Abstract
The safe and effective delivery of RNA therapeutics remains the major barrier to their broad clinical application. Here we develop a new nanoparticulate delivery system based on inorganic particles and biodegradable polycations. First, gold nanoparticles were modified with the hydrophilic polymer poly(ethylene glycol) (PEG), and then small interfering RNA (siRNA) was conjugated to the nanoparticles via biodegradable disulfide linkages, with ~30 strands of siRNA per nanoparticle. The particles were then coated with a library of end-modified poly(β-amino ester)s (PBAEs), previously identified as capable of facilitating intracellular DNA delivery. Nanoparticulate formulations developed here facilitate high levels of in vitro siRNA delivery, facilitating delivery as good or better than the commercially available lipid reagent, Lipofectamine 2000.
RNA interference (RNAi) is an endogenous process whereby double-stranded RNA (dsRNA) can mediate the catalytic destruction of its homologous mRNA target. Short 22 nt dsRNA fragments called small interfering RNA (siRNA), are intermediates in the process and have shown their potential as therapeutics.1–4 The development of RNAi based upon synthetic siRNA has led to a variety of potential therapeutic applications for diseases whose conventional treatments are limited.5–9 The safe and effective intracellular delivery of siRNA remains the most challenging barrier to the broad application of siRNA in the clinic.10–14 To date a number of carriers have been investigated for their potential as siRNA delivery agents15 including cationic polymers,16,17 lipids8 or lipid-like materials,18 iron oxide nanoparticles,19 gold particles,20–22 and semiconductor nanocrystals.23,24 Alternatively, siRNA has been chemically modified and conjugated to small organic molecules25,26 or polymeric materials12,27,28 to enhance its stability and cellular uptake.15
Poly(β-amino ester)s (PBAEs) have shown potential as delivery agents for DNA in various cell lines and therapeutic models.29–36 To the best of our knowledge, however, these materials have not yet demonstrated their ability to deliver siRNA. The disorderly interactions of siRNA with polymer are likely to result in incomplete condensation of the polymer into a particulate due to the stiffer nature of an RNA molecule relative to DNA (Figure 1A).37 We hypothesized that “assembled” multiple siRNA strands attached to a particle could facilitate PBAE interaction and nanoparticle formation.
Figure 1.
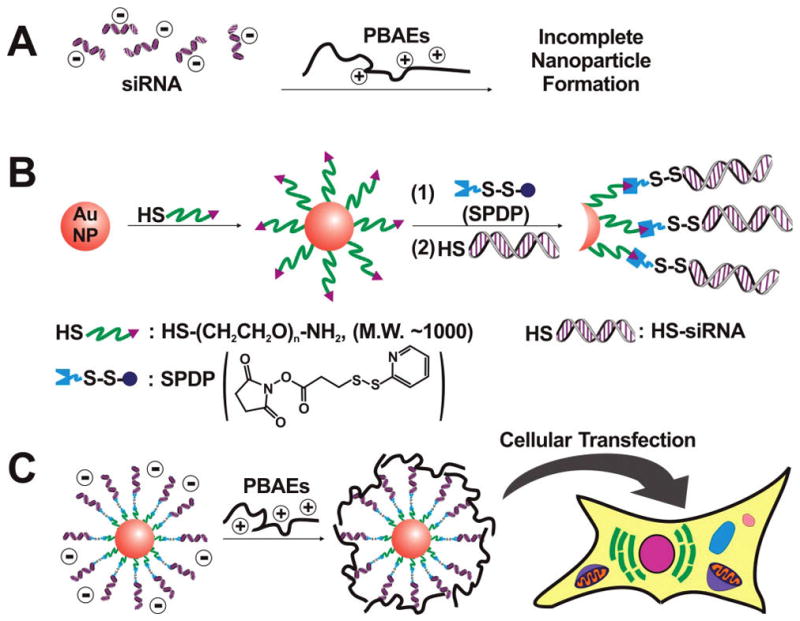
(A) The scheme illustrating incomplete nanoparticle formation of siRNA combined with PBAEs. (B) The synthesis of siRNA-modified AuNPs (siRNA–AuNPs) with a disulfide linkage. (C) The complexation of siRNA–AuNPs with PBAEs, followed by cellular transfection.
Herein, we present a multicomponent nanoparticulate siRNA into cells using PBAEs as a delivery enhancer, and gold nanoparticles (AuNPs) as a scaffold to assemble siRNA strands (Figure 1B,C). AuNPs have a number of desirable properties, including low cytotoxicity,38 easy size control,39 and well-developed surface chemistry for dense loading of functionalities.40–42 siRNA strands were assembled onto the AuNPs (siRNA–AuNPs) through a cleavable disulfide linkage. The presence of a disulfide bond between siRNA and gold was chosen due to its potential to facilitate cytoplasmic release of the siRNA from AuNPs under the reductive cytosolic conditions.43 To enhance the cellular uptake and further endosomal escape, a number of different PBAEs were chosen based upon structures previously identified as useful for DNA delivery,29–36 and assembled onto the particles.
The synthesis of the siRNA–AuNP conjugates (Figure 1B) begins with modifying AuNPs with HS–PEG–NH2 (Mw 1000 Da, see Supporting Information). PEG was used to isolate the gold surface from disulfide bonds, since AuNPs could react with the disulfide bonds and induce release of siRNA. These NH2–PEG-modified AuNPs (NH2–PEG–AuNPs) exhibit excellent salt stability and can be salted up to 2.5 M NaCl. This high salt stability was important to facilitate siRNA conjugation procedures and will be discussed in detail below. To introduce disulfide bonds on the particles, we conjugated N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP) as a disulfide-containing linker to the terminal NH2 group of PEG on AuNPs (Figure 1B, see Supporting Information). After SPDP was conjugated to NH2–PEG–AuNPs (SPDP–PEG–AuNP), we conjugated thiolated siRNA (antifirefly luciferase, HS-siRNA) to SPDP by the addition of excess HS–siRNA at pH 8.5 to displace the 2-pyridyldithio group of SPDP, still maintaining the disulfide linkage.
To maximize the siRNA loading, we optimized the reaction conditions by monitoring the number of siRNA per particle as a function of the salt concentration and the reaction time (Figure 2A,B). The number of siRNA strands per particle was quantitatively analyzed by cleaving the disulfide bond with dithiothreitol, followed by RNA analysis using commercially available RNA assay kits (RiboGreen, Invitrogen, see Supporting Information). The loading increased as the salt concentration was increased, presumably due to decreased repulsion between the negatively charged siRNA strands (Figure 2A).44 Figure 2B shows that as the reaction time of the conjugation of HS-siRNA to the SPDP–PEG–AuNPs increases, the loading of siRNA per AuNP increases. Importantly, the loading of siRNA per AuNP without SPDP was low, indicating that HS–siRNA is conjugated primarily to SPDP, not to the AuNP surface by displacing NH2–PEG–SH from gold (Figure 2B). The maximum loading of siRNA per particle was found to be ~40 strands after 80 h at 2.5 M NaCl. As we observed slight formation of AuNP aggregates after 40 h, however, the optimum conjugation time for HS–siRNA to AuNPs was determined to be 40 h, resulting in ~30 strands of siRNA per particle (Figure 2B). These loadings are comparable to those of siRNA on a 13 nm AuNP without PEG and SPDP (~33 strands/AuNP at 0.3 M NaCl).21 The high loading of siRNA per AuNP was hypothesized to be important both to facilitate PBAE complexation and potentially to increase potency per particle. Over the entire synthetic procedure at each step, the conjugated AuNPs exhibited high stability and low aggregation under the conditions studied, as characterized by the unique surface plasmon resonance of AuNPs around 525 nm in the UV–vis spectra (Figure 2C).
Figure 2.
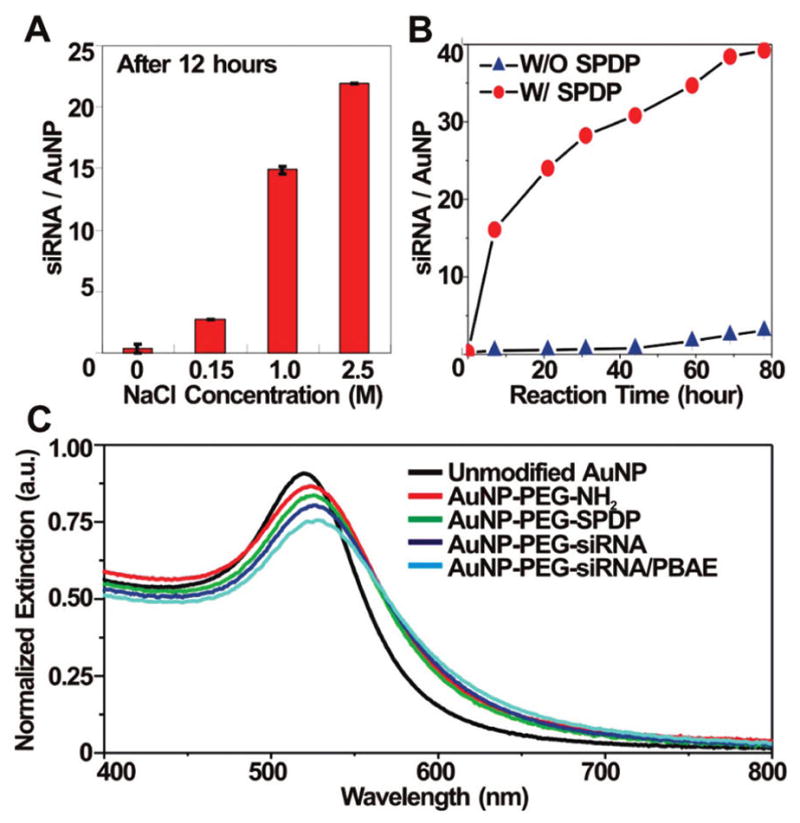
(A) The loading of siRNA on 15 nm AuNPs at each NaCl concentration after 12 h of incubation. (B) The loading of siRNA on 15 nm AuNPs at 2.5 M NaCl concentration with or without SPDP. (C) UV–vis spectra of unmodified AuNPs (black), NH2–PEG-modified AuNPs (red), SPDP–PEG-modified AuNPs (green), siRNA-modified AuNPs (siRNA–AuNPs, dark blue) and PBAE-complexed siRNA–AuNPs (light blue). The PBAE used to complex siRNA–AuNPs is C32-221 (see Figure 3). Note that the data in parts A and 2B were collected in triplicate.
After the synthesis of siRNA–AuNPs, we synthesized and screened a focused library of PBAEs for their ability to facilitate functional siRNA delivery, in vitro. Previous studies had identified poly(butane dioldiacrylate coaminopentanol) (C32, Figure 3A) as a leading DNA delivery polymer.34 Modification of the terminal groups of C32 can enhance delivery efficiency significantly, depending on the type of end-modifying amines.32,35,36 On the basis of these studies, we selected five of the best amines previously identified as useful for DNA delivery in previous studies (103, 116, 117, 118, and 122)32,35,36 and additionally seven new amines (208, 210, 212, 213, 221, 225, and 228), for a total of 12 primary diamines for the end modification of C32 (Figure 3B,C). Unmodified C32 and a more hydrophobic PBAE, D60, were also tested without the end modification for comparison (Figure 3D). The 14 PBAEs were synthesized following the synthetic schemes in our previous literature36 (Figure 3A,B) and combined with siRNA–AuNP for complexation (PBAE–siRNA–AuNPs, see Supporting Information). In all cases, no particle aggregation was observed after the addition of PBAEs (Figure 2C). The ζ-potential of the siRNA–AuNPs was ca. −34 mV but increased to ca. +13 mV when complexed with PBAEs, which was expected to facilitate delivery via interaction with the negatively charged cell membranes. The core–shell structure (AuNP@PBAE) of the PBAE–siRNA–AuNPs with a diameter of ~100 nm was also characterized by transmission electron microscopy (TEM) analysis (Figure 4).
Figure 3.
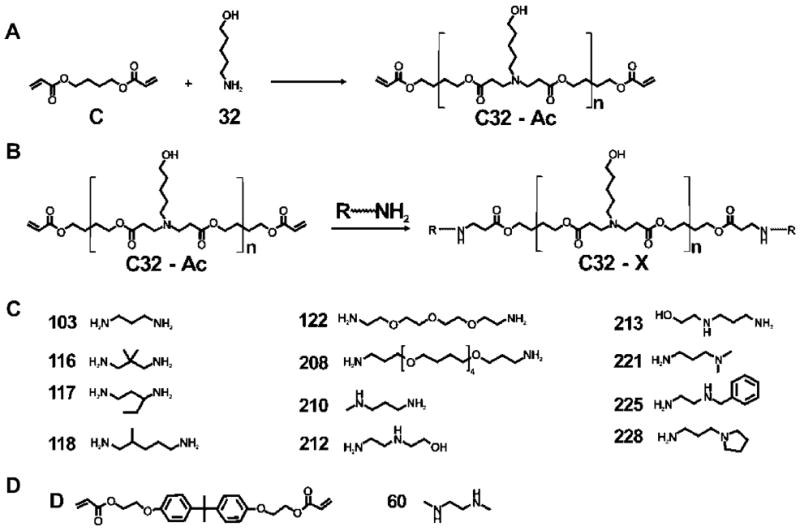
(A) The synthesis of poly(β-amino ester)s (PBAEs) for C32. (B) The end modification of PBAEs for C32. (C) Amines used for the end modification of C32. (D) The diacrylate and diamine monomers used for the synthesis of D60. Note that the alphabetical and numeric designations are adopted from our previous publications.29–36
Figure 4.
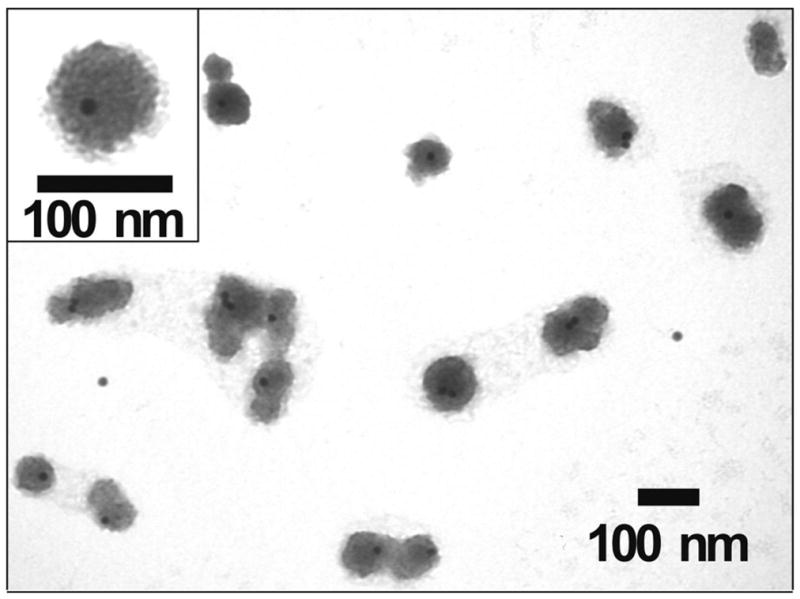
A representative TEM image of PBAE-complexed siRNA–AuNPs. The PBAE used to complex siRNA–AuNPs is C32-221 (see Figure 3). The image was taken by a Tecnai G2 Spirit at HV = 120 kV. The complex particles were observable without additional staining.
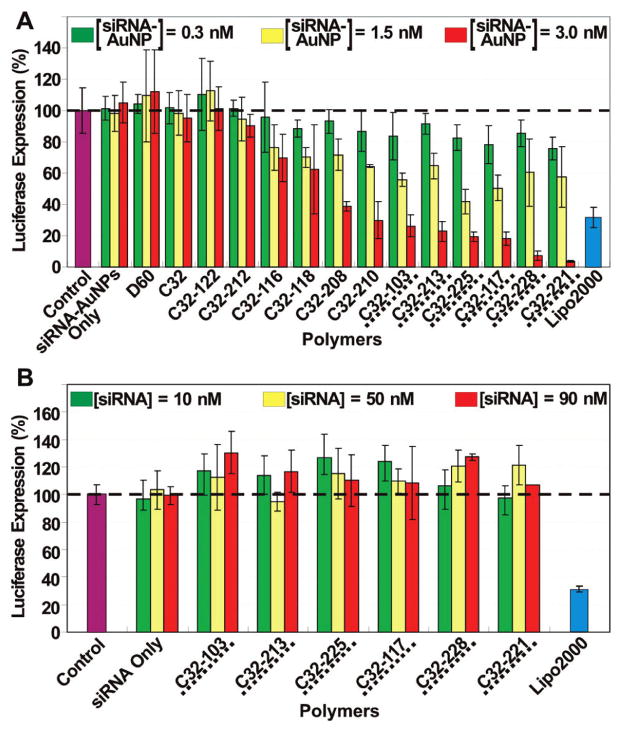
To verify the cellular transfection of the PBAE–siRNA–AuNPs, gene knockdown was evaluated in a modifed HeLa cell line, where the HeLa cells were genetically engineered to express both firefly luciferase and Renilla luciferase.18 As the sequence of siRNA was designed to selectively reduce the firefly luciferase expression, delivery efficacy can be identified by specific reduction in firefly luciferase, without reduction in the nonspecific Renilla luciferase signal. The particles complexed with different PBAEs (Figure 3) were applied to the HeLa cells in a 96-well format for screening and incubated for 4 h at 37 °C. After 24 h, the luminescence from each luciferase was measured, respectively, by a commercially available assay kit (Dual Glo Luciferase Assay System, see Supporting Information). Figure 5A shows the selective knockdown of the firefly luciferase expression depending on the type and the dose of PBAEs. Importantly, the best two PBAEs (C32-228 and 221) exhibit significantly better efficiency than the commercially available liposome-based delivery agent (Lipofectamine2000, Invitrogen) used as a positive control in accordance with manufacturer instructions (see Supporting Information). No significant toxicity was observed (see Supporting Information). In contrast to this result, the corresponding amount of unassembled siRNA combined with PBAEs did not exhibit any silencing effect regardless of the dose, indicating that assembling siRNA strands on AuNPs was required for the intracellular delivery of siRNA (Figure 5B). Note that siRNA–AuNPs without PBAEs (“siRNA–AuNP Only”) also did not exhibit any silencing effect (Figure 5A), despite their structural similarity to the AuNP-based gene silencing agent by Mirkin at el.21
Figure 5.
(A) In vitro screening of PBAEs for siRNA–AuNP delivery. Dose response of each PBAE–siRNA–AuNPs to gene knockdown in HeLa cells was obtained in triplicate. The data obtained from untreated cells (control), siRNA–AuNPs without PBAEs (siRNA–AuNPs only), and the commercially available gene delivery reagent (Lipo2000) are also shown for comparison. (B) In vitro screening of the best six PBAEs from Figure 5A for the delivery of the corresponding amount of naked siRNA without AuNPs. No gene knockdown in HeLa cells was observed. Data were obtained in triplicate in the presence of 10% fetal bovine serum (FBS).
Finally, the role of PBAEs for the cellular uptake of siRNA–AuNPs was further confirmed by TEM analysis. The PBAE–siRNA–AuNPs were prepared with C32-221, the PBAE showing the best delivery efficiency, and exposed to the cell culture media under the same conditions studied for the gene knockdown experiments. A typical TEM image of the HeLa cell transfected by the PBAE–siRNA–AuNPs is shown in Figure 6A. Numerous particles are observed as aggregates, in certain cases confined within endosomes in the cytosol. This indicates the cellular uptake of the particles in the presence of PBAEs. Interestingly, on the other hand, the siRNA–AuNPs and unmodified AuNPs, each in the absence of PBAEs, were not observed in the cells under the conditions studied (Figure 6B,C). The importance of the surface properties and their potential to facilitate uptake of AuNPs for cellular transfection has been investigated and reported in other formats.45–48
Figure 6.
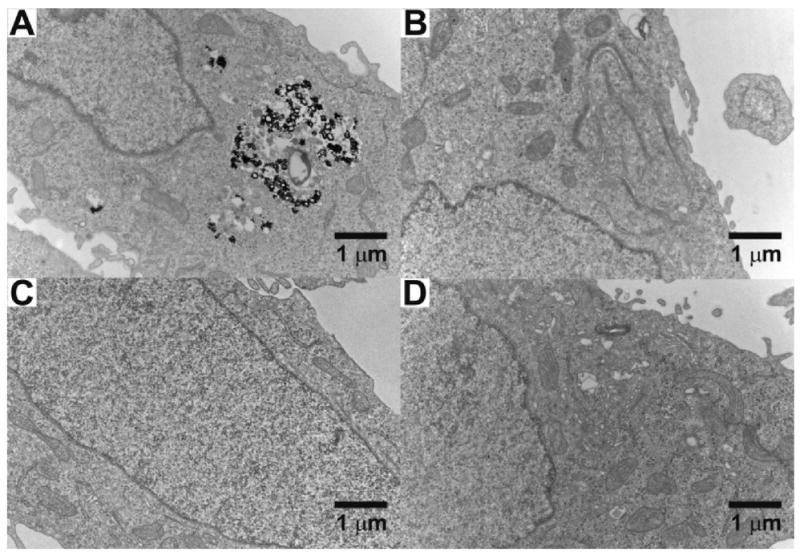
Representative TEM images of the HeLa cells exposed to 3 nM (A) PBAE–siRNA–AuNPs, (B) siRNA–AuNPs without PBAEs, (C) unmodified AuNPs, and (D) no nanoparticles (control). The TEM analysis was performed with more than 10 images from a large area of each sample. The PBAE used for the complexation with siRNA–AuNPs is C32-221 (see Figure 3). The images were taken with a Tecnai G2 Spirit at HV = 80 kV.
In conclusion, we describe the development, optimization, and characterization of a new nanoparticulate siRNA delivery vehicle using PBAEs and AuNPs. Future studies will address the potential of these particles in other cell types, as well as in vivo. It is possible that this method could be generalized to facilitate intracellular movement of other negatively charged particles associated with drugs or genes by the complexation with positively charged PBAEs
Supplementary Material
Acknowledgments
This work was supported by NIH grant EB00244 and Alnylam, Inc. The TEM analysis was conducted utilizing the W. M. Keck Foundation Biological Imaging Facility at the Whitehead Institute. Nicki Watson is acknowledged for the TEM operation and helpful discussions.
Footnotes
Supporting Information Available: Description of materials, experimental details for the synthesis of PBAE–siRNA–AuNPs, and the cell experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.McManus MT, Sharp PA. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ, Rossi JJ. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 4.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorsett Y, Tuschl T. Nat Rev Drug Discovery. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 6.Jacque JM, Triques K, Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DH, Rossi JJ. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann TS, Lee ACG, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 9.Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, Xu J, Liu Y, Zheng BJ, Woodle MC, Zhong N, Lu PY. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidel JD, Yu Z, Liu JYC, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Proc Natl Acad Sci USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Nature. 2007;448:39–45. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 12.Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hastrom JE, Wolff JA. Proc Natl Acad Sci USA. 2008;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong JH, Mok H, Oh YK, Park TG. Bioconjugate Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 16.Boussif O, Lezoualc’H F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr J-P. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jere D, Xu CX, Arote R, Yun CH, Cho MH, Cho CS. Biomaterials. 2008;29:2535–2547. doi: 10.1016/j.biomaterials.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, Fougerolles AD, Dorkin JR, Jayaprakash KN, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DWY, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DB. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. Nat Med. 2007:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 20.Oishi M, Nakaogami J, Ishii T, Nagasaki Y. Chem Lett. 2006;35:1046–1047. [Google Scholar]
- 21.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Nano Lett. 2009 doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 23.Qi L, Gao X. ACS Nano. 2008;2:1403–1410. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. J Am Chem Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Bioorg Med Chem Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Moschos SA, Jones SW, Perry MM, Williams AE, Erjefalt JS, Turner JJ, Barnes PJ, Sproat BS, Gait MJ, Lindsay MA. Bioconjugate Chem. 2007;18:1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oishi M, Nagasaki Y, Itaka K, Nishiyama N, Kataoka K. J Am Chem Soc. 2005;127:1624–1625. doi: 10.1021/ja044941d. [DOI] [PubMed] [Google Scholar]
- 28.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. J Controlled Release. 2006;116:123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Anderson MW, Holmes SM, Hanif N, Cundy CS. Angew Chem, Int Ed. 2000;39:2707–2710. doi: 10.1002/1521-3773(20000804)39:15<2707::aid-anie2707>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 30.Anderson DG, Lynn DM, Langer R. Angew Chem, Int Ed. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 31.Green JJ, Langer R, Anderson DB. Acc Chem Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zugates GT, Tedford NC, Zumbuehl A, Jhunjhunwala S, Kang CS, Griffith LG, Lauffenburger DA, Langer R, Anderson DG. Bioconjugate Chem. 2007;18:1887–1896. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]
- 33.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Nano Lett. 2007;7:874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 34.Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. Proc Natl Acad Sci USA. 2004;101:16028–16033. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Mol Ther. 2007;15:1306–1312. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- 36.Green JJ, Zugates GT, Tedford NC, Huang YH, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Adv Mater. 2007;19:2836–2842. [Google Scholar]
- 37.Gary DJ, Puri N, Won YY. J Controlled Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Connor EE, Mwamuka J, Gole A, Murphy CJ, Wyatt MD. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 39.Daniel MC, Astruc D. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 40.Love JC, Estroff JA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 41.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh PS, Kim CK, Han G, Forbes NS, Rotello VM. ACS Nano. 2008;2:2213–2218. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meister A, Anderson ME. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 44.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nativo P, Prior IA, Brust M. ACS Nano. 2008;2(8):1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 46.Verma A, Uzun O, Hu Y, Hu Y, Han HS, Watson N, Chen S, Irvine DJ, Stellacci F. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 48.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.