2004;72:190–199. doi: 10.1002/bdrc.20007. [DOI] [PubMed] [Google Scholar]
- Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development. 2004;131:5703–5716. doi: 10.1242/dev.01444. [DOI] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A, Soriano P. Ephrin signaling in vivo: look both ways. Dev Dyn. 2005;232:1–10. doi: 10.1002/dvdy.20200. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Reichman-Fried M, Stebler J, Koprunner M, Dorries J, Meyer D, Esguerra CV, Leung T, Raz E. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Mennecke R, Raz E. Signaling pathways controlling primordial germ cell migration in zebrafish. J Cell Sci. 2004;117:4787–4795. doi: 10.1242/jcs.01362. [DOI] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, Swartz ME, Crump JG, Kimmel CB. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Gonzalez C, Bronner-Fraser M. Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. J Neurobiol. 2006;67:47–56. doi: 10.1002/dneu.20326. [DOI] [PubMed] [Google Scholar]
- Graham A, Smith A. Patterning the pharyngeal arches. Bioessays. 2001;23:54–61. doi: 10.1002/1521-1878(200101)23:1<54::AID-BIES1007>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Holzschuh J, Wada N, Wada C, Schaffer A, Javidan Y, Tallafuss A, Bally-Cuif L, Schilling TF. Requirements for endoderm and BMP signaling in sensory neurogenesis in zebrafish. Development. 2005;132:3731–3742. doi: 10.1242/dev.01936. [DOI] [PubMed] [Google Scholar]
- Horuk R. Chemokines beyond inflammation. Nature. 1998;393:524–525. doi: 10.1038/31116. [DOI] [PubMed] [Google Scholar]
- Horuk R. Chemokine receptors. Cytokine Growth Factor Rev. 2001;12:313–335. doi: 10.1016/s1359-6101(01)00014-4. [DOI] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275:1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Eberhart JK. The midline, oral ectoderm, and the arch-0 problem. Integrative and Comparative Biology. 2008;48:668–680. doi: 10.1093/icb/icn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Blader P, Strahle U, Schier AF. Assembly of trigeminal sensory ganglia by chemokine signaling. Neuron. 2005;47:653–666. doi: 10.1016/j.neuron.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Knaut H, Werz C, Geisler R, Nusslein-Volhard C. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–282. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Nelson S, Zhang T, Schilling T. Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Dev Dyn. 2004;229:87–98. doi: 10.1002/dvdy.10494. [DOI] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development. 2005;132:3127–3138. doi: 10.1242/dev.01879. [DOI] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kopinke D, Sasine J, Swift J, Stephens WZ, Piotrowski T. Retinoic acid is required for endodermal pouch morphogenesis and not for pharyngeal endoderm specification. Dev Dyn. 2006;235:2695–26709. doi: 10.1002/dvdy.20905. [DOI] [PubMed] [Google Scholar]
- Krull CE. Inhibitory interactions in the patterning of trunk neural crest migration. Ann N Y Acad Sci. 1998;857:13–22. doi: 10.1111/j.1749-6632.1998.tb10103.x. [DOI] [PubMed] [Google Scholar]
- Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr Biol. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- Kucia M, Jankowski K, Reca R, Wysoczynski M, Bandura L, Allendorf DJ, Zhang J, Ratajczak J, Ratajczak MZ. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. 2004;35:233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Fraser SE. In ovo time-lapse analysis of chick hindbrain neural crest cell migration shows cell interactions during migration to the branchial arches. Development. 2000;127:1161–1172. doi: 10.1242/dev.127.6.1161. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Induction and patterning of the neural crest, a stem cell-like precursor population. J Neurobiol. 1998;36:175–189. doi: 10.1002/(sici)1097-4695(199808)36:2<175::aid-neu6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Langenberg T, Kahana A, Wszalek JA, Halloran MC. The eye organizes neural crest cell migration. Dev Dyn. 2008;237:1645–1652. doi: 10.1002/dvdy.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Li Q, Shirabe K, Thisse C, Thisse B, Okamoto H, Masai I, Kuwada JY. Chemokine signaling guides axons within the retina in zebrafish. J Neurosci. 2005;25:1711–1717. doi: 10.1523/JNEUROSCI.4393-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R, Kulesa PM. In vivo analysis reveals a critical role for neuropilin-1 in cranial neural crest cell migration in chick. Dev Biol. 2007;301:227–239. doi: 10.1016/j.ydbio.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Miyasaka N, Knaut H, Yoshihara Y. Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 2007;134:2459–2468. doi: 10.1242/dev.001958. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Verkade H, Heath JK, Kuroiwa A, Kikuchi Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development. 2008;135:2521–2529. doi: 10.1242/dev.020107. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Zinszner H, Kunwar PS, Schaible K, Stebler J, Sunshine MJ, O'Brien W, Raz E, Littman D, Wylie C, Lehmann R. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- Nair S, Schilling TF. Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science. 2008;322:89–92. doi: 10.1126/science.1160038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda RH, Svoboda KR, Wright MA, Taylor AD, Novak AE, Gamse JT, Eisen JS, Ribera AB. Knockdown of Nav1.6a Na+ channels affects zebrafish motoneuron development. Development. 2006;133:3827–3836. doi: 10.1242/dev.02559. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Nusslein-Volhard C. The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio) Dev Biol. 2000;225:339–356. doi: 10.1006/dbio.2000.9842. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Schilling TF, Brand M, Jiang YJ, Heisenberg CP, Beuchle D, Grandel H, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Nusslein-Volhard C. Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development. 1996;123:345–356. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodsdon W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Rehimi R, Khalida N, Yusuf F, Dai F, Morosan-Puopolo G, Brand-Saberi B. Stromal-derived factor-1 (SDF-1) expression during early chick development. Int J Dev Biol. 2008;52:87–92. doi: 10.1387/ijdb.072374rr. [DOI] [PubMed] [Google Scholar]
- Ruest LB, Dager M, Yanagisawa H, Charite J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasado T, Yasuoka A, Abe K, Mitani H, Furutani-Seiki M, Tanaka M, Kondoh H. Distinct contributions of CXCR4b and CXCR7/RDC1 receptor systems in regulation of PGC migration revealed by medaka mutants kazura and yanagi. Dev Biol. 2008;320:328–339. doi: 10.1016/j.ydbio.2008.05.544. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Piotrowski T, Grandel H, Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Hammerschmidt M, Kane DA, Mullins MC, van Eeden FJ, Kelsh RN, Furutani-Seiki M, Granato M, Haffter P, Odenthal J, Warga RM, Trowe T, Nusslein-Volhard C. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Henion TR, Nugent JD, Caplan B, Tobet S. Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J Neurosci. 2006;26:6834–6840. doi: 10.1523/JNEUROSCI.1728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Developmental potential of trunk neural crest cells in the mouse. Development. 1994;120:1709–1718. doi: 10.1242/dev.120.7.1709. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Fraser SE, Bronner-Fraser M. Pathways of trunk neural crest cell migration in the mouse embryo as revealed by vital dye labelling. Development. 1990;108:605–612. doi: 10.1242/dev.108.4.605. [DOI] [PubMed] [Google Scholar]
- Sperber SM, Dawid IB. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Dev Biol. 2008;321:101–110. doi: 10.1016/j.ydbio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetic V, Hollway GE, Elworthy S, Chipperfield TR, Davison C, Adams RJ, Eisen JS, Ingham PW, Currie PD, Kelsh RN. Sdf1a patterns zebrafish melanophores and links the somite and melanophore pattern defects in choker mutants. Development. 2007;134:1011–1022. doi: 10.1242/dev.02789. [DOI] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission. 2001 [Google Scholar]
- Thorpe JL, Doitsidou M, Ho SY, Raz E, Farber SA. Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev Cell. 2004;6:295–302. doi: 10.1016/s1534-5807(04)00032-2. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Specification and patterning of neural crest cells during craniofacial development. Brain Behav Evol. 2005;66:266–280. doi: 10.1159/000088130. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- Ungos JM, Karlstrom RO, Raible DW. Hedgehog signaling is directly required for the development of zebrafish dorsal root ganglia neurons. Development. 2003;130:5351–5362. doi: 10.1242/dev.00722. [DOI] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17:1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- Yelick PC, Schilling TF. Molecular dissection of craniofacial development using zebrafish. Crit Rev Oral Biol Med. 2002;13:308–322. doi: 10.1177/154411130201300402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CNCC migration beginning at 14 hpf in a WT tg{sox10::eGFP}embryo.
Lateral views, anterior to the left. (A,B) tg{fli1::eGFP} (green) embryo marking postmigratory CNCCs double labeled with the endoderm marker Zn-8 (red). (A) Control embryo treated from 24–28 hpf with DMSO shows normal arch and pharyngeal pouch development. (B) Embryo treated with SU5402 from 24–28 hpf shows reduced arches 3 and 4, as well as broad and less elongated posterior pharyngeal pouches as compared to DMSO treated control sibling.
Whole mount (A–C, E–G) and dissected (D,H) alcian blue stained cartilage at 4.5dpf, anterior to the top. Ventral cartilages of a wildtype or heterozygous larva (A). floating head (flt) (B) and notail (ntl) mutant larvae (F) show normal anterior ventral cartilage development, but are missing posterior ceratobranchials. floating head (C) and notail (G) mutant larvae form an ethmoid plate and trabeculae, but are missing the posterior neurocranium as compared to control neurocranium (E). Dissected neurocranium of floating head (D) and notail mutant larvae (H) showing a normal ethmoid plate. wt/het, wildtype or heterozygous; m, Meckel’s cartilage; ch, ceratohyal; cb1, 2, 3, ceratobranchial 1, 2, 3; ep, ethmoid plate; tr, tribeculae
(A) Sagittal section of 25 hpf wild type embryo stained for cxcr4a mRNA expression, arrow points to expression throughout the mesenchyme and arrowheads shows exclusion from the ectoderm. cxcr4a is also significantly reduced in the mesodermal core (B) Transverse section of 25 hpf wild type embryo stained for sdf1b expression. Expression is absent from the ectoderm (arrowhead) but present in the endoderm (arrow). Nt, neural tube; m, mesoderm
Lateral views, anterior to the left. Confocal micrographs of double fluorescent in situ hybridization of cxcr4a, cxcr7, sdf1b, dlx2a and barx1 in the pharyngeal arch region. (A–C) cxcr4a (red) is largely excluded from sdf1b (green) in the arches, but is slightly coexpressed with sdf1b at the endodermal border (D–F) cxcr7b (red) is coexpressed with sdf1b (green) throughout the medial endoderm and anterior pharyngeal pouches at 28hpf (arrowheads). (G–I) cxcr4a (green) is mostly excluded from cxcr7b (red) expressing cells, but does show some coexpression in the posterior pharyngeal region. (J–L) cxcr7b (red) is excluded from the dlx2a expressing CNCCs in the anterior arches but does show partial overlap with dlx2a expressing cells of the posterior pharyngeal arches. (M–O) barx1 is coexpressed with dlx2a in the ventral and medial portion of the pharyngeal arches but is reduced or absent in the dorsal arch (arrows).
CNCC migration beginning at 14hpf in a cxcr4a morphant embryo shows CNCC aberrantly migrating over the yolk at the end of migration.
Lateral views, anterior to the left. Expression of hand2, edn-1 and sox9a in wildtype (A, C, E) and cxcr4a morphant embryos (B, D, F) shows that expression is reduced but not absent after cxcr4a knockdown. (G) Wildtype larva at 6.5 dpf. (H) cxcr4a morphant larva at 6.5 dpf has a reduced forebrain and “dolphin” like phenotype (arrow and arrowhead).
Whole mount (A–D) and dissected (E–H) alcian blue stained cartilage at 6.5dpf, anterior to the top. Wildtype (A) and gfp mRNA injected controls (E,G) and examples of cxcr4a mRNA injected embryos (B–D,F,H). Injection of 100pg cxcr4a mRNA results in small ectopic cartilages often located near the Meckel’s and ceratohyal cartilages (B, D; arrow). We also often observe unilateral loss of Meckel’s cartilages and abnormal palatoquadrate elements (C; arrowheads). Injection of 250pg cxcr4a mRNA results in abnormal hyosymplectic elements (hs), fusions of the Meckel’s and ceratohyal cartilages, as well as fusions of the posterior ceratobranchial elements (H, arrow) as compared to control embryos injected with gfp mRNA (G). Injection of 100pg of cxcr4a mRNA causes ectopic cartilages and neurocranium defects with a reduction of the trabeculae (t) and a cleft within the ethmoid plate (ep; F; arrow) as compared to control embryos injected with gfp mRNA (E; arrow at same location). bc, basicapsular commissure; cb1–5, ceratobranchial cartilage 1–5; ch, ceratohyal; hs, hyosymplectic; pq, palatoquadrate; ep, ethmoid plate; t, trabeculae.
Dissected alcian blue stained cartilage at 6.5dpf, anterior to the top. Wildtype control larva (A, B) and sdf1b mRNA injected larvae (C, D). Injection of 250pg sdf1b mRNA results in small ectopic cartilages (C asterisk, arrow, D), abnormal hyosymplectic elements (C, arrow), fusions of the Meckel’s and palatoquadrate cartilages (C, arrowhead) as compared to control larvae (A). sdf1b mRNA overexpression causes ectopic cartilages and neurocranium defects with a reduction of the trabeculae, a clefted palate and loss of the ethmoid plate (D), as compared to control larvae (B).
Lateral views, anterior to the left of 28 hpf embryos (A,B). (A) WT tg{sox10::eGFP} embryos show normal pharyngeal arch patterning, while embryos injected with cxcr4a mRNA (B) show pharyngeal arch defects, with much of arch 1 missing and ectopic CNCCs ventro-lateral to the arch region along the yolk (arrows). Embryos injected with sdf1b mRNA (C) show similar arch defects to cxcr4a mRNA overexpression with fusion of arch 1 and 2. Lower (10x) magnification of embryo injected with sdf1b mRNA showing ectopic CNCCs along the yolk ventral to the eye and pharyngeal arches (D, arrows). Inset shows 20x magnification of ectopic CNCCs along yolk. eye is outlined with white line in panels A–C.
(A) Wildtype embryo stained for cxcr4a mRNA expression 25hpf. (B) lockjaw mutant embryos have reduced cxcr4a expression within the pharyngeal arches at 25hpf.