Abstract
At the developing vertebrate neuromuscular junction, the acetylcholine receptor becomes aggregated at high density in the postsynaptic muscle membrane. Receptor localization is regulated by the motoneuron-derived factor, agrin, and requires an intracellular, scaffolding protein called rapsyn. However, it remains unclear where rapsyn binds on the acetylcholine receptor and how their interaction is regulated. In this study, we identified rapsyn’s binding site on the acetylcholine receptor using chimeric constructs where the intracellular domain of CD4 was substituted for the major intracellular loop of each mouse acetylcholine receptor subunit. When expressed in heterologous cells, we found that rapsyn clustered and cytoskeletally anchored CD4- α, β and ε subunit loops but not CD4-δ loop. Rapsyn-mediated clustering and anchoring was highest for β loop, followed by ε and then α, suggesting that rapsyn interacts with the loops with different affinities. Moreover, by making deletions within the β subunit intracellular loop, we show that rapsyn interacts with the α-helical region, a secondary structural motif present in the C-terminal portion of the subunit loops. When expressed in muscle cells, rapsyn co-immunoprecipitated together with a CD4-α helical region chimera, independent of agrin signaling. Together, these findings demonstrate that rapsyn interacts with the acetylcholine receptor via an α-helical structural motif conserved between the α, β and ε subunits. Binding at this site likely mediates the critical rapsyn interaction involved in localizing the acetylcholine receptor at the neuromuscular junction.
Keywords: neuromuscular junction, synaptogenesis, agrin, postsynaptic membrane
At the developing neuromuscular junction, presynaptic nerve terminals become precisely aligned with high-density aggregates of acetylcholine receptor (AChR) in the postsynaptic membrane, ensuring the high fidelity of transmission at this synapse. This process is regulated in part by agrin, a motoneuron-derived factor that plays an essential role in stably aggregating the AChR at nascent synapses. Agrin induces and/or stabilizes AChR clusters by signaling via a receptor complex consisting of the low-density lipoprotein receptor related protein 4 (LRP4) and muscle-specific kinase (MuSK) (Glass et al., 1996, Kim et al., 2008, Zhang et al., 2008), and nerve-associated receptor clusters are absent in agrin, MuSK or LRP4 null mice at birth, resulting in perinatal lethality (DeChiara et al., 1996, Gautam et al., 1996, Weatherbee et al., 2006). MuSK signaling requires the adaptor proteins Dok7 and Tid1 (Beeson et al., 2006, Okada et al., 2006, Linnoila et al., 2008), and includes activation of the kinase PAK1 (Luo et al., 2002), geranylgeranyltransferase I (Luo et al., 2003), the small GTPases Rac, cdc42, and Rho (Weston et al., 2000, Weston et al., 2003), and the cytoplasmic tyrosine kinases src/fyn (Mittaud et al., 2001) and abl/arg (Finn et al., 2003), which may phosphorylate the AChR. However, the specific signaling events and protein interactions that mediate clustering of the AChR are still not clear (Strochlic et al., 2005).
One critical mediator of AChR localization is the intracellular, membrane-associated scaffolding protein, rapsyn. Rapsyn colocalizes precisely with AChR clusters in vitro and in vivo (Froehner et al., 1981, Burden, 1985, Noakes et al., 1993), and is estimated to be in approximately 1:1 stoichiometry with the AChR (Burden et al., 1983, LaRochelle and Froehner, 1986). Moreover, rapsyn clusters the AChR (and several other synaptic proteins) when they are co-expressed in heterologous cells (Froehner et al., 1990, Phillips et al., 1991b), and AChR clusters fail to form at neuromuscular contacts in rapsyn null mice (Gautam et al., 1995). In addition, rapsyn mutations in humans result in decreased AChR levels at the synapse, producing a severe myasthenic syndrome with impaired transmission and debilitating muscle weakness (Ohno et al., 2002, Maselli et al., 2003). In fact, up to 10% of congenital myasthenic syndromes may be due to rapsyn mutations (Engel et al., 2003, Maselli et al., 2003).
Surprisingly, the molecular mechanism by which rapsyn localizes the AChR remains unknown. Functional studies on rapsyn have revealed an N-terminal myristylation site required for rapsyn’s targeting to the plasma membrane (Phillips et al., 1991a), 7 tetratricopeptide repeats (aa 6–279) that mediate rapsyn self-association (Ramarao and Cohen, 1998, Ramarao et al., 2001), and a coiled-coil domain (aa 298–331) and cysteine-rich RING structure (aa 363–402) required for interaction with AChR and β-dystroglycan, respectively (Ramarao and Cohen, 1998, Bartoli et al., 2001, Ramarao et al., 2001). Rapsyn’s site of interaction on the AChR is unclear, however, and it is unknown whether rapsyn binds one or more subunits of the pentameric AChR (2α, β, δ and γ (fetal) or ε (adult) subunits) and if binding is constitutive or regulated. Indeed, each subunit contains a large intracellular loop between the third and fourth transmembrane domains that could potentially interact with rapsyn, and consistent with this, structural studies on the AChR have revealed an accessory protein, that is presumably rapsyn, lying immediately beneath the channel in association with the loops (Miyazawa et al., 1999). One candidate interaction site is the β subunit loop, as cross-linking studies have shown that rapsyn is in close proximity to this subunit (Burden et al., 1983), and rapsyn also interacted with the β subunit intracellular loop in a modified yeast two-hybrid assay (Bartoli et al., 2001). On the other hand, it has been reported that rapsyn can associate with all of the AChR subunits when individually expressed in heterologous cells, at least in intracellular aggregates (Maimone and Merlie, 1993, Huebsch and Maimone, 2003). In this study, we have used chimeric proteins in which CD4 is fused to the large intracellular loop of each of the AChR subunits to define the subunits and domains responsible for interaction with rapsyn.
EXPERIMENTAL PROCEDURES
CD4-subunit loop constructs
To generate the CD4- subunit loop chimeras, a Bgl II restriction site was introduced by site directed mutagenesis at the end of the transmembrane domain of the mouse CD4 cDNA. The intracellular domain of CD4 was then excised and the large intracellular loops of the AChR subunits were ligated into this site. The loops were obtained by PCR of cDNAs for each of the mouse AChR subunits, and all the constructs were then sequenced. All the CD4-loop constructs were expressed in the mammalian expression vector pcDNA3.
To generate CD4-β loop α-helix constructs, a Bgl II site was introduced at the end of the CD4 intracellular domain and PCR fragments comprising segments of the α-helix were ligated into this site (denoted CD4ct-β). This CD4 tail spacer was used to position the short α-helical segment further from plasma membrane. We also generated CD4-α and ε loop α-helix constructs, by replacing the β loop α-helix with the analogous region of the α and ε subunits.
Cell culture and transfection
COS cells were grown in Dulbecco’s modified eagle medium with high glucose (DMEM-HI), supplemented with 10 % fetal bovine serum and 100 U/ml penicillin-streptomycin. For biochemical experiments, cells growing in 10 cm dishes were transfected using the CaPO4 method. For immunostaining experiments, cells growing in eight well chamber slides (Nalge Nunc Itl. Naperville, IL) were transfected in parallel.
Sol8 mouse muscle cells were maintained in DMEM-HI, supplemented with 20 % fetal bovine serum, 100 U/ml of penicillin-streptomycin, and 2 mM L-glutamine. The myoblasts were transfected at 85 % confluency using the CaPO4 method, and then when confluent, the cells were incubated with fusion medium (DMEM-HI supplemented with 5 % horse serum and 2 mM L-glutamine) to induce formation of myotubes.
Protein extraction, immunoprecipitation and western blotting
To assay detergent extractability, transfected COS cells were rinsed, scraped off and pelleted in ice-cold PBS. They were then re-suspended in extraction buffer (25 mM Tris, 25 mM glycine, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100 and the protease inhibitors PMSF, benzamidine and Na2S4O6) and incubated for 10 min on ice, after which the insoluble proteins were pelleted by centrifugation (13,000 rpm for 5 min). The CD4 chimeric proteins were then immunoprecipitated from the soluble fraction with monoclonal antibody GK1.5 (BD Biosciences, San Jose, CA) and protein G-agarose (Invitrogen, Carlsbad, CA). In addition, the insoluble pellet was resuspended in 200 µl of loading buffer (SDS, glycerol, 5 % β-mercaptoethanol and bromophenol blue). The immunoprecipitated proteins and 20 % of the pellet fraction were separated on 10 % polyacrylamide gels. They were then immunoblotted with monoclonal antibody H129.19 to CD4 (BD Biosciences), followed by an anti-rat HRP-conjugated secondary antibody and visualized using enhanced chemiluminescence. The intensity of the western blot signals was quantified using Sci-Scan 5000 Bioanalysis (USB; Cleveland, OH) or ImageGauge V4.22 software.
To assay rapsyn association in transfected COS cells or Sol8 myotubes, the cells were extracted and CD4 immunoprecipitations performed as above. In the case of Sol8 myotubes, the AChR was also isolated using biotin-conjugated α-bungarotoxin and streptavidin agarose (Invitrogen-Molecular Probes; Carlsbad, CA). Co-immunoprecipitated rapsyn was then detected by immunobotting with polyclonal antibody B5668, which we generated against a peptide encompassing rapsyn amino acids 133–153. The blots were reprobed for the AChR α-subunit using mAb210 (Babco; Berkley, CA).
Immunostaining of transfected cells
Transfected COS cells were fixed with 2 % paraformaldehyde/ PBS, blocked with 10 % horse serum/PBS and stained for surface CD4 with rat monoclonal antibodies GK1.5 or H129.19 (BD Biosciences). After washing with PBS, the cells were permeabilized in 0.5 % Triton X-100/PBS for 10 min, and incubated with rapsyn monoclonal antibody 1234 (gift of Dr. Froehner, U. Washington) or rabbit polyclonal antibody B6766 (Lee et al., 2008) for 45 min. The cells were then incubated with Alexa 488- and 594- conjugated secondary antibodies (Invitrogen). In some experiments we confirmed that rapsyn clustered surface-expressed CD4 chimeras by incubating live cells with anti-CD4 antibodies at 4°C for 15 min, and then fixing, permeabilizing and immunostaining for rapsyn.
To quantify rapsyn-induced clustering of CD4- subunit loops, we selected random fields and then scored all rapsyn-positive cells according to whether they had strong, weak or no clustering of CD4. Cells were defined as having strong clusters when CD4 staining overlapped precisely with rapsyn aggregates with little staining elsewhere, and weak clusters when significant CD4 staining was evident elsewhere on the cell surface.
RESULTS
Construction and expression of CD4-subunit loop chimeras
The muscle AChR is a pentamer composed of 2 α, β, δ and γ (fetal) or ε (adult) subunits. The subunits share ~31% homology (~16% sequence identity) and have the same membrane topology, with a large intracellular loop between transmembrane domains 3 and 4 that is the most likely site of interaction with rapsyn. To define rapsyn’s binding site on the AChR, therefore, we generated chimeric constructs where the intracellular domain of CD4 was substituted for the major intracellular loop of each AChR subunit (Fig. 1A,B). This allowed surface expression and circumvents the problem that individually expressed subunits are retained in the endoplasmic reticulum, with only fully assembled AChR being trafficked to the cell surface. Thus, it enabled us to test rapsyn’s interaction with each subunit loop and map the binding domain.
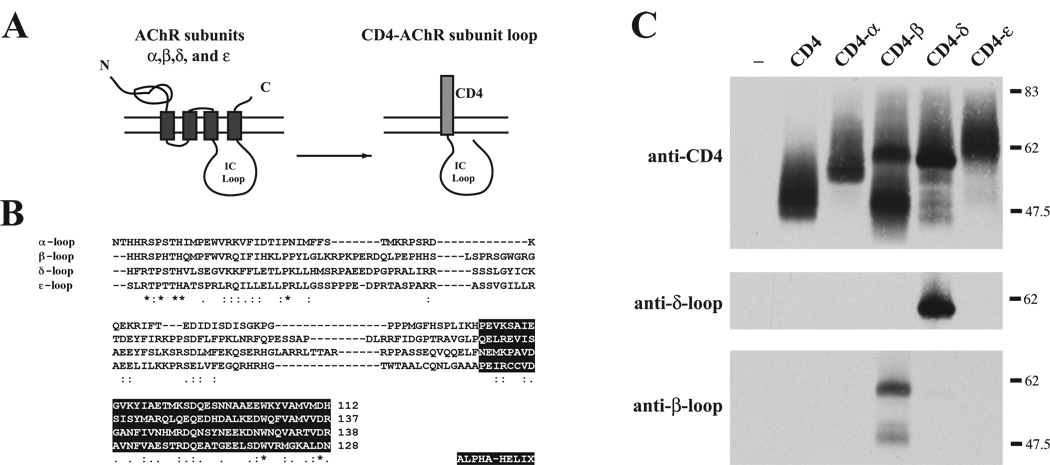
Figure 1. Construction and expression of CD4-AChR subunit loop chimeras.
(A) Schematic showing construction of the chimeric proteins consisting of CD4 extracellular and transmembrane domains fused to the major intracellular loop of the AChR subunits. (B) Alignment of the amino acid sequences for each of the subunits shows little sequence similarity in the intracellular loop region (* denotes identical residues; : & . denote conserved and semi-conserved substitutions, respectively). Sequences in the black boxes show the predicted α-helices. (C) CD4-subunit loop constructs expressed in COS cells were immunoprecipitated and immunoblotted with anti-CD4 antibody. All CD4-subunit loop chimeras were expressed at similar levels. CD4-β loop was also detected by mAb124 that recognizes an epitope in the β loop, and CD4-δ by mAb88b that recognizes the δ loop.
To confirm expression of the CD4-subunit loop chimeras, we transiently transfected the constructs into COS cells and then immunoprecipitated and immunoblotted the chimeric proteins using an antibody to the CD4 extracellular domain. We found that all the CD4-loop chimeras were highly expressed and that the proteins ran at the expected molecular weights (Fig. 1C). Reprobing with antibodies specific to the β (mAb124) and δ (mAb88b) intracellular loops also identified the respective CD4-subunit loop constructs.
Rapsyn selectively clusters CD4-α, -β and -ε subunit loops
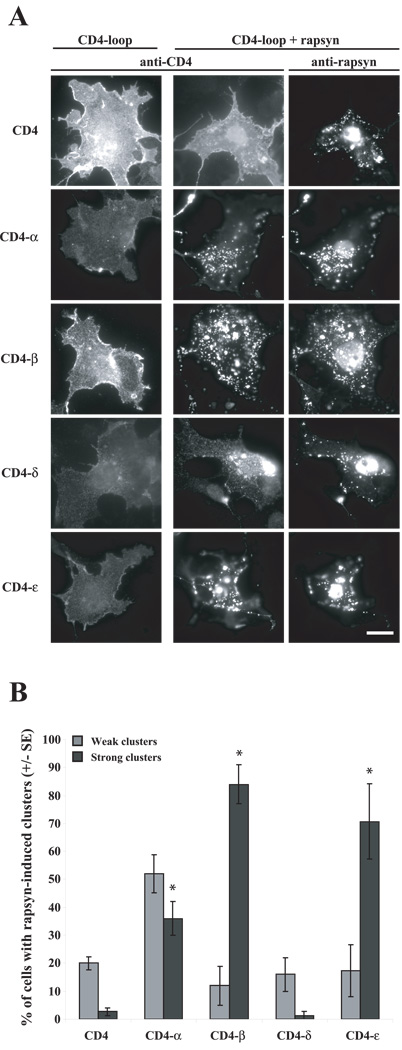
To test whether rapsyn interacts with one or more AChR subunit, we first assayed rapsyn-induced clustering of each CD4-subunit loop in heterologous cells. To do this, we expressed each CD4-subunit loop chimera in COS cells, either alone or together with rapsyn, and then immunostained specifically for surface CD4 loop chimeras and then for intracellular rapsyn. When expressed alone, we found that CD4 and all the CD4 chimeras were expressed on the cell surface in a diffuse pattern (Fig. 2A). When co-expressed with rapsyn, however, we found that CD4-α, -β, and -ε chimeras formed surface clusters that colocalized with rapsyn aggregates, whereas CD4 and CD4-δ remained diffusely distributed (Fig. 2A). To compare the efficiency of rapsyn-induced clustering of the different subunit loop chimeras, we quantified the percentage of rapsyn-expressing cells that had clusters of CD4 loop chimeras and classified the clustering as weak or strong (Fig. 2B; see Materials and Methods). We found that rapsyn induced little clustering of CD4 and CD4-δ, with these proteins being diffusely distributed in the majority of the cells (~80%). In contrast, rapsyn induced strong clustering of CD4-β and -ε loops in most cells (70–84%) (Fig. 2B), and clustering of CD4-α loop occurred at intermediate levels. Thus, we find that rapsyn clusters a subset of the AChR subunit loops (α,β,ε). Moreover, individual subunit loops are sufficient for rapsyn interaction.
Figure 2. Rapsyn clusters CD4-α, -β, and -ε loops in COS cells.
(A) CD4-loop chimeras were expressed in COS cells alone or together with rapsyn, and then the cells were immunostained for surface-expressed CD4-loops and for rapsyn. Expressed alone, CD4 and all of the CD4- subunit loops were diffusely distributed on the cell surface. When co-expressed with rapsyn, CD4-α, -β, and -ε loops coclustered with rapsyn aggregates, but CD4 and CD4-δ loop remained diffusely distributed. Scale bar = 50 µm. (B) Quantification of rapsyn-induced clustering of the CD4- chimeras, showing the percentage of co-expressing cells that had strong or weak CD4-loop clusters (see methods). Rapsyn induced robust clustering of CD4-β and ε loops and weaker clustering of CD4-α loop (mean±SEM, * p < 0.05 by Student’s t-test; n=3 independent experiments).
Rapsyn anchors CD4-α, -β and -ε loops
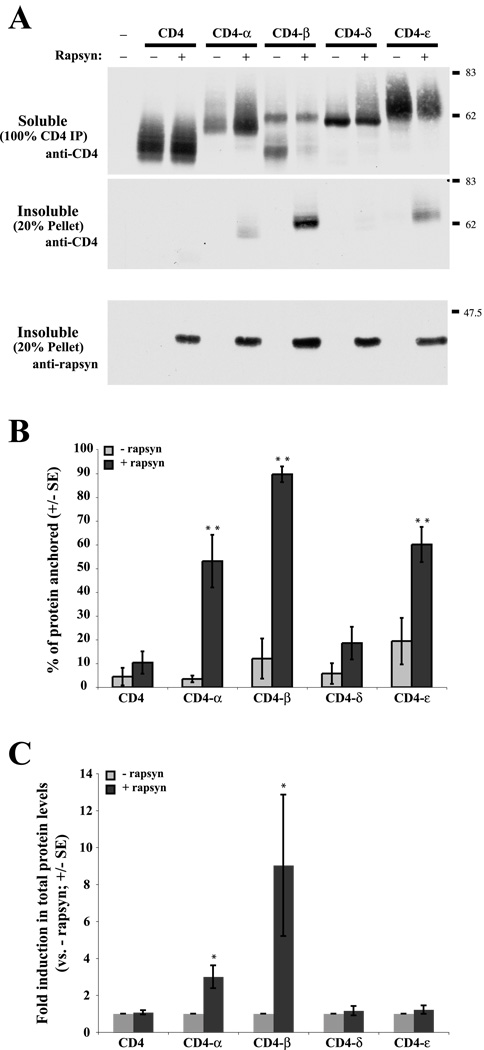
Rapsyn was previously shown to anchor the AChR to the cytoskeleton in heterologous cells (Phillips et al., 1993, Mohamed and Swope, 1999). Therefore, we tested whether rapsyn also induced anchoring of the CD4 subunit loop chimeras, assayed by monitoring their detergent extractability as described previously (Kassner et al., 1998, Mohamed and Swope, 1999). Briefly, transfected cells were extracted with buffer containing 1% Triton X-100 for 10 minutes on ice, and the distribution of the chimeras in the soluble versus insoluble (cytoskeleton-containing pellet) fraction was determined by immunoblotting. When expressed alone, we found that CD4 and all the CD4-subunit loop chimeras were almost completely solubilized in 1% Triton X-100 buffer (Fig. 3A,B). When co-expressed with rapsyn, however, we found that the detergent extractability of CD4-α, -β, and -ε loops were significantly reduced and they were now retained to varying degrees in the cytoskeletal fraction. Notably, rapsyn induced anchorage of ~90% of CD4-β loop, 60% of CD4-ε loop and 50% of CD4-α loop protein (Fig. 3B). These results indicate that rapsyn interacts selectively with 3 subunit loops, but with different apparent affinities (β > α, ε subunit loops; p < 0.05; ANOVA with Tukey post-test).
Figure 3. Rapsyn cytoskeletally anchors CD4-α, -β and -ε loops.
(A) CD4- subunit loops were expressed in COS cells alone or together with rapsyn. The transfected cells were then extracted in 1% Triton-containing buffer, and the distribution of the CD4-loop chimeras in the soluble and insoluble fractions was determined by immunoblotting with anti-CD4 antibody. Note that the immunoblots show 100% of the CD4 immunoprecipitation (soluble fraction) but only 20% of the pellet (insoluble fraction). Expressed alone, CD4 and all the CD4-loops were readily extracted and recovered in the soluble fraction. When co-expressed with rapsyn, however, a significant amount of CD4-β loop, and to a lesser extent CD4-α and -ε loops, were retained in the insoluble, cytoskeletal fraction. The pellet fraction was also immunoblotted for rapsyn, showing that equal levels were present in each of the transfections. (B) Quantification of the percentage of each CD4-loop protein retained in the insoluble fraction. Rapsyn-induced anchoring was highest for CD4-β loop (~90%) followed by CD4-α and -ε loops (50–60%) (α,β,ε > CD4, ** p < 0.001; and β > α,ε; p < 0.05; ANOVA with Tukey post-test; n=5 independent experiments). In contrast, the extractability of CD4 and CD-δ were not significantly affected by rapsyn. (C) Quantification of total protein levels, showing that rapsyn induced three- and nine-fold increases in CD4-α and -β loop protein levels, respectively (* p<0.05, Student’s t-test; n=5 independent experiments). Total levels of CD4, CD4-δ loop or CD4-ε loop were not significantly altered by rapsyn co-expression.
Interestingly, in these experiments, we also observed that the total level of CD4-β loop protein was increased ~9 fold in the presence of rapsyn (Fig. 3C); this most likely stems from decreased turnover (Phillips et al., 1997, Wang et al., 1999) associated with the strong rapsyn-induced clustering and anchoring of the β loop but this remains to be confirmed.
Rapsyn interacts with α-helix region of β loop
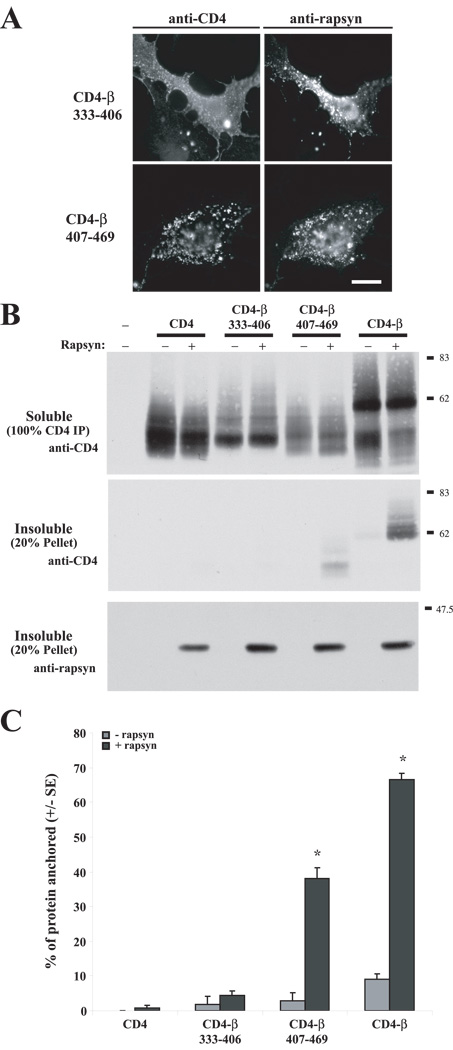
Next, we defined the region of the intracellular loop that interacts with rapsyn by assaying additional CD4-loop deletion constructs, focusing on the β-subunit as it displayed the strongest interaction with rapsyn. First, we tested two loop fragments; CD4-β333–406, which contains the first half of the β-loop encoded primarily by exon 9, and CD4-β407–469, which contains the second half of the β-loop encoded by exon 10. When co-expressed in COS cells, we found that rapsyn induced robust clustering of CD4-β407–469 but not CD4-β333–406, which remained evenly distributed on the cell surface (Fig. 4A). Similarly, rapsyn reduced the detergent extractability of CD4-β407–469, with ~40% of the chimeric protein becoming cytoskeletally-anchored, but had no effect on the detergent extractability of CD4-β333–406 (Fig. 4 B,C). Thus, rapsyn interacts with a region in the second half of the loop.
Figure 4. Rapsyn interacts with the β loop 407–469 region.
(A) CD4-β333–406 and CD4-β407–469 chimeras were expressed in COS cells alone or together with rapsyn. Rapsyn induced robust clustering of CD4-β407–469, but not CD4-β333–406. (B) Rapsyn-induced anchoring of CD4-β333–406 and -β407–469 was assayed by extracting transfected COS cells in 1% Triton-containing buffer and immunoblotting the soluble and pellet fractions with anti-CD4 antibody. With rapsyn co-expression, significant amounts of CD4-β loop and CD4-β407–469 were retained in the insoluble, cytoskeletal fraction, whereas CD4 and CD-β333–406 were recovered entirely in the soluble fraction. The pellet fraction was immunoblotted for rapsyn to show equivalent protein levels. (C) Quantification of the percentage of each protein retained in the insoluble fraction. Rapsyn anchored ~70% of CD4-β loop and ~40% of CD4-β407–469 (*p<0.05 by Student’s t-test; n=3). In contrast, the extractability of CD4 and CD4-β333–406 were not significantly affected by rapsyn.
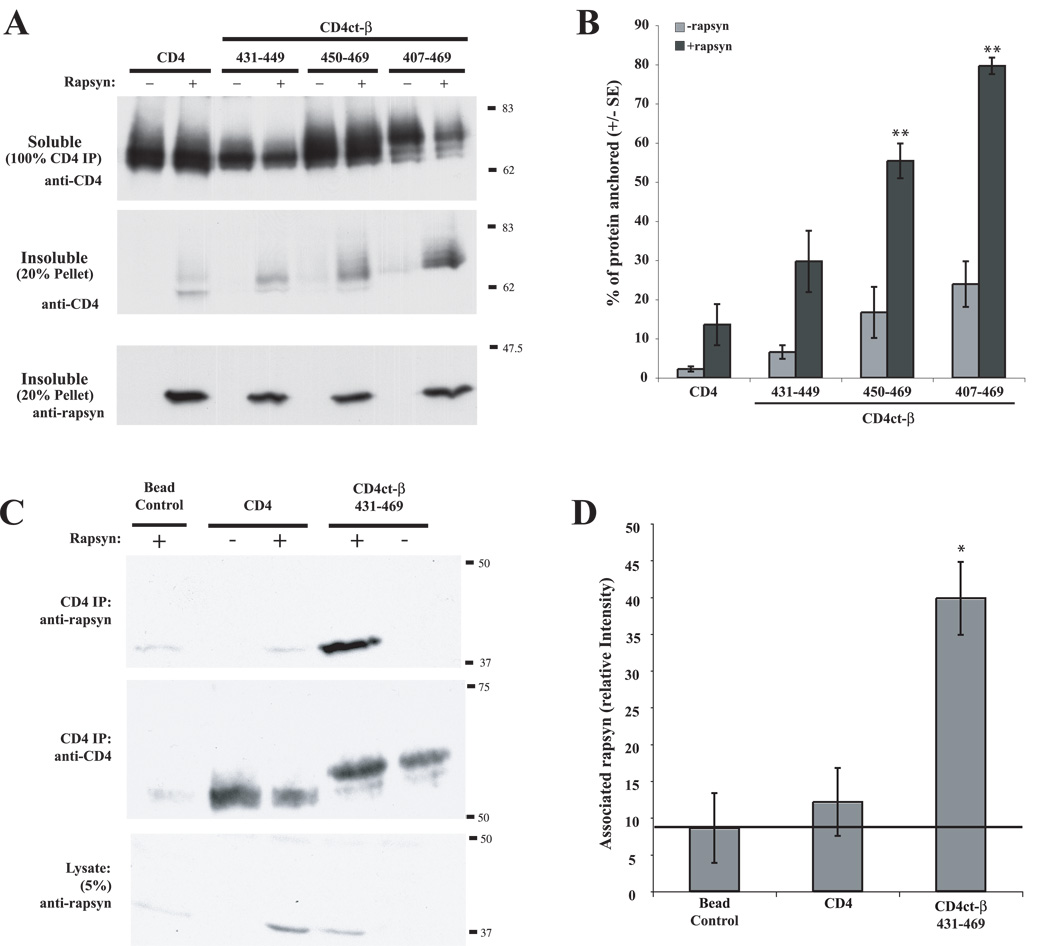
Although the AChR subunit loops share little sequence homology (~5% identity and ~14% similarity), they all contain predicted α-helical structures in the C-terminal loop region analogous to β407–469 (Fig. 1B). The precise position and length of the helices is unclear, with different studies (Finer-Moore and Stroud, 1984, Le Novere et al., 1999) or secondary-structure prediction programs (SSThread; (Ito et al., 1997)) giving variable results; however, α-helices are apparent in each of the subunit loops in a 4Å resolution structure derived from electron microscopy of purified Torpedo AChR-rich membrane (Unwin, 2005). We tested, therefore, whether the β loop α-helix (~amino acids 431–469; Fig. 1B) was sufficient for rapsyn interaction and whether a specific region or sequence within the helix was involved. Interestingly, we found that rapsyn clustered both CD4ct-β431–449 and CD4ct-β450–469 when co-expressed in COS cells (data not shown), and that it anchored ~30% of CD4ct-β431–449 and ~55% of CD4ct-β450–469 in the detergent-insoluble fraction (Fig. 5A,B). Neither the first or second halves of the helix were anchored as efficiently as the entire, helix, however (~80% of CD4ct-β407–469 > β431–449 and β450–469; p < 0.001 and 0.01, respectively, ANOVA with Tukey post-test). To further confirm the interaction, we immunoprecipitated CD4ct-β431–469 from COS cells cotransfected with rapsyn and immunoblotted for rapsyn (Fig. 5C,D). We readily detected rapsyn in association with the β loop α-helix (CD4ct-β431–469) but not the CD4 control. Reprobes also confirmed that rapsyn and both CD4 proteins were expressed at similar levels in each of the transfections. Together, these findings demonstrate that rapsyn interacts with the α-helical domain of the β subunit. Similarly, we found that rapsyn induced robust clustering of CD4-loop chimeras where the β α-helical domain had been replaced by the analogous region of the α or ε subunits (Suppl. Fig. 1). As this staining was performed on live cells, it further confirms that rapsyn clustered CD4 chimeras on the cell surface rather than in intracellular aggregates. Thus, rapsyn interacts with α-helical domains in the α, β and ε subunit loops, likely via direct binding although we were unable to demonstrate this in overlay blots (data not shown). This may reflect a requirement for native conformation of the proteins, as binding to the β subunit has been shown in a modified yeast two-hybrid assay that identifies protein interactions at the plasma membrane (Bartoli et al., 2001); however we cannot discount the possibility that the rapsyn/α-helix interaction involves an additional protein.
Figure 5. Rapsyn interacts with the α-helical region of β loop.
CD4-chimeras encompassing the β loop α-helical domain were expressed in COS cells alone or together with rapsyn. (A) Transfected cells were extracted in 1% Triton-containing buffer and the soluble and pellet fractions were immunoblotted with anti-CD4 antibody. Rapsyn anchored both the first and second halves of the α-helix (CD4ct-β430–449 and CD4ct-β450–469), although to a lesser extent than the complete helix (CD4ct-β407–469). (B) Quantification of the percentage of each protein retained in the pellet fraction. Rapsyn anchored ~30% of CD4ct-β430–449 (not significant), 55% of β450–469 and 80% of β407–469 (** p < 0.001; ANOVA with Tukey post-test; n=6–7 independent experiments). Anchoring of β407–469 > β430–449 and β450–469 (p < 0.01, ANOVA with Tukey post-test). (C) CD4 and CD4ct-β430–469 were immunoprecipitated and immunoblotted with anti-CD4 and anti-rapsyn antibodies. Rapsyn was readily detected in association with CD4ct-β430–469 whereas only background levels (equivalent to beads alone) associated with CD4. The immunoblots also show that similar levels of rapsyn and CD4 / CD4ct-β430–469 were present in the different transfection conditions. (D) Quantification of co-immunoprecipitation experiments showing that rapsyn specifically associates with the α-helical domain of β loop.
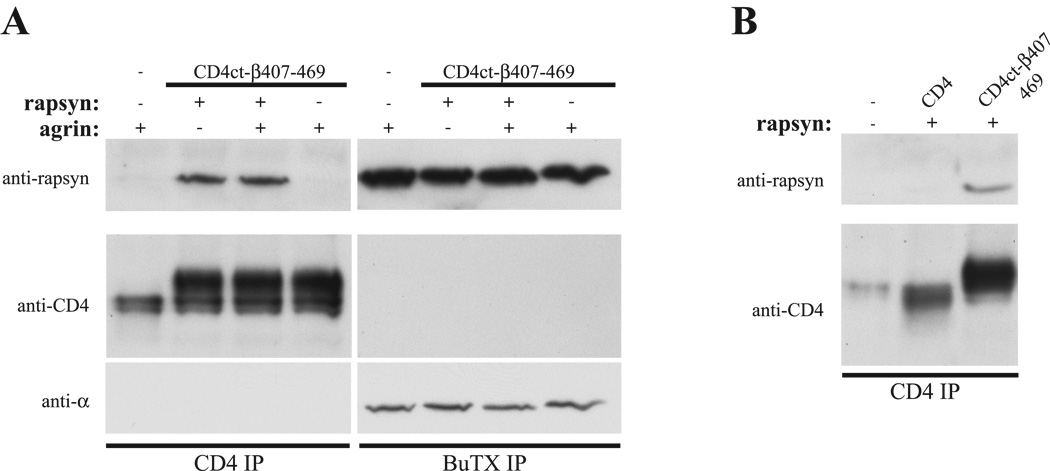
Finally, we tested whether rapsyn interacts with CD4ct-β407–469 in muscle and if agrin signaling regulates their binding. To do this, we expressed CD4ct-β407–469 in Sol8 myotubes by transient transfection, either alone or together with additional rapsyn, and treated with agrin (C-Ag4,8; 100 pM, 1 hr); we then immunoprecipitated CD4ct-β407–469 and the AChR from cell extracts and immunoblotted for rapsyn (Fig. 6A). Rapsyn was readily detected in association with the AChR, but only trace amounts co-immunoprecipitated together with CD4ct-β407–469 when expressed alone. This may be due to rapsyn association with the endogenous AChR, with little free pool available for binding of the introduced construct, or to a lower affinity interaction with the isolated helix compared to the intact AChR. Indeed, we found that rapsyn was readily co-immunoprecipitated with CD4ct-β407–469 when they were co-transfected into the muscle cells. This was not mediated indirectly via the AChR, as no association was observed between CD4ct-β407–469 and the receptor (Fig. 6A). Moreover, no rapsyn association was observed with control constructs such as CD4 (Fig. 6B). Interestingly, agrin treatment increased rapsyn association with the endogenous AChR (Borges et al., 2008) but had no effect on rapsyn interaction with CD4ct-β407–469. Thus, we did not observe an agrin-regulated interaction of rapsyn with the isolated β407–469 region.
Figure 6. Rapsyn interacts with CD4-β407–469 in muscle.
(A) CD4ct-β407–469 was expressed in Sol8 myotubes, either alone or with additional rapsyn. CD4ct-β407–469 and AChR were then isolated from muscle cell lysates and rapsyn association assayed by immunoblotting with a rapsyn polyclonal antibody. Rapsyn was readily co-immunoprecipitated together with CD4ct-β407–469 when they were co-expressed, but only trace amounts of endogenous rapsyn associated with CD4ct-β407–469 expressed alone. Treatment with agrin also increased the interaction of rapsyn and the endogenous AChR, but had no effect on rapsyn/CD4ct-β407–469 association. Reprobing with CD4 and AChR α-subunit (mAb210) antibodies confirms that CD4ct-β407–469 does not associate with the AChR. (B) Rapsyn co-immunoprecipitated with CD4ct-β407–469 but not CD4.
DISCUSSION
The localization of the AChR at the developing neuromuscular junction is mediated by the scaffolding protein rapsyn but their mode of interaction is poorly understood. Here, we have mapped rapsyn’s interaction site on the AChR using CD4-subunit loop chimeric proteins and find that rapsyn selectively clusters and anchors the major intracellular loop of the α, β, and ε subunits. As the isolated loops of each subunit were sufficient for rapsyn interaction, this suggests that there are up to 4 independent rapsyn binding sites on the assembled AChR (ie. 2α/1β/ 1ε loop). The 4 sites are not equivalent, however, as we observed consistent differences in the relative levels of interaction of rapsyn with the three subunit loops, with rapsyn-mediated clustering and anchoring being highest for β subunit loop, followed by ε and α loops. The simplest interpretation of these findings is that rapsyn binds the loops with differing affinities, preferentially interacting with the β subunit loop. These findings are consistent with cross-linking experiments showing rapsyn association with the β (Burden et al., 1983) and perhaps γ subunits (fetal homolog of the ε subunit) (Shoji et al., 1992).
Our finding that rapsyn can interact with multiple loops suggests that they share a common binding motif, however little homology is evident in the primary sequences of the 3 interacting loops. Rather, we find that rapsyn interacts with the α-helical domain of β loop, a predicted secondary structure conserved between the subunits. Indeed, α-helices of approximately 40 amino acids each are predicted in the C-terminal portion of the α, β and ε subunits (Le Novere et al., 1999). One notable exception is the δ subunit loop that is predicted to contain a shorter helix (SSThread; (Ito et al., 1997)), and we failed to detect rapsyn interaction with this subunit. Rapsyn binding to the α-helix likely involves recognition of secondary structure of the helix rather than primary sequence given the relatively low similarity between the subunit loops. This is also suggested by our finding that rapsyn interacts weakly with both the first and second halves of the α-helix, but more strongly with the complete α-helix. Rapsyn likely binds directly to the β loop α-helical domain as they were efficiently co-immunoprecipitated from heterologous cells. An interaction between rapsyn and the β subunit loop has also been demonstrated in a modified yeast two-hybrid assay that identifies protein interactions at the plasma membrane (Bartoli et al., 2001), and may be mediated via rapsyn’s coiled-coil domain (Ramarao and Cohen, 1998, Ramarao et al., 2001). We were unable to confirm direct binding of rapsyn or its coiled-coil domain to the α-helix in overlay blots, however, and cannot discount the possibility that their interaction involves an additional protein.
It is not possible to delete or mutate the α-helix of multiple loops as this prevents receptor assembly. However, a 3 codon deletion in the β loop α-helix has been linked to a congenital myasthenic syndrome with severe AChR deficiency that is consistent with both defective interaction with rapsyn and impaired assembly (Quiram et al., 1999). An α-helical region has also been implicated in the synaptic localization of the glycine receptor, forming the binding site for gephyrin on the GlyR beta2 subunit intracellular loop (Meyer et al., 1995, Kneussel et al., 1999). Our findings raise the possibility, therefore, that this structural motif may be broadly involved in the localization of this superfamily of ligand-gated ion channels.
Rapsyn-mediated clustering of the AChR is regulated by MuSK in muscle cells, as both muscle-autonomous and nerve-associated AChR clusters fail to form in MuSK knockout mice (DeChiara et al., 1996, Lin et al., 2001). Although the signaling pathway is unclear, MuSK appears to regulate both the interaction of pre-existing rapsyn/AChR complexes with the postsynaptic scaffold and the stoichiometry of rapsyn/AChR complexes. A constitutive rapsyn/AChR interaction is evident prior to clustering as rapsyn associates with the AChR in the late secretory pathway and is co-transported to the cell surface (Bignami et al., 1998, Marchand et al., 2000, Marchand et al., 2002). Moreover, rapsyn associates with unclustered surface AChR in denervated muscle or cultured myotubes (Moransard et al., 2003), and is downregulated following antibody-induced internalization and degradation of AChR (Marangi et al., 2001). We propose that this constitutive interaction is mediated by rapsyn binding to the loop α-helical domain, as we found that they interacted in muscle independent of agrin signaling. Our findings do not discount an additional, regulated interaction at this site, however, which might be undetected due to overexpression of rapsyn or use of a single β loop fragment, isolated from other potential rapsyn binding or regulatory regions present in the intact AChR (Maimone and Enigk, 1999, Huebsch and Maimone, 2003). Constitutive or regulated rapsyn binding to the α-helical domain may have several functions. First, constitutive rapsyn/AChR complexes may facilitate the rapid aggregation of AChR at developing synapses, via MuSK regulated linkage to the postsynaptic scaffold. Second, constitutive rapsyn association may be required for some aspects of MuSK signaling. Surprisingly, MuSK-induced tyrosine phosphorylation of the AChR β and δ subunits is significantly reduced in rapsyn null myotubes (Apel et al., 1997) and several findings suggest that rapsyn localizes or activates cytoplasmic tyrosine kinases that phosphorylate the receptor (Lee et al., 2008) and references therein). Third, additional rapsyn could bind to free α-helix sites on the AChR during maturation of postsynaptic receptor clusters, to increase AChR stability. Indeed, the ratio of rapsyn to AChR increases during maturation of the NMJ (Gervasio et al., 2007, Brockhausen et al., 2008), and overexpression of rapsyn stabilizes and increases the packing density of the AChR (Gervasio and Phillips, 2005). Similarly, overexpression of rapsyn protects against antibody-mediated loss of AChRs in experimental autoimmune myasthenia gravis induced by passive transfer of anti-AChR antibody (Losen et al., 2005).
Agrin/ MuSK signaling also increases rapsyn/AChR association (Moransard et al., 2003) via a phosphorylation-dependent binding site in the β subunit loop, distinct from the α-helical domain (Borges et al., 2008). Interestingly, agrin-induced phosphorylation of tyrosine 390 in this 20 amino acid motif recruits additional rapsyn, thus increasing the stoichiometry of rapsyn binding to the AChR. This helps stably cluster and anchor the receptor in the postsynaptic membrane, and mice with targeted mutation of this phosphorylation site have neuromuscular junctions that are small and that have significantly decreased levels of AChR (Friese et al., 2007).
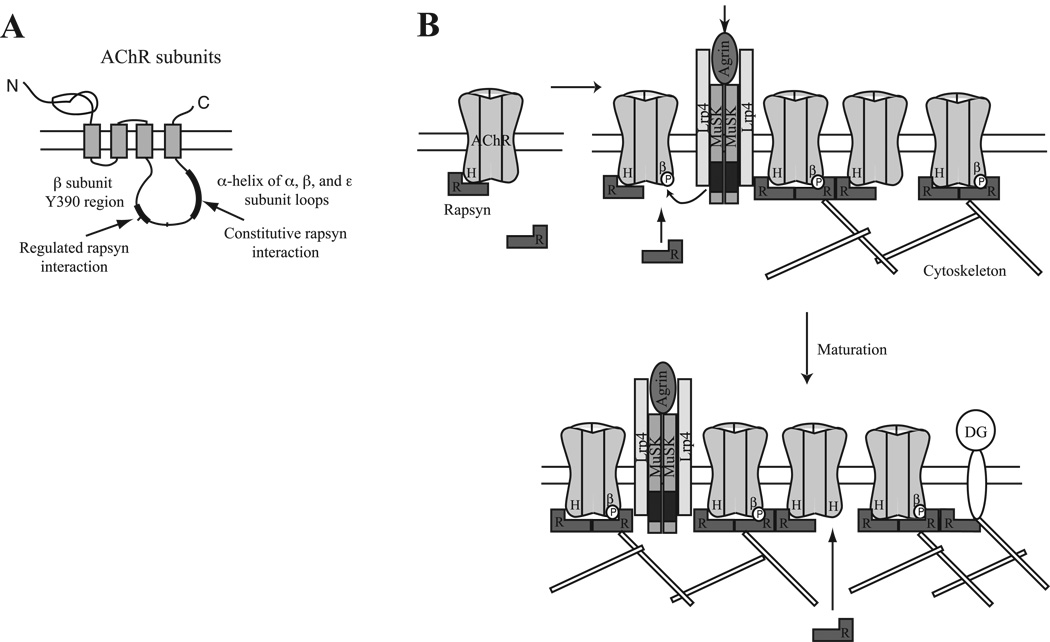
Taken together, these findings suggest that rapsyn interacts in both a constitutive and regulated manner with the AChR, mediated by α-helical domains on the α, β and ε subunit loops and the β loop tyrosine 390 region. How many of these potential binding sites are utilized in a given AChR is unclear. The stoichiometry of rapsyn to AChR in synaptic membranes from Torpedo electric organ is in the range of 0.5 – 2 (LaRochelle and Froehner, 1986). A 4Å structure derived from electron images of tubular Torpedo membranes also shows a two-fold symmetry for rapsyn, implying that more than 1 rapsyn molecule associates with a single AChR (Mitra et al., 1989, Miyazawa et al., 1999). Thus, we propose that at least two rapsyn binding sites are involved in clustering AChR. In our model (Fig. 7B), unclustered AChR is constitutively associated with one molecule of rapsyn. Upon MuSK activation at nascent synapses, these preformed complexes become linked to the postsynaptic scaffold and a second molecule of rapsyn binds AChRs with a phosphorylated β loop. At later stages of development, additional rapsyn might also bind via unoccupied α-helix binding sites on other AChR subunit loops. In both cases, the increase in stoichiometry of rapsyn/AChR binding appears to increase the anchoring and stability of the receptor in the postsynaptic membrane (Borges and Ferns, 2001, Friese et al., 2007, Gervasio et al., 2007). Thus, different forms of rapsyn interaction might have distinct functions that combine to localize the AChR at appropriate levels at the neuromuscular junction.
Figure 7. Model for AChR localization involving constitutive and regulated interactions with rapsyn.
(A) Schematic showing the subunit major intracellular loop regions involved in constitutive and regulated interactions with rapsyn. (B) We propose a model where unclustered AChR is constitutively associated with one rapsyn (R), via binding to the α-helical domain (H) in the β, ε or α subunit intracellular loop. Upon MuSK activation, tyrosine phosphorylation of the AChR β subunit (βP) induces binding of a second rapsyn to some receptors. Additional rapsyn binding may also occur during synapse maturation, via unoccupied sites on AChRs, dystroglycan (DG) or other cytoskeletal proteins. Changes in the stoichiometry of rapsyn binding regulate anchoring and stability of the AChR, modulating receptor levels and density in the postsynaptic membrane.
Supplementary Material
COS cells transfected with CD4 chimeras and rapsyn were incubated live with anti-CD4 antibodies at 4°C and then fixed, permeabilized and immunostained for rapsyn. Rapsyn clustered CD4ct-β407–469 but not CD4 on the cell surface. Similarly, rapsyn clustered CD4 chimeras where the β loop helix had been replaced by the helical domain of the α and β subunits (CD4-β/ α391–428 and ε419–456; n = 2–3 independent experiments).
ACKNOWLEDGEMENTS
This work was supported by a grant to M.F. from the National Institutes of Health (NS049354) and was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR17348-01 from the National Center for Research Resources, National Institutes of Health. We thank Dr S. Froehner (Univ. Washington) for providing anti-rapsyn antibodies and other members of the Ferns laboratory for their helpful comments on the manuscript.
ABBREVIATIONS
- aa
amino acid
- AChR
acetylcholine receptor
- BuTX
α-bungarotoxin
- C-terminal
carboxyl terminal
- LRP4
low-density lipoprotein receptor related protein 4
- MuSK
muscle specific kinase
- N-terminal
amino terminal
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Apel ED, Glass DJ, Moscoso LM, Yancopoulos GD, Sanes JR. Rapsyn is required for MuSK signaling and recruits synaptic components to a MuSK-containing scaffold. Neuron. 1997;18:623–635. doi: 10.1016/s0896-6273(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Bartoli M, Ramarao MK, Cohen JB. Interactions of the rapsyn RING-H2 domain with dystroglycan. J Biol Chem. 2001;276:24911–24917. doi: 10.1074/jbc.M103258200. [DOI] [PubMed] [Google Scholar]
- Beeson D, Higuchi O, Palace J, Cossins J, Spearman H, Maxwell S, Newsom-Davis J, Burke G, Fawcett P, Motomura M, Muller JS, Lochmuller H, Slater C, Vincent A, Yamanashi Y. Dok-7 mutations underlie a neuromuscular junction synaptopathy. Science. 2006;313:1975–1978. doi: 10.1126/science.1130837. [DOI] [PubMed] [Google Scholar]
- Bignami F, Camus G, Marchand S, Bailly L, Stetzkowski-Marden F, Cartaud J. Targeting of acetylcholine receptor and 43 kDa rapsyn to the postsynaptic membrane in Torpedo marmorata electrocyte. J Physiol Paris. 1998;92:177–181. doi: 10.1016/s0928-4257(98)80006-5. [DOI] [PubMed] [Google Scholar]
- Borges LS, Ferns M. Agrin-induced phosphorylation of the acetylcholine receptor regulates cytoskeletal anchoring and clustering. J Cell Biol. 2001;153:1–12. doi: 10.1083/jcb.153.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges LS, Yechikhov S, Lee YI, Rudell JB, Friese MB, Burden SJ, Ferns MJ. Identification of a motif in the acetylcholine receptor beta subunit whose phosphorylation regulates rapsyn association and postsynaptic receptor localization. J Neurosci. 2008;28:11468–11476. doi: 10.1523/JNEUROSCI.2508-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen J, Cole RN, Gervasio OL, Ngo ST, Noakes PG, Phillips WD. Neural agrin increases postsynaptic ACh receptor packing by elevating rapsyn protein at the mouse neuromuscular synapse. Developmental neurobiology. 2008;68:1153–1169. doi: 10.1002/dneu.20654. [DOI] [PubMed] [Google Scholar]
- Burden SJ. The subsynaptic 43-kDa protein is concentrated at developing nerve-muscle synapses in vitro. Proc Natl Acad Sci U S A. 1985;82:8270–8273. doi: 10.1073/pnas.82.23.8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden SJ, DePalma RL, Gottesman GS. Crosslinking of proteins in acetylcholine receptor-rich membranes: association between the beta-subunit and the 43 kd subsynaptic protein. Cell. 1983;35:687–692. doi: 10.1016/0092-8674(83)90101-0. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Bowen DC, Valenzuela DM, Simmons MV, Poueymirou WT, Thomas S, Kinetz E, Compton DL, Rojas E, Park JS, Smith C, DiStefano PS, Glass DJ, Burden SJ, Yancopoulos GD. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- Engel AG, Ohno K, Sine SM. Sleuthing molecular targets for neurological diseases at the neuromuscular junction. Nat Rev Neurosci. 2003;4:339–352. doi: 10.1038/nrn1101. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J, Stroud RM. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci U S A. 1984;81:155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn AJ, Feng G, Pendergast AM. Postsynaptic requirement for Abl kinases in assembly of the neuromuscular junction. Nat Neurosci. 2003;6:717–723. doi: 10.1038/nn1071. [DOI] [PubMed] [Google Scholar]
- Friese MB, Blagden CS, Burden SJ. Synaptic differentiation is defective in mice lacking acetylcholine receptor {beta}-subunit tyrosine phosphorylation. Development. 2007;134:4167–4176. doi: 10.1242/dev.010702. [DOI] [PubMed] [Google Scholar]
- Froehner SC, Gulbrandsen V, Hyman C, Jeng AY, Neubig RR, Cohen JB. Immunofluorescence localization at the mammalian neuromuscular junction of the Mr 43,000 protein of Torpedo postsynaptic membranes. Proc Natl Acad Sci U S A. 1981;78:5230–5234. doi: 10.1073/pnas.78.8.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehner SC, Luetje CW, Scotland PB, Patrick J. The postsynaptic 43K protein clusters muscle nicotinic acetylcholine receptors in Xenopus oocytes. Neuron. 1990;5:403–410. doi: 10.1016/0896-6273(90)90079-u. [DOI] [PubMed] [Google Scholar]
- Gautam M, Noakes PG, Moscoso L, Rupp F, Scheller RH, Merlie JP, Sanes JR. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- Gautam M, Noakes PG, Mudd J, Nichol M, Chu GC, Sanes JR, Merlie JP. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature. 1995;377:232–236. doi: 10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- Gervasio OL, Armson PF, Phillips WD. Developmental increase in the amount of rapsyn per acetylcholine receptor promotes postsynaptic receptor packing and stability. Dev Biol. 2007;305:262–275. doi: 10.1016/j.ydbio.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Gervasio OL, Phillips WD. Increased ratio of rapsyn to ACh receptor stabilizes postsynaptic receptors at the mouse neuromuscular synapse. J Physiol. 2005;562:673–685. doi: 10.1113/jphysiol.2004.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ, Bowen DC, Stitt TN, Radziejewski C, Bruno J, Ryan TE, Gies DR, Shah S, Mattsson K, Burden SJ, DiStefano PS, Valenzuela DM, DeChiara TM, Yancopoulos GD. Agrin acts via a MuSK receptor complex. Cell. 1996;85:513–523. doi: 10.1016/s0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- Huebsch KA, Maimone MM. Rapsyn-mediated clustering of acetylcholine receptor subunits requires the major cytoplasmic loop of the receptor subunits. J Neurobiol. 2003;54:486–501. doi: 10.1002/neu.10177. [DOI] [PubMed] [Google Scholar]
- Ito M, Matsuo Y, Nishikawa K. Prediction of protein secondary structure using the 3D-1D compatibility algorithm. Comput Appl Biosci. 1997;13:415–424. doi: 10.1093/bioinformatics/13.4.415. [DOI] [PubMed] [Google Scholar]
- Kassner PD, Conroy WG, Berg DK. Organizing effects of rapsyn on neuronal nicotinic acetylcholine receptors. Mol Cell Neurosci. 1998;10:258–270. doi: 10.1006/mcne.1998.0664. [DOI] [PubMed] [Google Scholar]
- Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M, Hermann A, Kirsch J, Betz H. Hydrophobic interactions mediate binding of the glycine receptor beta-subunit to gephyrin. J Neurochem. 1999;72:1323–1326. doi: 10.1046/j.1471-4159.1999.0721323.x. [DOI] [PubMed] [Google Scholar]
- LaRochelle WJ, Froehner SC. Determination of the tissue distributions and relative concentrations of the postsynaptic 43kDa protein and the acetylcholine receptor in Torpedo. J Biol Chem. 1986;261:5270–5274. [PubMed] [Google Scholar]
- Lee Y, Rudell J, Yechikhov S, Taylor R, Swope S, Ferns M. Rapsyn carboxyl terminal domains mediate muscle specific kinase-induced phosphorylation of the muscle acetylcholine receptor. Neuroscience. 2008;153:997–1007. doi: 10.1016/j.neuroscience.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Corringer PJ, Changeux JP. Improved secondary structure predictions for a nicotinic receptor subunit: incorporation of solvent accessibility and experimental data into a two-dimensional representation. Biophys J. 1999;76:2329–2345. doi: 10.1016/S0006-3495(99)77390-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, Lee KF. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature. 2001;410:1057–1064. doi: 10.1038/35074025. [DOI] [PubMed] [Google Scholar]
- Linnoila J, Wang Y, Yao Y, Wang ZZ. A mammalian homolog of Drosophila tumorous imaginal discs, Tid1, mediates agrin signaling at the neuromuscular junction. Neuron. 2008;60:625–641. doi: 10.1016/j.neuron.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losen M, Stassen MH, Martinez-Martinez P, Machiels BM, Duimel H, Frederik P, Veldman H, Wokke JH, Spaans F, Vincent A, De Baets MH. Increased expression of rapsyn in muscles prevents acetylcholine receptor loss in experimental autoimmune myasthenia gravis. Brain. 2005;128:2327–2337. doi: 10.1093/brain/awh612. [DOI] [PubMed] [Google Scholar]
- Luo ZG, Je HS, Wang Q, Yang F, Dobbins GC, Yang ZH, Xiong WC, Lu B, Mei L. Implication of geranylgeranyltransferase I in synapse formation. Neuron. 2003;40:703–717. doi: 10.1016/s0896-6273(03)00695-0. [DOI] [PubMed] [Google Scholar]
- Luo ZG, Wang Q, Zhou JZ, Wang J, Luo Z, Liu M, He X, Wynshaw-Boris A, Xiong WC, Lu B, Mei L. Regulation of AChR clustering by Dishevelled interacting with MuSK and PAK1. Neuron. 2002;35:489–505. doi: 10.1016/s0896-6273(02)00783-3. 2002. [DOI] [PubMed] [Google Scholar]
- Maimone MM, Merlie JP. Interaction of the 43 kd postsynaptic protein with all subunits of the muscle nicotinic acetylcholine receptor. Neuron. 1993;11:53–66. doi: 10.1016/0896-6273(93)90270-2. [DOI] [PubMed] [Google Scholar]
- Marangi PA, Forsayeth JR, Mittaud P, Erb-Vogtli S, Blake DJ, Moransard M, Sander A, Fuhrer C. Acetylcholine receptors are required for agrin-induced clustering of postsynaptic proteins. Embo J. 2001;20:7060–7073. doi: 10.1093/emboj/20.24.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand S, Bignami F, Stetzkowski-Marden F, Cartaud J. The myristoylated protein rapsyn is cotargeted with the nicotinic acetylcholine receptor to the postsynaptic membrane via the exocytic pathway. J Neurosci. 2000;20:521–528. doi: 10.1523/JNEUROSCI.20-02-00521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand S, Devillers-Thiery A, Pons S, Changeux JP, Cartaud J. Rapsyn escorts the nicotinic acetylcholine receptor along the exocytic pathway via association with lipid rafts. J Neurosci. 2002;22:8891–8901. doi: 10.1523/JNEUROSCI.22-20-08891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maselli RA, Dunne V, Pascual-Pascual SI, Bowe C, Agius M, Frank R, Wollmann RL. Rapsyn mutations in myasthenic syndrome due to impaired receptor clustering. Muscle Nerve. 2003;28:293–301. doi: 10.1002/mus.10433. [DOI] [PubMed] [Google Scholar]
- Meyer G, Kirsch J, Betz H, Langosch D. Identification of a gephyrin binding motif on the glycine receptor beta subunit. Neuron. 1995;15:563–572. doi: 10.1016/0896-6273(95)90145-0. [DOI] [PubMed] [Google Scholar]
- Mitra AK, McCarthy MP, Stroud RM. Three-dimensional structure of the nicotinic acetylcholine receptor and location of the major associated 43-kD cytoskeletal protein, determined at 22 A by low dose electron microscopy and x-ray diffraction to 12.5 A. J Cell Biol. 1989;109:755–774. doi: 10.1083/jcb.109.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittaud P, Marangi PA, Erb-Vogtli S, Fuhrer C. Agrin-induced activation of acetylcholine receptor-bound Src family kinases requires Rapsyn and correlates with acetylcholine receptor clustering. J Biol Chem. 2001;276:14505–14513. doi: 10.1074/jbc.M007024200. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. J Mol Biol. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- Mohamed AS, Swope SL. Phosphorylation and cytoskeletal anchoring of the acetylcholine receptor by Src class protein-tyrosine kinases. Activation by rapsyn. J Biol Chem. 1999;274:20529–20539. doi: 10.1074/jbc.274.29.20529. [DOI] [PubMed] [Google Scholar]
- Moransard M, Borges LS, Willmann R, Marangi PA, Brenner HR, Ferns MJ, Fuhrer C. Agrin regulates rapsyn interaction with surface acetylcholine receptors, and this underlies cytoskeletal anchoring and clustering. J Biol Chem. 2003;278:7350–7359. doi: 10.1074/jbc.M210865200. [DOI] [PubMed] [Google Scholar]
- Noakes PG, Phillips WD, Hanley TA, Sanes JR, Merlie JP. 43K protein and acetylcholine receptors colocalize during the initial stages of neuromuscular synapse formation in vivo. Dev Biol. 1993;155:275–280. doi: 10.1006/dbio.1993.1025. [DOI] [PubMed] [Google Scholar]
- Ohno K, Engel AG, Shen XM, Selcen D, Brengman J, Harper CM, Tsujino A, Milone M. Rapsyn Mutations in Humans Cause Endplate Acetylcholine-Receptor Deficiency and Myasthenic Syndrome. Am J Hum Genet. 2002;70:4. doi: 10.1086/339465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Inoue A, Okada M, Murata Y, Kakuta S, Jigami T, Kubo S, Shiraishi H, Eguchi K, Motomura M, Akiyama T, Iwakura Y, Higuchi O, Yamanashi Y. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- Phillips WD, Maimone MM, Merlie JP. Mutagenesis of the 43-kD postsynaptic protein defines domains involved in plasma membrane targeting and AChR clustering. J Cell Biol. 1991a;115:1713–1723. doi: 10.1083/jcb.115.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips WD, Noakes PG, Roberds SL, Campbell KP, Merlie JP. Clustering and immobilization of acetylcholine receptors by the 43-kD protein: a possible role for dystrophin-related protein. J Cell Biol. 1993;123:729–740. doi: 10.1083/jcb.123.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips WD, Vladeta D, Han H, Noakes PG. Rapsyn and agrin slow the metabolic degradation of the acetylcholine receptor. Mol Cell Neurosci. 1997;10:16–26. doi: 10.1006/mcne.1997.0634. [DOI] [PubMed] [Google Scholar]
- Phillips WP, Kopta C, Blount P, Gardner PD, Steinbach JH, Merlie JP. ACh receptor-rich membrane domains organized in fibroblasts by recombinant 43-kilodalton protein. Science. 1991b;251:568–570. doi: 10.1126/science.1703661. [DOI] [PubMed] [Google Scholar]
- Quiram PA, Ohno K, Milone M, Patterson MC, Pruitt NJ, Brengman JM, Sine SM, Engel AG. Mutation causing congenital myasthenia reveals acetylcholine receptor beta/delta subunit interaction essential for assembly. The Journal of clinical investigation. 1999;104:1403–1410. doi: 10.1172/JCI8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarao MK, Bianchetta MJ, Lanken J, Cohen JB. Role of rapsyn tetratricopeptide repeat and coiled-coil domains in self- association and nicotinic acetylcholine receptor clustering. J Biol Chem. 2001;276:7475–7483. doi: 10.1074/jbc.M009888200. [DOI] [PubMed] [Google Scholar]
- Ramarao MK, Cohen JB. Mechanism of nicotinic acetylcholine receptor cluster formation by rapsyn. Proc Natl Acad Sci U S A. 1998;95:4007–4012. doi: 10.1073/pnas.95.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji H, Nomoto H, Ohta M, Hayashi K. Crosslinking of proteins in acetylcholine receptor-rich membranes from Torpedo californica: relation of 43-kD protein and Torpedo dystrophin to acetylcholine receptor. Biochem Int. 1992;28:1071–1077. [PubMed] [Google Scholar]
- Strochlic L, Cartaud A, Cartaud J. The synaptic muscle-specific kinase (MuSK) complex: new partners, new functions. Bioessays. 2005;27:1129–1135. doi: 10.1002/bies.20305. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Wang ZZ, Mathias A, Gautam M, Hall ZW. Metabolic stabilization of muscle nicotinic acetylcholine receptor by rapsyn. J Neurosci. 1999;19:1998–2007. doi: 10.1523/JNEUROSCI.19-06-01998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee SD, Anderson KV, Niswander LA. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development. 2006;133:4993–5000. doi: 10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- Weston C, Gordon C, Teressa G, Hod E, Ren XD, Prives J. Cooperative regulation by Rac and Rho of agrin-induced acetylcholine receptor clustering in muscle cells. J Biol Chem. 2003;278:6450–6455. doi: 10.1074/jbc.M210249200. [DOI] [PubMed] [Google Scholar]
- Weston C, Yee B, Hod E, Prives J. Agrin-induced acetylcholine receptor clustering is mediated by the small guanosine triphosphatases Rac and Cdc42. J Cell Biol. 2000;150:205–212. doi: 10.1083/jcb.150.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
COS cells transfected with CD4 chimeras and rapsyn were incubated live with anti-CD4 antibodies at 4°C and then fixed, permeabilized and immunostained for rapsyn. Rapsyn clustered CD4ct-β407–469 but not CD4 on the cell surface. Similarly, rapsyn clustered CD4 chimeras where the β loop helix had been replaced by the helical domain of the α and β subunits (CD4-β/ α391–428 and ε419–456; n = 2–3 independent experiments).