Abstract
Purpose
Breast cancer treatment often employs DNA double-strand breaks (DSBs), such as that induced by irradiation or anticancer agents. Ubiquitination is required at the site of DNA damage and plays a crucial role in the DSB repair pathway. We investigated the effect of proteasome inhibitors on the pathway after exposure to chemotherapeutic agents and examined its correlation with cytotoxicity.
Methods
Cells were exposed for 1 h to DNA damage-inducing chemotherapeutic agents. After DNA damage, nuclear foci formation of conjugated ubiquitin (Ub-foci) and cell viability were examined in the absence or presence of proteasome inhibitors MG132 and epoxomicin.
Results
Proteasome inhibitors trapped conjugated ubiquitin in the cytosol and blocked irinotecan (CPT-11)- and epirubicin-induced Ub-foci formation in MCF10A cells and HeLa cells, but not in MCF7 cells. MG132 sensitized MCF10A cells to CPT-11 and epirubicin treatment, demonstrating a synergistic effect. This synergistic effect is likely due to the failure to repair DNA, because a significant rise in unrepaired DNA damage was observed in the cells treated with MG132. On the other hand, no synergy was observed in MCF7 cells or when MG132 was combined with docetaxel.
Conclusions
The synergistic effect of proteasome inhibitors in combination with DNA damage-inducing agents warrants further investigating into its effectiveness in the treatment of breast cancer.
Keywords: Chemosensitivity, Ubiquitin, Proteasome inhibitor, Nuclear foci formation, DNA damage
Introduction
Induction of DNA double-strand breaks (DSBs) by irradiation or anticancer agents is a major strategy employed for breast cancer treatment. DSBs cause breast cancer cells to undergo apoptosis when appropriate repair pathways, such as those mediated by BRCA1, are perturbed. Therefore, chemosensitivity and the competence of DNA damage repair pathways are closely correlated. For example, it was recently reported that acquired resistance to cisplatin or PARP inhibitor in BRCA1- or BRCA2-mutated tumors can be mediated by secondary mutations in these genes that restore the wild-type reading frame [1–3].
Recently, the cascade of events in response to DSBs has been significantly uncovered. The sequential recruitment of repair proteins at the site of DNA damage includes two RING finger type E3 ubiquitin ligases, RNF8 and BRCA1. RNF8 catalyzes lysine 63-linked polyubiquitin (K63-Ub) chains on H2AX [4–7]. Ubiquitinated H2AX then recruits the BRCA1/Abraxas/RAP80 complex through the RAP80 subunit, an adaptor that contains UIM (ubiquitin interacting motif) domains [8–10]. BRCA1 forms a RING heterodimer E3 ubiquitin ligase with BARD1 [11] and is required for the recruitment of BRCA2 and Rad51 to damaged sites for homologous recombination repair [12]. Thus, ubiquitination is involved in key steps that properly conduct the homologous recombination repair pathway after DSBs. Indeed, inhibition of IR-induced nuclear foci (IRIF) formation of conjugated ubiquitin results in defective downstream events, including BRCA1 IRIF formation and IR hypersensitivity [5–7, 13].
Ubiquitin modification regulates a wide range of cellular pathways such as removal of misfolded or aged housekeeping proteins, protein trafficking, the cellular immune response by antigenic peptide processing, the cell cycle and the DNA damage response. Ubiquitin modification requires several critical enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin carrier protein (E2), and a ubiquitin ligase (E3) [14]. The E3 catalyzes the formation of polyubiquitin chains (and sometimes monoubiquitination), utilizing ubiquitins that have been activated by the E1 and E2 enzymes, and transfers them onto specific substrate(s). While ubiquitin modifications signal a variety of processes depending upon the type of ubiquitin chains, the most common pathway is the ubiquitin-proteasome system (UPS) that is mediated by Lys48-linked polyubiquitin chains [14, 15]. Substrates conjugated with Lys48-linked chains are recognized by the 19S regulatory cap subunits of the 26S proteasome and are degraded by the 20S catalytic core subunits [16]. These reactions can be inhibited by proteasome inhibitors such as MG132, epoxomicin or the clinically used bortezomib (PS-341, Velcade®).
The effect of proteasome inhibitors on the response to DNA damage is not fully understood. Because the known major ubiquitin chains built at the damaged site in response to DSBs are Lys63- and Lys6-linked [9, 15, 17], the direct effect of the proteasome inhibitor could be limited. Interestingly, recent studies showed that inhibition of the 26S proteasome by MG132 depleted the pool of available nuclear ubiquitin because undegraded polyubiquitinated proteins accumulated in the cytosol [5, 18]. The depletion of free nuclear ubiquitin resulted in the loss of IRIF formation of conjugated ubiquitin, accompanied by loss of BRCA1- and 53BP1-IRIF formations [5]. This suggests the possibility that proteasome inhibitors may also inhibit the repair pathway of DNA damage caused by treatment with DNA damage-inducing chemotherapeutic agents, thereby having an additive or synergistic effect on cytotoxicity. In this regard, we investigated the effect of proteasome inhibitors on the cellular distribution of conjugated ubiquitin and its correlation with chemotherapeutic agent-induced nuclear foci formation and cytotoxicity. The results suggest that the effect of the proteasome inhibitors on ubiquitin distribution varies among cell lines and that it correlates with the DNA damage response and chemosensitivity.
Materials and methods
Cell culture
MCF7 breast carcinoma cells and HeLa cervical carcinoma cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% antibiotic–antimycotic agent (Life Technologies, Inc, Grand Island, NY) in 5% CO2 at 37°C. MCF10A normal human breast epithelial cells were grown in DMEM/Ham’s F12 (1:1) medium supplemented with 2.5% FCS, 100 ng/ml cholera toxin, 20 ng/ml epidermal growth factor, 500 ng/ml hydrocortisone, 10 mg/ml insulin and 1% antibiotic–antimycotic agent. Cells were seeded at a concentration of 1.5 × 103 cells per well in flat-bottom 96-well microplates or 5.0 × 104 cells per well in Lab-Tek eight-well chamber slides (Nalge Nunc) for the study of cell viability or immunofluorescence, respectively, 24 h before treatment with proteasome inhibitor and/or chemotherapeutic agents. For induction of DNA damage with chemotherapeutic agents, cells were incubated for 1 h with the indicated doses of either CPT-11 (Sigma, Saint Louis, MO) or epirubicin (Pfizer, New York, NY). Cells were washed with phosphate buffered saline (PBS) and further incubated in the culture medium with or without proteasome inhibitors. In some experiments, docetaxel (TAXOLTM, Sanofi-aventis K.K. Tokyo, Japan) was added instead of the DNA-damaging agents. Proteasome inhibitors MG132 (Calbiochem, San Diego, CA) or epoxomicin (Millennium Pharma, Cambridge, MA) or solvent DMSO was added at the indicated doses simultaneously with the chemotherapeutic agents and was maintained after the removal of the chemotherapeutic agents.
Indirect immunocytochemistry
Indirect immunocytochemistry was performed as previously described with one modification [19]. Briefly, proliferating cells were fixed with 3% formalin for 15 min and permeabilized with 0.2% Triton X-100 for 5 min. Cells were washed with PBS, blocked with 0.5% bovine serum albumin in PBS and incubated with a monoclonal antibody to conjugated ubiquitin (FK2, Nippon Bio-Test, Japan) diluted in the blocking buffer (10 μg/ml). FITC-conjugated anti-mouse IgG (Jackson Immuno Research, West Grove, PA) was used as the secondary antibody at a 1:25 dilution. For double staining with phosphorylated H2AX (γH2AX), a monoclonal antibody to γH2AX (upstate, Lake Placid, NY) was directly fluorescently labeled with Alexa Fluor® 555 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and was added to the cells previously stained with anti-conjugated ubiquitin. Cells were washed and re-fixed with 3% formalin for 15 min. After washing three times, the nucleus was counterstained with DAPI-containing fluorescent mounting medium (Invitrogen, Carlsbad, CA), and the cells were examined with a confocal laser scanning microscope (LSM 510, Carl Zeiss).
Measurements of chemosensitivity
Twenty-four hours after release from transient exposure to chemotherapeutic agents, cell viabilities were determined with the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega Corp., Madison, WI) according to the manufacturer’s instructions. Briefly, 6 μM CellTiter 96® Aqueous One Solution Reagent was directly added to the culture medium of cells in 96-well plates, incubated for 2 h, and the quantity of formazan product was measured by light absorbance at 570 nm primary wavelength and 600 nm reference wavelength by scanning with a 96-well plate reader (multi-spectrophotometer, DAINIPPON, Osaka, Japan). The percentages of cell viability are exhibited as the mean of triplicate experiments, where cells not exposed to chemotherapeutic agents or MG132 represent 100% viability.
Comet assay
Neutral comet assays were performed using the Trevigen’s CometAssay kit (4250-050-K) according to the manufacturer’s instructions. DNA was stained with Trevigen SYBR green, and Comet images were examined with a confocal laser scanning microscope (LSM 510, Carl Zeiss). Tail moments were analyzed by the TriTek CometScore Freeware program. The average value of tail moments was determined by counting at least 100 cells per sample.
Results
Effect of proteasome inhibitors on the cellular distribution of conjugated ubiquitin
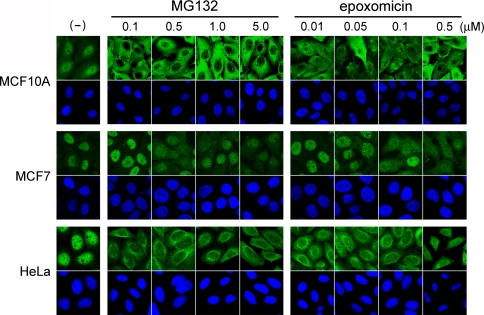
Previously it was shown that treatment of cells with the proteasome inhibitor MG132 resulted in accumulation of conjugated ubiquitin in the cytosol [5]. To investigate whether this phenomenon is commonly observed among various cell types and with different proteasome inhibitors, we incubated MCF10A, MCF7, and HeLa cells with MG132 or epoxomicin at different doses for 4 h. Epoxomicin is currently the most specific proteasome inhibitor, reacting irreversibly with the chymotrypsin-like site of the proteasome [20–22]. Cells were then stained with FK2 antibody that recognizes conjugated ubiquitin. Interestingly, whereas conjugated ubiquitin staining accumulated in the cytosol in MCF10A cells and HeLa cells at the lowest dose of MG132 (0.1 μM) examined, it remained nuclear in MCF7 cells even at the highest dose (5 μM) (Fig. 1). The same results were observed when MG132 was substituted with epoxomicin (Fig. 1). Thus, MCF7 is more resistant to the proteasome inhibitors in regard to the cytosol trap of ubiquitin than other cells tested. The results indicate that there are different competencies among cell lines for proteasome inhibitor-induced cytosol trapping of ubiquitin.
Fig. 1.
Effect of proteasome inhibitors on cellular distribution of conjugated ubiquitin. MCF10A, MCF7 and HeLa cells were incubated with or without (–) the indicated dose of MG132 or epoxomicin for 4 h. Cells were fixed and stained with antibody to conjugated ubiquitin (upper panels). The nucleus was counterstained with DAPI (lower panels)
Proteasome inhibitors block CPT-11-induced Ub-foci formation at the site of DNA damage
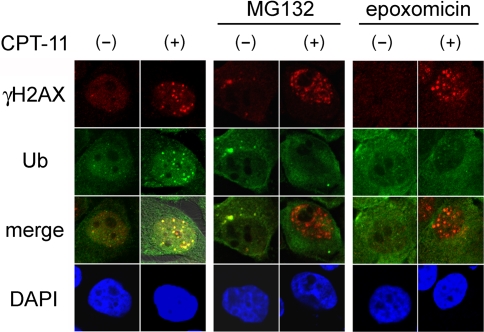
MG-132-induced cytosol trapping of ubiquitin inhibited IR-induced Ub-foci formation as well as accumulation of downstream proteins at the damaged foci [5]. This prompted us to test whether proteasome inhibitors also inhibit Ub-foci formation caused by DNA-damaging chemotherapeutic agents. MCF10A cells were treated for 1 h with CPT-11, a topoisomerase I inhibitor that interacts with cellular topoisomerase I—DNA complexes and provokes single strand DNA breaks or S-phase specific DSBs in cells[23]. The Ub-foci were analyzed 4 h after incubation in the presence or absence of the proteasome inhibitors. Nuclear foci formation of γH2AX was also analyzed as a marker for sites of DNA damage. CPT-11 treatment resulted in Ub-foci formation that merged with γH2AX foci in the absence of proteasome inhibitors (Fig. 2, left panels). On the other hand, in the presence of either MG132 or epoxomicin, CPT-11 treatment resulted in γH2AX foci, but not Ub-foci formation (Fig. 2, middle and right panels). The same results were observed with HeLa cells (data not shown). Thus, the proteasome inhibitors blocked CPT-11-induced Ub-foci formation at the sites of DNA damage.
Fig. 2.
Proteasome inhibitors perturb DNA damage-induced Ub-foci formation after exposure to CPT-11. MCF10A cells were incubated with (+) or without (–) 45 μM CPT-11 for 1 h in the absence (left panels) or presence of 0.5 μM MG132 or 0.05 μM epoxomicin, as indicated. Cells were further incubated with the proteasome inhibitors for 3 h, fixed and stained with the indicated antibodies. Merge indicates overlaid images of the two detected proteins. The nucleus was counterstained with DAPI. Ub: anti-conjugated ubiquitin antibody
The effect of proteasome inhibitors on Ub-foci formation varies among cell lines
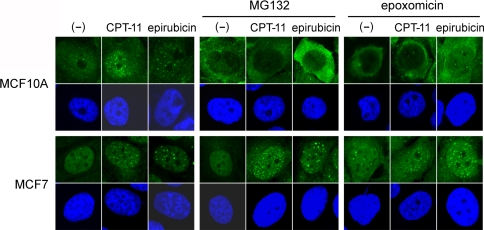
Because cytosol trapping of ubiquitin by proteasome inhibitors varied among cell lines (Fig. 1), we next tested whether the inhibitors also cause variation of Ub-foci formation after CPT-11 treatment. MCF10A and MCF7 cells were treated with CPT-11 in the presence or absence of the proteasome inhibitors MG132 or epoxomicin, and the Ub-foci were analyzed. Consistent with the cellular distribution of conjugated ubiquitin, both MG132 and epoxomicin blocked Ub-foci formation in MCF10A and HeLa cells, though they did not affect the foci in MCF7 cells (Fig. 3 and data not shown). We next tested epirubicin, a topoisomerase II inhibitor that also provokes DSBs and is often used in breast cancer treatments. Ub-foci formation induced by epirubicin was again blocked by MG132 and epoxomicin in MCF10A cells, but not in MCF7 cells (Fig. 3). These results suggest that the ability of proteasome inhibitors to trap ubiquitin in the cytosol is directly linked to the ability to inhibit DNA damage-induced Ub-foci formation, which varies among cell lines.
Fig. 3.
The effect of proteasome inhibitors on Ub-foci formation varies among cell lines. MCF10A and MCF7 cells were incubated with or without (–) 45 μM CPT-11 or 1.8 μg/ml epirubicin for 1 h in the absence (left panels) or presence of 0.5 μM MG132 or 0.05 μM epoxomicin, as indicated. Cells were further incubated with the proteasome inhibitors for 3 h, fixed and stained with antibody to conjugated ubiquitin (upper panels). The nucleus was counterstained with DAPI (lower panels)
A proteasome inhibitor modulates chemosensitivity of cells in correlation with its effect on Ub-foci formation
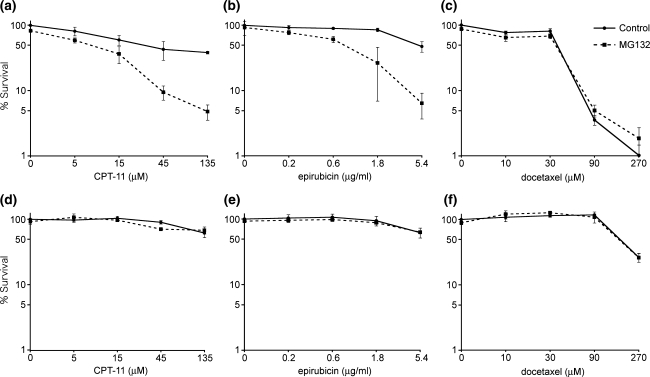
Defects in Ub-foci formation after DNA damage cause the failure to initiate proper DNA damage responses including the DNA repair process, resulting in hypersensitivity of cells to the damage. Therefore, treatment of cells with proteasome inhibitor in combination with DNA damage-inducing chemotherapeutic reagents could result in additive or synergistic effects on cytotoxicity. To test this hypothesis, MCF10A cells were incubated with CPT-11 or epirubicin for 1 h to induce DNA damage in the presence or absence of MG132 (Fig. 4a, b). Cells were further cultured with or without MG132 for 24 h and cell viability was analyzed. Without CPT-11 or epirubicin, exposure of cells to MG132 alone did not affect viability. However, MG132 treatment dramatically reduced cell viability after CPT-11- or epirubicin-induced DNA damage when compared to that without MG132. Because MG132 treatment does not cause cytotoxicity at the dosage (0.5 μM) and time length (25 h) employed in the experiments (Fig. 4 and data not shown), the result suggests that MG132 has a synergistic effect on the cytotoxicity of MCF10A cells when used in combination with CPT-11 or epirubicin. On the other hand, combination of MG132 with docetaxel, an anti-mitotic agent often used in breast cancer treatment that binds to and stabilizes microtubules but does not directly provoke DNA damage, did not show a synergistic or additive effect (Fig. 4c). Furthermore, the synergistic effect of MG132 and CPT-11 or epirubicin was not observed in MCF7 cells (Fig. 4d, e), where epirubicin-induced Ub-foci formation was retained regardless of the presence of MG132 (Fig. 3). Together, the results suggest that the proteasome inhibitor causes cells to become sensitive to DNA-damaging agents through inhibition of Ub-foci formation. Cells resistant to this effect caused by proteasome inhibitors also could be resistant to the synergistic killing effect of MG132 and DNA-damaging agents.
Fig. 4.
The effect of proteasome inhibitors on cell viability of cells treated with chemotherapeutic agents MCF10A cells (a–c) and MCF7 cells (d–f) were incubated with the indicated doses of CPT-11 (a, d), epirubicin (b, e) or docetaxel (c, f) for 1 h in the absence (control, solid lines) or presence (dashed line) of 0.5 μM MG132, as indicated. Cells were further incubated with or without MG132, and cell viability was analyzed 24 h after exposure to the chemotherapeutic agents
A proteasome inhibitor increases damaged DNA after treatment with CPT-11 or epirubicin
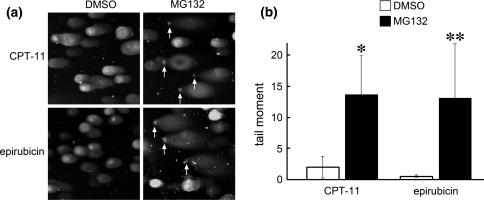
The absence of Ub-foci required for homologous recombination repair of DSB accompanied by increased chemosensitivity suggested that cells treated with MG132 were susceptible to DNA damage-inducing agents because of failure to repair DNA. To further address this possibility, we next performed single-cell electrophoresis analysis under neutral conditions (neutral comet assay) to specifically measure DSBs and to directly estimate unrepaired DNA damage in these cells. MCF10A cells were treated with CPT-11 or epirubicin for 1 h followed by incubation in the presence or absence of the proteasome inhibitor MG132. Unrepaired DNA fragments were measured 18 h post-incubation. Consistent with our hypothesis, cells treated with MG132 exhibited significantly more DNA damage than control cells (Fig. 5). The standard deviations of tail moments in MG132-treated cells were relatively large (Fig. 5b) because only a portion of cells from each sample contained fragmented DNA. This may be a result of incubating the cells with DNA damage-inducing agents for too short of a time (1 h) causing only a select portion of cells, for example cells in S-phase, to be sensitive to DNA damage. The percentage of cells that exhibited tail moments higher than a value of 5 was 32.9% for CPT-11/MG132- and 39.4% for epirubicin/MG132-treated cells. None (0%) of the solvent DMSO-treated cells incubated with either CPT-11 or epirubicin showed a value higher than 5. These results suggest that cells are unable to repair DSB caused by CPT-11 or epirubicin in the presence of a proteasome inhibitor.
Fig. 5.
Cells are deficient in repairing DSB caused by CPT-11 or epirubicin in the presence of proteasome inhibitors. (a) MCF10A cells treated with CPT-11 (135 μM) or epirubicin (5.4 μg/ml) were further incubated for 18 h in the absence (DMSO) or presence of MG132 (0.5 μM) and analyzed for the presence of DSB by the neutral comet assay. SYBR green staining of DNA shows comet tails migrating out of the nucleus. Images are representative samples. Arrows indicate the positions of remnant undamaged nuclear DNA. (b) Comet tail moments of cells from (a). Data represent the mean of 100 cells. Error bars represent standard deviation. Significantly different from control: *P = 0.021, **P = 0.035
Discussion
The UPS contributes to a wide range of cellular events and, therefore, the components of this pathway have become attractive, novel targets for therapeutic intervention. Breakthroughs have been achieved with the proteasome inhibitor bortezomib (Velcade, PS-341, Millennium) [24]. Approximately a third of relapsed, refractory multiple myeloma patients showed a significant response to bortezomib [25], and the US FDA approved bortezomib for use as a therapy for multiple myeloma. Preclinical and early clinical studies suggested bortezomib is likely to be effective in solid tumors, and clinical trials in non-hematologic malignancies including breast cancer are ongoing. Phase II studies showed that bortezomib failed to show any objective response on metastatic advanced breast cancer when used as a single agent [26, 27]. However, the effect for selected patients, such as those with tumors expressing a particular hormone receptor, HER2 status or those in earlier stages of breast cancer, remain to be determined. In addition, recent clinical data have proven the efficacy of bortezomib in combination with several chemotherapeutic agents for breast cancer treatment [28–30].
The mechanisms underlying the therapeutic effect of bortezomib have been investigated intensively. In multiple myeloma, inhibition of the transcription factor NFκB by blocking the degradation of its inhibitory partner IκB is one such putative model [31]. However, recent studies suggest that multiple factors might contribute to the efficacy of the drug. For DNA damage responses, depletion of ubiquitination at sites of DNA damage could be one of the major mechanisms leading cells to apoptosis. Proteasome inhibitors sensitize tumor cells to DNA damage-inducing chemotherapeutic agents [32, 33]. Particularly relevant for breast cancer, proteasome inhibitors sensitize myeloma cells to doxorubicin [32]. One potent mechanism for this effect is that bortezomib down-regulates the expression of effectors involved in the cellular response to DNA damage [32]. However, our result and a recent study reported from other laboratories [34, 35] suggest that there is an alternative mechanism where proteasome inhibitors perturb the ubiquitination-mediated DNA repair cascade. Proteasome inhibitors as well as depletion of 19S and 20S proteasome subunits inhibited DNA damage–signaling processes, including monoubiquitination and nuclear foci formation of FANCD2 and foci formation of phosphorylated ATM, 53BP1, NBS1, BRCA1 and RAD51 [34, 35]. The mechanisms responsible for inhibition remain unclear. Because PCNA monoubiquitination in response to DNA damage in cells treated with proteasome inhibitors was retained, it was speculated that free ubiquitin was still available to modify nuclear proteins at sites of DNA damage [34]. However, it was recently reported that K63- or K6-linked polyubiquitination at DNA damaged sites is required for the recruitment of BRCA1 and 53BP1 [8–10] and that proteasome inhibitors inhibit nuclear foci formation of conjugated ubiquitin after IR [5]. Our results show that proteasome inhibitors also inhibit foci formation after exposure to chemotherapeutic agents (Fig. 2, 3). Together, it is possible that proteasome inhibitors reduce the free ubiquitin pool in the nucleus to a level where monoubiquitination of PCNA is not affected but where polyubiquitination required for BRCA1/53BP1 recruitment is perturbed. The observation that FANCD2 foci formation was more severely affected than FANCD2 monoubiquitination in cells depleted for proteasome subunits [34] is consistent with this explanation.
Our results indicate that the effectiveness of proteasome inhibitors in terms of the cytosol trap of ubiquitin is remarkably different among cell lines. These differences influence the capacity of Ub-foci formation and chemosensitivity. It was previously reported that bortezomib has differential cellular and molecular effects in human breast cancer cells [36]. However, no relationship was observed between the effect of bortezomib on cell viability and expression of HER-2, epidermal growth factor receptor, AKT or ERK1/2 [36]. From our results, it is possible that the capacity of Ub-foci formation may cause the observed differential effects of bortezomib on cytotoxicity.
Recent studies in gene expression profiling identified specific subtypes of breast cancer with biologic and therapeutic implications. One particular subset, basal-like breast cancer expressing basal/myoepithelial cell markers such as cytokeratin 5/6, 14, 17 or Vimentin but not expressing estrogen receptor, progesterone receptor or HER2, exhibited an aggressive phenotype and a particularly poor prognosis [37–39]. Evidence from multiple sources strongly indicates that impairment of BRCA1 pathways is responsible for this phenotype [40–43]. Therefore, it is expected that this phenotype could be sensitive to DNA damage-inducing agents such as topoisomerase inhibitors, DNA cross-linkers or PARP inhibitors. Because proteasome inhibitors sensitize cells to DNA damage-inducing agents, they could affect the efficacy of these agents in the treatment of basal-like breast cancer. Alternatively, proteasome inhibitors may perturb the ubiquitin-mediated BRCA1 cascade in luminal type breast cancers, leading the DNA repair pathway to mimic that of basal-like breast cancer.
Conclusion
Treatment of cells with proteasome inhibitors trapped conjugated ubiquitin in the cytosol causing failure of Ub-foci formation and failure to repair DNA in response to DNA damage-inducing chemotherapeutic agents. The degree of this effect varied among cell lines. Cells that exhibited cytosol trapping of ubiquitin upon treatment with proteasome inhibitors were sensitized to DNA damage-inducing agents, exhibiting lower rates of cell viability. In contrast, cells not exhibiting the trapping effect were resistant to the synergy between proteasome inhibitors and DNA damage-inducing agents. Our results suggest that the synergistic effect of proteasome inhibitors in combination with DNA damage-inducing agents warrants further investigation into its effectiveness in the treatment of breast cancer.
Acknowledgments
This study was supported by grants from Japan Society for the Promotion of Science and the Japanese Ministry of Education, Culture, Sports, Science and Technology. We thank Mrs. Rie Ogawa for secretarial assistance.
Conflict of interest statement The authors declare that they have no competing interests.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ et al (2008) Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451:1116–1120 [DOI] [PMC free article] [PubMed]
- 2.Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T (2008) Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res 68:2581–2586 [DOI] [PMC free article] [PubMed]
- 3.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451:1111–1115 [DOI] [PubMed]
- 4.Wang B, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 104:20759–20763 [DOI] [PMC free article] [PubMed]
- 5.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900 [DOI] [PubMed]
- 6.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM et al (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637–1640 [DOI] [PMC free article] [PubMed]
- 7.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914 [DOI] [PMC free article] [PubMed]
- 8.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316:1194–1198 [DOI] [PMC free article] [PubMed]
- 9.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316:1198–1202 [DOI] [PMC free article] [PubMed]
- 10.Kim H, Chen J, Yu X (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202–1205 [DOI] [PubMed]
- 11.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276:14537–14540 [DOI] [PubMed]
- 12.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM (2006) Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev 20:34–46 [DOI] [PMC free article] [PubMed]
- 13.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K et al (2007) A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell 25:663–675 [DOI] [PubMed]
- 14.Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479 [DOI] [PubMed]
- 15.Pickart CM (2001) Ubiquitin enters the new millennium. Mol Cell 8:499–504 [DOI] [PubMed]
- 16.Tanaka K, Chiba T (1998) The proteasome: a protein-destroying machine. Genes Cells 3:499–510 [DOI] [PubMed]
- 17.Morris JR, Solomon E (2004) BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13:807–817 [DOI] [PubMed]
- 18.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol 173:19–26 [DOI] [PMC free article] [PubMed]
- 19.Sato K, Hayami R, Wu W, Nishikawa T, Nishikawa H, Okuda Y, Ogata H, Fukuda M, Ohta T (2004) Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J Biol Chem 279:30919–30922 [DOI] [PubMed]
- 20.Hanada M, Sugawara K, Kaneta K, Toda S, Nishiyama Y, Tomita K, Yamamoto H, Konishi M, Oki T (1992) Epoxomicin, a new antitumor agent of microbial origin. J Antibiot (Tokyo) 45:1746–1752 [DOI] [PubMed]
- 21.Sin N, Kim KB, Elofsson M, Meng L, Auth H, Kwok BH, Crews CM (1999) Total synthesis of the potent proteasome inhibitor epoxomicin: a useful tool for understanding proteasome biology. Bioorg Med Chem Lett 9:2283–2288 [DOI] [PubMed]
- 22.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA 96:10403–10408 [DOI] [PMC free article] [PubMed]
- 23.Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP (2000) Mechanism of action of camptothecin. Ann N Y Acad Sci 922:1–10 [DOI] [PubMed]
- 24.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC (2001) The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res 61:3071–3076 [PubMed]
- 25.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, Rajkumar SV, Srkalovic G, Alsina M, Alexanian R et al (2003) A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med 348:2609–2617 [DOI] [PubMed]
- 26.Engel RH, Brown JA, Von Roenn JH, O’Regan RM, Bergan R, Badve S, Rademaker A, Gradishar WJ (2007) A phase II study of single agent bortezomib in patients with metastatic breast cancer: a single institution experience. Cancer Invest 25:733–737 [DOI] [PubMed]
- 27.Yang CH, Gonzalez-Angulo AM, Reuben JM, Booser DJ, Pusztai L, Krishnamurthy S, Esseltine D, Stec J, Broglio KR, Islam R et al (2006) Bortezomib (VELCADE) in metastatic breast cancer: pharmacodynamics, biological effects, and prediction of clinical benefits. Ann Oncol 17:813–817 [DOI] [PubMed]
- 28.Awada A, Albanell J, Canney PA, Dirix LY, Gil T, Cardoso F, Gascon P, Piccart MJ, Baselga J (2008) Bortezomib/docetaxel combination therapy in patients with anthracycline-pretreated advanced/metastatic breast cancer: a phase I/II dose-escalation study. Br J Cancer 98:1500–1507 [DOI] [PMC free article] [PubMed]
- 29.Dees EC, O’Neil BH, Lindley CM, Collichio F, Carey LA, Collins J, Riordan WJ, Ivanova A, Esseltine D, Orlowski RZ (2008) A phase I and pharmacologic study of the combination of bortezomib and pegylated liposomal doxorubicin in patients with refractory solid tumors. Cancer Chemother Pharmacol [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 30.Schmid P, Kuhnhardt D, Kiewe P, Lehenbauer-Dehm S, Schippinger W, Greil R, Lange W, Preiss J, Niederle N, Brossart P et al (2008) A phase I/II study of bortezomib and capecitabine in patients with metastatic breast cancer previously treated with taxanes and/or anthracyclines. Ann Oncol 19:871–876 [DOI] [PubMed]
- 31.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, Munshi N, Dang L, Castro A, Palombella V et al (2002) NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem 277:16639–16647 [DOI] [PubMed]
- 32.Mitsiades N, Mitsiades CS, Richardson PG, Poulaki V, Tai YT, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M et al (2003) The proteasome inhibitor PS-341 potentiates sensitivity of multiple myeloma cells to conventional chemotherapeutic agents: therapeutic applications. Blood 101:2377–2380 [DOI] [PubMed]
- 33.Yunmbam MK, Li QQ, Mimnaugh EG, Kayastha GL, Yu JJ, Jones LN, Neckers L, Reed E (2001) Effect of the proteasome inhibitor ALLnL on cisplatin sensitivity in human ovarian tumor cells. Int J Oncol 19:741–748 [DOI] [PubMed]
- 34.Jacquemont C, Taniguchi T (2007) Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res 67:7395–7405 [DOI] [PubMed]
- 35.Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H et al (2007) Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res 67:8536–8543 [DOI] [PubMed]
- 36.Codony-Servat J, Tapia MA, Bosch M, Oliva C, Domingo-Domenech J, Mellado B, Rolfe M, Ross JS, Gascon P, Rovira A et al (2006) Differential cellular and molecular effects of bortezomib, a proteasome inhibitor, in human breast cancer cells. Mol Cancer Ther 5:665–675 [DOI] [PubMed]
- 37.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA et al (2000) Molecular portraits of human breast tumours. Nature 406:747–752 [DOI] [PubMed]
- 38.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100:8418–8423 [DOI] [PMC free article] [PubMed]
- 39.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L et al (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10:5367–5374 [DOI] [PubMed]
- 40.Turner N, Tutt A, Ashworth A (2004) Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4:814–819 [DOI] [PubMed]
- 41.McCarthy A, Savage K, Gabriel A, Naceur C, Reis-Filho JS, Ashworth A (2007) A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol 211:389–398 [DOI] [PubMed]
- 42.Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, Kerkhoven RM, van Vliet MH, Wessels LF, Peterse JL et al (2007) Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA 104:12111–12116 [DOI] [PMC free article] [PubMed]
- 43.Shakya R, Szabolcs M, McCarthy E, Ospina E, Basso K, Nandula S, Murty V, Baer R, Ludwig T (2008) The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc Natl Acad Sci USA 105(19):7040–7045 [DOI] [PMC free article] [PubMed]