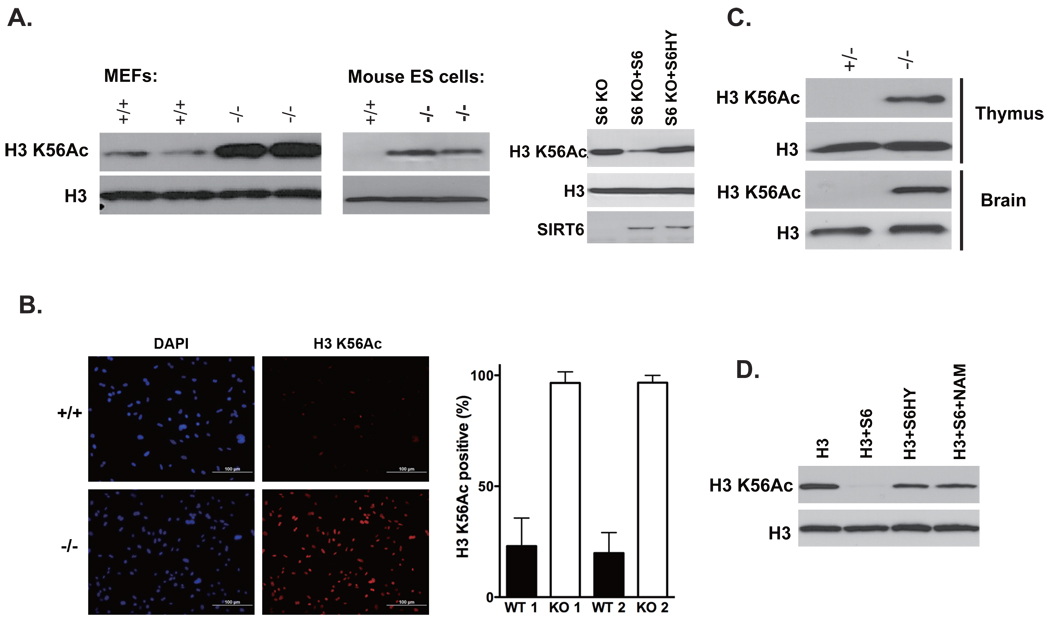
Sirtuin deacetylases promote longevity in invertebrates. Mammals possess seven sirtuin homologs, which are involved in a diverse array of processes including development, metabolism, and stress resistance1. Among these, the sirtuin SIRT6 plays a particularly important role in the maintenance of cellular and organismal homeostasis. Mouse embryonic fibroblasts (MEFs) and embryonic stem (ES) cells lacking SIRT6 show poor growth, sensitivity to genotoxins (MMS, hydrogen peroxide, and ionizing radiation), and an increased frequency of genomic aberrations, including chromosomal gaps, breaks, and fusions2. The molecular basis for the genomic instability observed in SIRT6 deficiency remains obscure. SIRT6 is tightly associated with nuclear chromatin2. It has been reported that in human cells, SIRT6 maintains telomeric stability via deacetylation of histone H3 acetylated on lysine 9 (H3 K9Ac)3; and that SIRT6 promotes recruitment of DNA-PKcs, a protein involved in the non-homologous end-joining (NHEJ) DNA repair pathway, to DNA lesions4. However, SIRT6-deficient mice show neither telomeric instability, nor defects observed in NHEJ mutants5. Therefore we sought to define alternative mechanisms by which SIRT6 maintains chromosomal integrity. Acetylation of histone H3 on lysine 56 (H3 K56Ac) is a recently described chromatin modification linked to genomic stability and genotoxin resistance. H3 K56Ac is deacetylated by the sirtuins Hst3 and Hst4 in yeast6, and SIRT1 and SIRT2 in mammalian cells7, 8. Based on knockdown experiments, it has been reported that SIRT6 does not play a significant role in H3 K56Ac deacetylation7, 9. In yeast and mammals, failure to regulate H3 K56Ac levels, or mutations mimicking such a defect, leads to impaired cell cycle progression, genotoxin sensitivity and spontaneous DNA damage6, 7, all phenotypes associated with SIRT6 deficiency in mammalian cells. To examine a potential role for SIRT6 in deacetylating H3 K56Ac in the context of SIRT6 null mutants, levels of this modification were assessed in SIRT6-deficient MEFs and littermate controls. Levels of H3 K56Ac were dramatically increased in SIRT6 knockout cells (Fig. 1A, left panel). H3 K56Ac levels were also elevated in two independent SIRT6-deficient ES cell lines compared with a wild-type control (Fig. 1A, middle panel). Reconstitution with wild-type SIRT6, but not a catalytically inactive SIRT6 mutant (H133Y)3, 10, rescued H3 K56 hyperacetylation in SIRT6-deficient fibroblasts (Fig. 1A, right panel), indicating that H3 K56 hyperacetylation in these cells is a direct consequence of loss of SIRT6 function. For assessment of H3 K56Ac levels in individual cells, immunofluorescence analysis was performed on SIRT6-deficient primary MEFs and littermate controls. H3 K56Ac staining was markedly more intense in SIRT6-deficient MEFs, and was also present in a much greater fraction of cells (Fig. 1B, left panel). H3 K56Ac was detectable in 97% of SIRT6-deficient cells, but in only 22% of controls (Fig. 1B, right panel). To test the role of SIRT6 in deacetylating H3 K56Ac in vivo, H3 K56Ac levels were assessed in tissues from SIRT6-deficient mice and littermate controls. We focused on thymus and brain, both sites of prominent SIRT6 expression2. SIRT6 deficiency was associated with dramatically higher levels of H3 K56Ac in both tissues (Fig. 1C). In order to rule out the possibility that hyperacetylation of H3 K56 in SIRT6 deficiency might represent an indirect effect of the genomic instability in this mutant background, the ability of SIRT6 to deacetylate H3 K56Ac in vitro was assessed. Wild-type SIRT6, but not the SIRT6 catalytic mutant, robustly deacetylated H3 K56Ac, a reaction blocked by the sirtuin inhibitor nicotinamide (Fig. 1D). We conclude that SIRT6 directly deacetylates H3 K56Ac to regulate global levels of this histone modification in vivo. H3 K56Ac levels are tightly regulated in yeast, flies, and mammals; increased levels of this modification lead to genomic instability and genotoxin sensitivity6, 7. In this context, there are conflicting reports regarding the impact of DNA damage induction on H3 K56Ac levels in metazoans7–9. In MEFs, we do not observe any major effect of DNA damaging agents on global H3 K56Ac levels (Fig. S1). Precise mechanisms by which H3 K56Ac modulates genomic stability and genotoxin sensitivity in mammals are unclear, and likely multi-factorial. The ability of SIRT6 to deacetylate H3 K56Ac likely represents a principal means by which this sirtuin promotes genomic stability.
Fig. 1.
SIRT6 deacetylates H3 K56Ac. (A) Hyperacetylation of H3 K56Ac in the absence of SIRT6. (Left panel) Hyperacetylation of H3 K56Ac in SIRT6-deficient MEFs. (Middle panel) Hyperacetylation of H3 K56Ac in SIRT6-deficient ES cells. (Right panel) Reconstitution of a SIRT6-deficient fibroblast cell line with catalytically active SIRT6 reduces H3 K56Ac levels. (B) Immunofluorescence analysis of SIRT6-deficient MEFs and littermate controls. (C) Hyperacetylation of H3 K56Ac in thymus (top panels) and brain (bottom panels) of a SIRT6-deficient mouse and littermate control. (D) Recombinant SIRT6 deacetylates H3 K56Ac in vitro.
Supplementary Material
Acknowledgements
The authors would like to thank Frederick Alt, Raul Mostoslavsky and Regeneron Pharmaceuticals, Inc. for SIRT6-deficient mice and ES cells; Yali Dou and David Ferguson for comments on the manuscript; and Yali Dou for p300 enzyme. Work in the authors’ laboratory is supported by a K08 award (NIA), pilot funds from the University of Michigan Cancer Research Committee, and startup funds from the University of Michigan.
References
- 1.Schwer B, et al. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Mostoslavsky R, et al. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 3.Michishita E, et al. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCord RA, et al. Aging. 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombard DB. Aging. 2009;1:12–16. doi: 10.18632/aging.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozdemir A, et al. Cell Cycle. 2006;5:2602–2608. doi: 10.4161/cc.5.22.3473. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J, et al. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das C, et al. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjeertes JV, et al. EMBO J. 2009 doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liszt G, et al. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.