Abstract
During the annual cycle, oysters are exposed to seasonal slow changes in temperature, but during emersion at low tide on sunny summer days, their internal temperature may rise rapidly, resulting in acute heat stress. We experimentally exposed oysters to a 1-h acute thermal stress and investigated the transcriptional expression level of some genes involved in cell stress defence mechanisms, including chaperone proteins (heat shock proteins Hsp70, Hsp72 and Hsp90 (HSP)), regulation of oxidative stress (Cu-Zn superoxide dismutase, metallothionein (MT)), cell detoxification (glutathione S-transferase sigma, cytochrome P450 and multidrug resistance (MDR1)) and regulation of the cell cycle (p53). Gene mRNA levels were quantified by reverse transcription-quantitative polymerase chain reaction and expressed as their ratio to actin mRNA, used as a reference. Of the nine genes studied, HSP, MT and MDR1 mRNA levels increased in response to thermal stress. We compared the responses of oysters exposed to acute heat shock in summer and winter and observed differences in terms of magnitude and kinetics. A larger increase was observed in September, with recovery within 48 h, whereas in March, the increase was smaller and lasted more than 2 days. The results were also compared with data obtained from the natural environment. Though the functional molecule is the protein and information at the mRNA level only has limitations, the potential use of mRNAs coding for cell stress defence proteins as early sensitive biomarkers is discussed.
Keywords: Gene expression, Oyster, Seasonal variations, Thermal stress
Introduction
As a marine poikilotherm bivalve, the oyster is exposed to large changes in body temperature. One reason is the annual seasonal cycle which results in differences of more than 10°C between the lowest and highest seawater temperatures along the French coasts. Seasonal cyclic changes are slow, and they are well known to influence oyster physiology (reproduction, growth). But seawater temperature is not the only parameter changing during the seasonal cycle, and the relationship between the temperature and oyster physiology is complex. Because they live attached to rocks on the foreshore or in pouches when being raised in oyster farms, oysters are subjected to tidal emersion/immersion cycles. During emersion, their internal temperature reaches that of the atmosphere, which may vary over a far greater range than that of seawater (Hamdoun et al. 2003). This study showed that, at low tide on sunny summer days, midday sunlight raises oyster body temperature to about 35°C, resulting in acute thermal stress.
Molecular cell stress markers are used to an increasing extent in ecotoxicology, with the aim of detecting the effects of the presence of contaminants in the environment. However, their use is hindered by several difficulties: they may show a basal expression level in non-contaminated conditions, and this level may be not constant; an increased level of molecular markers does not necessarily indicate a physiological degradation; they are generally not specific because all types of environmental stress induce complex patterns of cellular damage, some of which are similar to each other. Cells react by up-regulating categories of defence functions including detoxification of xenobiotics, oxidative stress regulation, management of denatured molecules by chaperone proteins, DNA damage repair machinery and, if DNA repair fails, regulation of the cell cycle. Despite these difficulties, it is clear that significant increases in molecular marker levels indicate that cells react to a stimulus (internal or external) and, interpreted with caution, may be considered as very early and sensitive indicators of stressing conditions.
A prerequisite to using molecular cell stress markers in the study of oyster response to stress is to determine their natural range of expression levels, including inter-individual variability as well as possible seasonal changes. The transcription of several cell stress marker genes was previously monitored in oyster during the annual cycle (Farcy et al. 2007). This study showed that the mRNA levels exhibited a seasonal pattern with maximal observed values generally coinciding with lowest seawater temperature during winter and, for some genes, a secondary peak during summer. The minimal values were generally observed in September. On the basis of this large preliminary dataset acquired under environmental conditions, these genes can be used as molecular parameters to test the response of oysters to various experimental conditions. However, the seasonal pattern of the baseline expression of these genes raises the question of whether their capacity to respond to stress is the same in winter and in summer. In this study, we exposed Pacific oysters (Crassostrea gigas) to acute thermal stress in these two extreme periods of the annual cycle. In the natural environment, at low tide on sunny summer days, oyster body temperature may rise to more than 35°C for an hour or so (Hamdoun et al. 2003). In view of this result, Pacific oysters were exposed to a heat stress of 1 h at 37°C. This acute and intense non-lethal stress was also designed as a positive control to determine how these cell stress marker genes might respond to experimental stress conditions. In order to examine different mechanisms of cell stress defence and based on available DNA sequence information for C. gigas, the expression levels of the following genes were monitored by mRNA quantification: heat shock chaperone proteins (HSP); superoxide dismutase (SOD) and metallothionein (MT), which are both involved in the regulation of oxidative stress (although MT proteins are better known for their role in metal metabolism); glutathione S-transferase (GST), cytochrome P450 and multidrug resistance (MDR), which take part in cell detoxification of organic compounds; and p53, a member of the cell cycle regulation machinery.
Materials and methods
Animals and protocol
The experiments were performed at the two extreme periods of the seasonal cycle, in September 2005 and in March 2006. In the English Channel, minimal and maximal seawater temperatures are observed in March and September, respectively (see Farcy et al. 2007 for a detailed seawater temperature monitoring profile). Oysters were obtained from a local oyster farm (Normandy, France); all oysters used were diploid, 3–4 years old and had spent their entire growing life at the farm. They ranged from 66 to 85 g in total weight (including intervalval fluid). Oysters were shucked, the shell was discarded and all oyster soft tissues were kept, including attached mantle, visceral mass, gills and attached adductor muscle.
In order to match laboratory control values with environmental data in March and in September, a first batch was processed on the spot on removal from the natural medium. The gills of 20 oysters and the whole soft tissues of another 20 oysters were dissected and immediately frozen in liquid nitrogen.
A second batch was acclimated to laboratory condition for 1 week in 120-l tanks (≤36 oysters per tank) in fully aerated seawater at the same temperature as in the environment: 7°C in March and 18°C in September. Seawater was changed every day for 3 days, then every other day up to the end of the experiment. Along the French coast of the English Channel, low tide lasts about 6.5 h, and oyster tables are placed in the medium level of the intertidal zone so they are only partly emerged, depending on the tide coefficient. Oysters may be exposed to more than 1 h of heat shock, but we applied a 1-h stress on the basis of the study by Hamdoun et al. (2003) in order to ensure non-lethal conditions. After the laboratory acclimation time, the oysters to be stressed were transferred to another 120-l tank containing seawater at 37°C for 1 h and then returned to the acclimation tank. Unstressed control oysters were kept in the acclimation tank. We observed that the oysters stayed closed during immersion at 37°C, and no mortality occurred up to 3 days after the heat shock. Control and heat-stressed oysters were dissected at 0, 3, 8, 24 or 48 h after the end of the heat shock. For each measurement, the gills and the remaining soft tissues of the oysters were dissected and immediately frozen in liquid nitrogen.
All frozen tissues were crushed in a ball crusher (Verder MM301) in buckets cooled by liquid nitrogen. The tissues from the 40 oysters from the natural medium were pooled to give eight samples each including tissue from five individuals (four pools of five gills and four pools of five whole tissues), while the tissues from the oysters exposed to thermal stress in the laboratory and their controls were processed individually (six oysters at each time point).
RNA extraction and cDNA synthesis
Total RNA was extracted with TRI Reagent (Sigma-Aldrich) according to the manufacturer’s instructions. The amount and quality of RNA were estimated by measuring absorbance at 260 and 280 nm in a UV spectrophotometer. Total RNA was then digested with DNase I (Amplification Grade, Sigma-Aldrich) to remove genomic DNA, and an aliquot of DNase-treated RNA was then tested by polymerase chain reaction (PCR) with actin primers to check the absence of genomic DNA. DNase-treated total RNA (500 ng) was then reverse-transcribed using 500 ng random primers (Promega), 200 U Moloney murine leukaemia virus reverse transcriptase (Promega), 12.5 μmol RNase-free dNTP and 25 U recombinant RNasin (Promega).
Gene-specific mRNA quantification
The sequences of the forward and reverse primers used in real-time PCR were designed with the aid of Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and synthesized by Eurogentec. All sequences were specific for Crassostrea gigas, and GenBank accession numbers are given in Table 1. The primers were designed to have the following characteristics: primer length from 18 to 20 bp, Tm from 59°C to 61°C and GC% from 40% to 60%. The sequences of the primers are given in Table 1 with the length of each expected amplicon. The efficiency of each pair of primers was tested using the standard curve method. For this purpose, a range of dilutions of the total amount of starting RNA (reverse-transcribed) was prepared, and the cycle threshold was plotted as a function of the log of starting RNA concentration. Primer pairs showing a good efficiency level (100 ± 5%) over a four orders of magnitude range were kept for quantification. Real-time PCR was performed in a MyiQ™ Cycler (Biorad). Amplification was carried out on 96-well plates, in a total reaction volume of 15 μl containing 7.5 μl of 2X iQ SYBR® Green supermix (Biorad), each primer (500 nM final) and cDNA samples obtained from reverse transcription of 5 ng of DNase-treated total RNA (except for the quantification of 18S RNA, where 1,000-fold diluted cDNA samples were used). Amplification conditions were 40 cycles of 15 s at 95°C and 45 s at 60°C, followed by the protocol for the melting curve consisting of 80 cycles of 10 s with an increase of 0.5°C between each cycle from 55°C to 95°C. The specificity of the reaction was confirmed by observing a single peak at the expected Tm on the melting curve analysis and by sequencing the PCR product once. All determinations were carried out in duplicate. Controls without template cDNA were included on PCR plates.
Table 1.
GenBank accession numbers of cDNA sequences from C. gigas used to design the primers
| Gene | GenBank accession # | Forward primer | Reverse primer | Length of the amplicon (pb) |
|---|---|---|---|---|
| Actina | AF026063 | 5′ GCCCTGGACTTCGAACAA 3′ | 5′ CGTTGCCAATGGTGATGA 3′ | 100 |
| GAPDHa | AJ544886 | 5′ TTGTCTTGCCCCTCTTGC 3′ | 5′ CGCCAATCCTTGTTGCTT 3′ | 126 |
| 18Sa | AB064942 | 5′ CGGGGAGGTAGTGAC GAA 3′ | 5′ ACCAGACTTGCCCTCCAA 3′ | 110 |
| Hsp70 | AJ305315 | 5′ AGCAAGCCAGCACAGCA 3′ | 5′ GCGATGATTTCCACCTTC 3′ | 92 |
| Hsp72 | AF144646 | 5′ GAGGATCGCAGCCAAGAA 3′ | 5′ TATCGCCCTCGCTGATCT 3′ | 98 |
| Hsp90 | AJ431681 | 5′ GGAGAGCAAAACCCTCACC 3′ | 5′ TGGCAATGGTTCCAAGGT 3′ | 83 |
| Cu-Zn SOD | AJ496219 | 5′ AACCCCTTCAACAAAGAGCA 3′ | 5′ TTTGGCGACACCGTCTTC 3′ | 96 |
| MTb | AJ243263 AJ242657 | 5′ GGACCGGAAAACTGCAAA 3′ | 5′ CCAGTGCATCCTTTACCACA 3′ | 98 |
| GST sigma | AJ557140 | 5′ AACGCCACCATTCACGAC 3′ | 5′ AAGACCCCACCCAATGCT 3′ | 118 |
| CYP2E1 | AF075692 | 5′ CCCTGGGAGTTCAAACCTG 3′ | 5′ CGACGCCAAATCCAATAAA 3′ | 94 |
| MDR1 | AJ422120 | 5′ CCGAGAACATCCGCTACG 3′ | 5′ GCCCTGTGGGAGTTCCTT 3′ | 104 |
| p53 | AM236465 | 5′ ACCCAGCTCCGACTCATTT 3′ | 5′ TCATGGGGGATGATGACAC 3′ | 97 |
Gene-specific mRNA quantification was performed by normalizing data to the amount of mRNA encoding for actin, a cytoskeleton protein taken as a housekeeping gene (Farcy et al. 2007). For each analysed specific DNA, real-time PCR provides a cycle threshold (Ct) value where the fluorescence signal is detectable above the background. This Ct value is reproducibly and accurately dependent on the starting amount of cDNA. An actin Ct value in the range [18.5–20.1] was used to confirm that the starting amount of cDNA was reproducible in all PCR quantifications. Quantification of specific stress gene mRNA (reverse-transcribed to cDNA) was performed using the Delta Cycle threshold method: gene of interest normalized mRNA 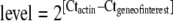 . This calculates the amount of mRNA of interest as its ratio to the amount of actin mRNA, so it has no unit. We also tested glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and 18S rRNA as reference RNAs. GAPDH is another commonly used reference mRNA, while 18S rRNA accounts for total RNA. The reported data are mean and standard deviation values from four pools of five oysters and six individual oysters for field and laboratory experiments, respectively. Statistical analysis was carried out using Statgraphics Centurion XV (Statpoint, Inc.) software. The hypothesis of normality was tested and accepted by a Shapiro Wilk’s W test. The hypothesis of homogeneity of variances was accepted using a Bartlett test. Heat-shocked and control oysters were compared using a t test (n = 6), and the significance of the difference was given as *p < 0.05 or **p < 0.01.
. This calculates the amount of mRNA of interest as its ratio to the amount of actin mRNA, so it has no unit. We also tested glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and 18S rRNA as reference RNAs. GAPDH is another commonly used reference mRNA, while 18S rRNA accounts for total RNA. The reported data are mean and standard deviation values from four pools of five oysters and six individual oysters for field and laboratory experiments, respectively. Statistical analysis was carried out using Statgraphics Centurion XV (Statpoint, Inc.) software. The hypothesis of normality was tested and accepted by a Shapiro Wilk’s W test. The hypothesis of homogeneity of variances was accepted using a Bartlett test. Heat-shocked and control oysters were compared using a t test (n = 6), and the significance of the difference was given as *p < 0.05 or **p < 0.01.
Results
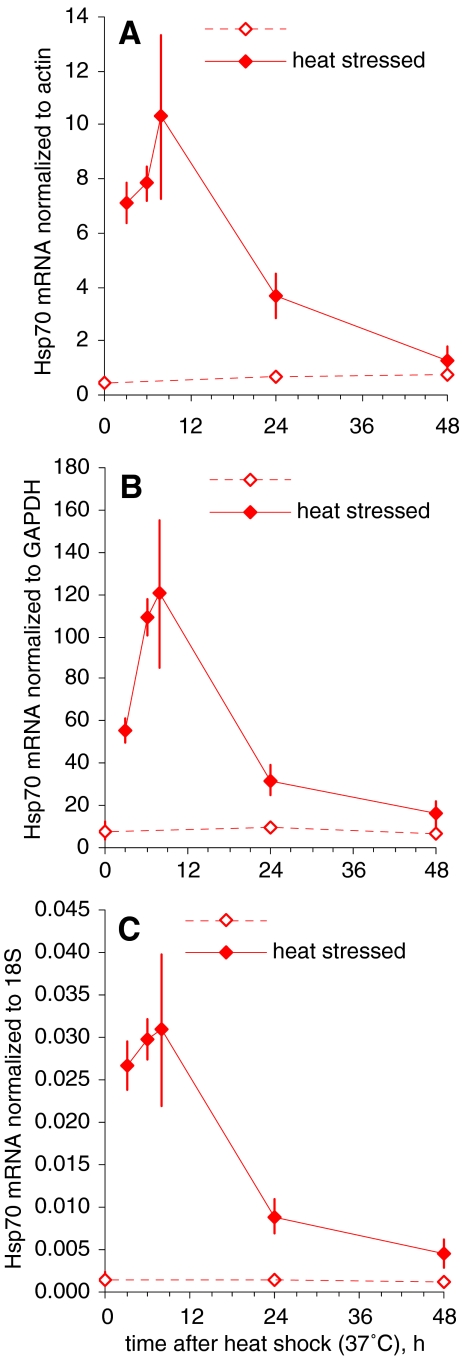
The expression levels of nine genes (hsp70, hsp72, hsp90, Cu-Zn SOD, MT, GST sigma, CYP2E1, MDR1 and p53) potentially involved in cell stress defence were calculated as the ratio of the amount of their mRNA to that of actin, used as a reference mRNA. The two other commonly used reference RNAs were also analysed in parallel. As an illustrative example, Fig. 1 shows the results obtained with actin mRNA, GAPDH mRNA and 18S rRNA as references for hsp70 mRNA levels in gills, measured in the September 2005 experiment. A similar pattern was observed with all three reference RNAs. This was confirmed for other studied genes, in the homogenates of the other soft tissues and in March 2006 (data not shown). All subsequent results presented below are expressed as normalized to actin mRNA.
Fig. 1.
An illustrative example of mRNA normalization to actin mRNA (a), GAPDH mRNA (b) and 18S rRNA (c) as a reference: Hsp70 mRNA levels in gills of oysters exposed to 1-h heat stress at 37°C in September (mean and standard deviation, n = 6)
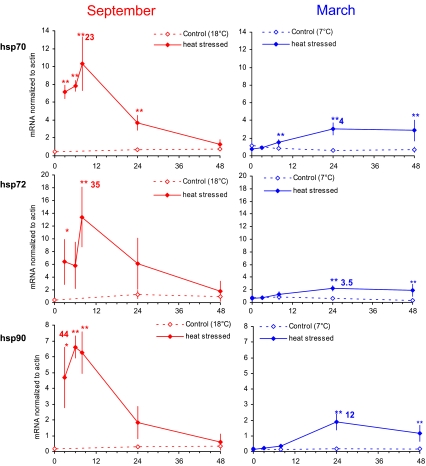
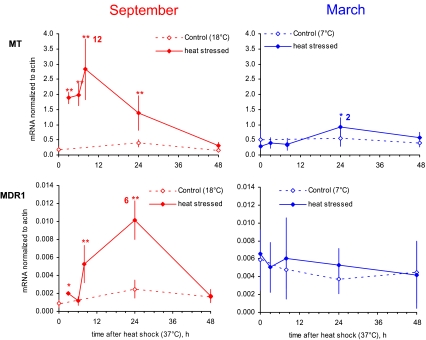
Data for gills as a function of time after exposure to 1 h at 37°C in March and in September are presented in Figs. 2 and 3. No significant change was observed in response to acute thermal stress for genes encoding Cu-Zn SOD, GST sigma, CYP2E1 or p53 (data not presented). Data obtained from the homogenates of the remaining soft tissues were found to parallel those from gills (data not shown). The expression patterns of five out of the nine studied genes in the gills of oysters exposed to thermal stress in September (left) or in March (right) were clearly different in terms of magnitude and kinetics. A higher increase was observed in summer oysters with recovery within 2 days after the heat stress. The increase in winter oysters was lower by an order of magnitude and lasted at least 2 days. In winter oysters, the increase in MDR1 expression level in response to thermal stress was not significant, whereas it was in summer.
Fig. 2.
Levels of hsp70, hsp72 and hsp90 mRNA in oysters exposed to 1-h heat stress at 37°C (mean and standard deviation, n = 6). Experiments were carried out in September (left panel) and in March (right panel)
Fig. 3.
Levels of MT and MDR1 mRNA in oysters exposed to 1-h heat stress at 37°C (mean and standard deviation, n = 6). Experiments were carried out in September (left panel) and in March (right panel)
Data were obtained from the natural environment in order to determine the range of variation between the coldest and warmest seawater seasons. It was also crucial to check whether maintaining the oysters in the laboratory resulted in any additional stress that could interfere with the artificial heat shock. For this purpose, we needed to estimate average levels, but we expected quite high variability (as is usually observed when dealing with shellfish from the natural medium), so we needed a high number of replicates. Pooling five individuals per sample allowed average levels to be obtained from 20 oysters with only four measurements. This procedure reduces the magnitude of the apparent variability and precludes any statistical comparison test between datasets from pools and individuals. In order to compare data from oysters collected in the environment and oysters acclimated to laboratory conditions, the ranges of expression levels of genes in the gills are summarized in Table 2. For the two periods (March and September), three columns are presented: the min–max range of environment values; the min–max range of laboratory control values; and the mean peak values in gills of oysters exposed to heat stress, with the ratio between the heat stress peak value and the control value in parentheses. It must be emphasized that values from the environment correspond to pools of five oysters, while values from the laboratory correspond to individual oysters. However, the comparison of min–max ranges from the natural medium and from laboratory control conditions did not give any reason to suspect additional stress resulting from the laboratory itself. Conversely, a very large increase was observed in response to heat shock in the laboratory (except for MDR1 in March), and the observed stress values were very clearly outside the ranges of the data from the natural medium.
Table 2.
Comparison of gene expression levels in the environment (n = 4 pools of five oysters) and in the laboratory (n = 6 oysters)
| Gene | March | September | ||||
|---|---|---|---|---|---|---|
| Environment min–max | Lab control min–max | Lab Heat Stress peak | Environment min–max | Lab control min–max | Lab Heat Stress peak | |
| Hsp70 | 0.55–1.27 | 0.72–1.13 | 3.1 (4) | 0.23–0.38 | 0.45–0.76 | 10.3 (23) |
| Hsp72 | 0.38–1.62 | 0.36–0.84 | 2.3 (3.5) | 0.25–0.49 | 0.36–1.26 | 13.4 (35) |
| Hsp90 | 0.20–0.57 | 0.14–0.18 | 1.9 (12) | 0.03–0.20 | 0.15–0.32 | 6.6 (44) |
| MT | 0.55–1.20 | 0.32–0.55 | 0.92 (1.7) | 0.13–0.24 | 0.16–0.39 | 2.8 (12) |
| MDR1 | 0.006–0.027 | 0.006–0.008 | 0.007 n.s. | 0.001–0.004 | 0.001–0.003 | 0.010 (6) |
For the two periods (March and September), three columns are presented: the min–max range of environment values; the min–max range of laboratory control values; the mean peak values in gills of oysters exposed to heat stress, with the ratio between the heat stress peak value and the control value in parentheses
n.s. no significant difference
Discussion
In the oyster, gills are not only involved in gas and ion exchanges but also constitute the filtering organ which collects suspended matter for feeding. As a primarily exposed organ, it is an early sensor of chemical environmental changes, and thus, it was assumed to be also sensitive to thermal stress. It is easy to isolate the gills from the visceral mass. Moreover, the data obtained from gills and from the remaining soft parts were similar, confirming that gills may be a convenient target to study gene expression in response to environmental stress in the oyster.
Before using biomarkers for environmental monitoring or for laboratory experiments, it is crucial to control or at least be aware of their natural and/or seasonal variability. The comparison of molecular cell stress markers measured in laboratory conditions and in the natural environment provided crucial information for the interpretation of the response to thermal stress. Firstly, the levels observed in laboratory control conditions were similar to those observed at the oyster farm, indicating that acclimation of the animals did not induce any additional stress due to handling in the laboratory. Secondly, the ranges of variation in field data, which were documented in greater detail in our previous study regarding seasonal expression patterns (Farcy et al. 2007), provided a valuable basis for assessing the significance of observed changes in response to experimental exposure to stress. In the present work, heat stress values were clearly outside the ranges of control environmental and laboratory values.
The present study was intended to monitor the expression levels of genes encoding components of cell stress defence mechanisms in oysters exposed to thermal stress. We wanted to screen the main categories of cellular functions involved in stress response using a limited number of genes. The genes addressed in the present work are involved in detoxification of organic compounds, oxidative stress regulation, management of denatured molecules by chaperone proteins and regulation of the cell cycle. Most data available in the literature regarding molecular markers in molluscs were obtained at the protein level. Here we investigated potential markers at the transcriptional level with the aim of detecting early sensitive changes. Five out of the nine tested genes were found to be clearly up-regulated in response to acute thermal stress; they belonged to different categories of cellular stress defence functions.
For some 40 years, it has been known that exposure of organisms or cells to elevated temperature induces the synthesis of very well-conserved proteins called heat shock proteins. These chaperone proteins guide renaturation of misfolded or partly denatured proteins resulting from thermal stress (see Feder and Hofmann 1999 and Kregel 2002 for reviews).
We focused on transcriptional gene expression measurements for several reasons. A response of heat shock proteins to thermal stress has already been demonstrated in the oyster (Shamseldin et al. 1997; Clegg et al. 1998; Hamdoun et al. 2003). Analysis by semi-quantitative methods at the mRNA level by Northern blot or at the protein level by Western blot clearly demonstrated complex regulation. Furthermore, more recent evidence showed that HSPs are involved in the cell response to most environmental and physiological stressors (reviewed by Wegele et al. 2004). This includes studies in molluscs (Cruz-Rodriguez et al. 2000; Snyder et al. 2001; Boutet et al. 2003; Piano et al. 2004). Here again, semi-quantitative methods were used to obtain evidence for an increase in various stressing conditions. In order to better understand the responses of the multiple HSP family members to cell stress, it is useful to analyse these markers with the most specific and accurate quantification methods available. Transcriptional gene expression analysis by qPCR meets these requirements, since it can discriminate isoforms on the basis of cDNA sequences and provide reproducible and accurate quantitative information. However, it is clear that the functional molecule is the protein, and information at the mRNA level only has limitations.
In our study, genes from the HSP family showed the strongest response, with mRNA level changes by up to 40-fold in oysters exposed to acute heat stress. Gene expression of the HSP class of proteins involves a very well-conserved sequence, the heat shock element (HSE), as a promoter. Induction of the gene starts with the binding of a cytoplasmic protein, heat shock factor (HSF), to the HSE. Mainly present as inactive cytosolic monomers in the absence of stress, HSF forms multimers in response to cytotoxic agents or heat shock, migrates to the nucleus and binds to the HSE (see Morimoto et al. 1992 for a review). It is noteworthy that HSF can also directly or indirectly activate the promoters of MT genes (Sorger 1991; Silar et al. 1991) as well as the MDR1 gene (Chin et al. 1990; Tchénio et al. 2006). HSP induction observed in our experiment is likely to be related to protein denaturation by heat, but it may also result from oxidative stress generated by the substantial temperature elevation.
An increase in MT protein following heat stress was previously reported by Piano et al. (2004) in the European flat oyster exposed for 1 h at 35°C. In the absence of metal contamination, our observations, at the transcriptional level, seem to confirm the possible interaction of HSF with the antioxidant responsive elements and the metal responsive elements in the promoter region of MT genes, as suggested by Dalton et al. (1996).
An increase in the activity of p-glycoprotein, a product of the MDR gene, in mussels exposed to thermal stress was reported by several authors (Eufemia and Epel 2000; Luedeking and Koehler 2004; Minier et al. 2000). Our data are the first report of MDR induction at the transcriptional level in molluscs. However, it should be pointed out that the increase of MDR transcript observed in September was not statistically significant in March, showing that this response may be influenced by the season.
In summary, HSF is known to activate the transcription of HSP, MT and MDR genes. Protein interactions were previously reported between p53 and MDR1 (Li et al. 1997) or p53 and Hsp70 (Fourie et al. 1997; Sturzbecher et al. 1987). All these observations suggest that the different cellular functions involved in the response to thermal stress (chaperone proteins, oxidative stress, detoxification of organic compounds and cell cycle regulation) are closely interconnected.
In response to acute heat stress, Pacific oysters exhibited different results in March and in September in terms of both magnitude and kinetics. The increase in the mRNA level of the studied genes, presented in Figs. 2 and 3, was much higher in September than in March. Conversely, it must be pointed out that the magnitude of the temperature shift was greater in March (from 7°C to 37°C) than in September (from 18°C to 37°C). Thus, the mRNA levels did not correlate with the magnitude of the change in seawater temperature, and there was definitely a seasonal effect on the experiment. The recovery from heat stress was also different. In September, mRNA levels returned to control values 48 h after stress, while in March, mRNA levels were still elevated 48 h after the heat shock. In the environment, because of natural seasonal variations, higher levels of mRNA encoding the studied genes were observed in oysters collected in March–April, compared to September (Farcy et al. 2007). Higher HSP70 protein concentrations were also observed in horse mussels during winter compared to summer (Lesser and Kruse 2004). Our laboratory results reported here showed that the response to heat stress was stronger and quicker in September than in March. In the absence of information on protein concentrations, it is difficult to interpret the significance of these kinetics at the transcriptional level. Nevertheless, the different responses of oysters to heat stress in March and September may be assumed to be related to some adaptation mechanism. In the natural medium, oysters experience higher temperatures and stronger thermal stress in summer than in winter. It was previously shown that a moderate elevation of temperature promotes the induction of members of the HSP gene family, which makes cells more resistant to any further challenge with heat stress (Li and Werb 1982). This was also investigated in the oyster (Shamseldin et al. 1997; Clegg et al. 1998; Hamdoun et al. 2003). This thermoresistant status (or acquired thermotolerance as described by Kregel 2002) not only protects the cells against heat (Mosser and Martin 1992; Mosser et al. 1997; Gabai et al. 1997) but also against many kinds of stress such as chemotherapeutic agents (Samali and Cotter 1996; Jaattela 1999), nitric oxide (Bellmann et al. 1996), UV (Simon et al. 1995) and irradiation (Park et al. 2000; Kang et al. 2002). The protective effects of HSP against free oxygen radicals are thought to explain their effectiveness against these different sources of cellular stress conditions. Nevertheless, the possible relationship between the different kinetics of HSP mRNA induction in oysters exposed to heat stress and seasonal thermoadaptation deserves further investigation. Moreover, it should be kept in mind that temperature is not the only parameter changing with the seasons and suspected to be responsible for stress in oysters. Other parameters include environmental factors such as trophic conditions, oxygen and nature of sediment (Delaporte et al. 2003; Soletchnik et al. 2005) as well as physiological and genetic factors such as reproduction, oxidative stress and parasitism (Le Roux et al. 2002; Huvet et al. 2004; Costil et al. 2005).
In conclusion, our study suggests that quantification of the expression of certain cell stress genes at the transcriptional level may be used to obtain sensitive indicators of oyster environmental stress, at least under acute exposure conditions. However, possible natural seasonal patterns of the transcriptional expression level of these genes may interfere considerably and should be investigated prior to their use in laboratory experiments. Moreover, the question arises of how the observed changes at the transcriptional level quantitatively affect the function of proteins encoded by these genes. Information at the transcriptional level only may be a limitation. Measurements of the amounts as well as the activities of the proteins, with the same specificity and quantitative accuracy as mRNA quantification, should be ideally carried out in parallel. Lastly, the responses at the cellular and molecular levels may correspond to routine, adaptative or detrimental situations for the cells, and their consequences should also be further investigated at the integrated level to determine whether they reflect a harmful effect on the health of individuals and/or populations.
Acknowledgements
Emilie FARCY was supported by fellowships from the Institute of Radioprotection and Nuclear Safety (IRSN) and “Région Basse-Normandie”.
Footnotes
Concise summary
We addressed a set of genes involved in cell stress defence mechanisms as biomarkers to investigate the response of the Pacific oyster to a one-hour acute heat shock in the laboratory. The transcriptional levels of the genes could be used as early sensitive biomarkers, provided their ranges of natural variability and potential seasonal cycles were taken into account. Different responses were observed depending on the season when the experiments were performed.
References
- Bellmann K, Jaattela M, Wissing D, Burkart V, Kolb H. Heat shock protein hsp70 overexpression confers resistance against nitric oxide. FEBS Lett. 1996;391:185–188. doi: 10.1016/0014-5793(96)00730-2. [DOI] [PubMed] [Google Scholar]
- Boutet I, Tanguy A, Rousseau S, Auffret M, Moraga D. Molecular identification and expression of heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70) genes in the Pacific oyster Crassostrea gigas. Cell Stress Chaperones. 2003;8:76–85. doi: 10.1379/1466-1268(2003)8<76:MIAEOH>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K-V, Tanaka S, Darlington G, Pastan I, Gottesman MM. Heat shock and arsenite increase expression of the multidrug resistance (MDR1) gene in human renal carcinoma cells. J Biol Chem. 1990;265:221–226. [PubMed] [Google Scholar]
- Clegg JS, Cher GN, Rifkin E, Friedman CS. Induced thermotolerance and the heat shock protein-70 family in the Pacific oyster Crassostrea gigas. Mol Mar Biol Biotechnol. 1998;7:21–30. [Google Scholar]
- Costil K, Royer J, Ropert M, Soletchnik P, Mathieu M. Spatio-temporal variations in biological performances and summer mortality of the Pacific oyster Crassostrea gigas in Normandy (France) Helgoland Marine Research. 2005;59:286–300. doi: 10.1007/s10152-005-0004-5. [DOI] [Google Scholar]
- Cruz-Rodríguez L, Baucum A, Soudant P, Chu FLE, Hale R. Effects of PCBs sorbed to algal paste and sediments on the stress protein response (HSP70 family) in the eastern oyster, Crassostrea virginica. Mar Environ Res. 2000;50:341–345. doi: 10.1016/S0141-1136(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Dalton TP, Li Q, Bittel D, Liang L, Andrews GK. Oxidative stress activates metal-responsive transcription factor-1 binding activity. J Biol Chem. 1996;271:26233–26241. doi: 10.1074/jbc.271.42.26233. [DOI] [PubMed] [Google Scholar]
- Delaporte M, Soudant P, Moal J, Lambert C, Quere C, Miner P, Choquet G, Paillard C, Samain JF. Effect of a mono-specific algal diet on immune functions in two bivalve species—Crassostrea gigas and Ruditapes philippinarum. J Exp Biol. 2003;206:3053–3064. doi: 10.1242/jeb.00518. [DOI] [PubMed] [Google Scholar]
- Eufemia NA, Epel D. Induction of the multixenobiotic defense mechanism (MXR), P-glycoprotein, in the mussel Mytilus californianus as a general cellular response to environmental stresses. Aquat Toxicol. 2000;49:89–100. doi: 10.1016/S0166-445X(99)00068-5. [DOI] [PubMed] [Google Scholar]
- Farcy E, Voiseux C, Lebel J-M, Fievet B. Seasonal changes in mRNA encoding for cell stress markers in the oyster Crassostrea gigas exposed to radioactive discharges in their natural environment. Sci Total Environ. 2007;374:328–341. doi: 10.1016/j.scitotenv.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fourie AM, Hupp TR, Lane DP, Sang BC, Barbosa MS, Sambrook JF, Gething MJ. HSP70 binding sites in the tumor suppressor protein p53. J Biol Chem. 1997;272:19471–19479. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- Gabai VL, Meriin AB, Mosser DD, Caron AW, Rits S, Shifrin VI, Sherman MY. Hsp70 prevents activation of stress kinases. A novel pathway of cellular thermotolerance. Biol Bull. 1997;272:18033–18037. doi: 10.1074/jbc.272.29.18033. [DOI] [PubMed] [Google Scholar]
- Hamdoun AM, Cheney DP, Cherr GN. Phenotypic plasticity of HSP70 and HSP70 gene expression in the pacific oyster (Crassostrea gigas): implications for thermal limits and induction of thermal tolerance. Biol Bull. 2003;205:160–169. doi: 10.2307/1543236. [DOI] [PubMed] [Google Scholar]
- Huvet A, Herpin A, Dégremont L, Labreuche Y, Samain JF, Cunningham C. The identification of genes from the oyster Crassostrea gigas that are differentially expressed in progeny exhibiting opposed susceptibility to summer mortality. Gene. 2004;343:211–220. doi: 10.1016/j.gene.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Jaattela M. Escaping cell death: survival proteins in cancer. Exp Cell Res. 1999;248:30–43. doi: 10.1006/excr.1999.4455. [DOI] [PubMed] [Google Scholar]
- Kang CM, Park KP, Cho CK, Seo JS, Park WY, Lee SJ, Lee YS. Hspa4 (HSP70) is involved in the radioadaptive response: results from mouse splenocytes. Radiat Res. 2002;157:650–655. doi: 10.1667/0033-7587(2002)157[0650:HHIIIT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Roux F, Gay M, Lambert C, Waechter M, Poubalanne S, Chollet B, Nicolas JL, Berthe F. Comparative analysis of Vibrio splendidus-related strains isolated during Crassostrea gigas mortality events. Aquat Living Resour. 2002;15:251–258. doi: 10.1016/S0990-7440(02)01176-2. [DOI] [Google Scholar]
- Lesser MP, Kruse VA. Seasonal temperature compensation in the horse mussel, Modiolus modiolus: metabolic enzymes, oxidative stress and heat shock proteins. Comp Biochem Physiol A Mol Integr Physiol. 2004;137:495–504. doi: 10.1016/j.cbpb.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Li GC, Werb Z. Correlation between synthesis of heat shock proteins and development of thermotolerance in Chinese hamster fibroblasts. Proc Natl Acad Sci U S A. 1982;79:3218–3222. doi: 10.1073/pnas.79.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-H, Zhu Y-J, Lit X-T. Wild-type p53 gene increases MDR1 gene expression but decreases drug resistance in an MDR cell line KBV200. Cancer Lett. 1997;119:177–184. doi: 10.1016/S0304-3835(97)00267-X. [DOI] [PubMed] [Google Scholar]
- Luedeking A, Koehler A. Regulation of expression of multixenobiotic resistance (MXR) genes by environmental factors in the blue mussel Mytilus edulis. Aquat Toxicol. 2004;69:1–10. doi: 10.1016/j.aquatox.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Minier C, Borghi V, Moore MN, Porte C. Seasonal variation of MXR and stress proteins in the common mussel, Mytilus galloprovincialis. Aquat Toxicol. 2000;50:167–176. doi: 10.1016/S0166-445X(99)00104-6. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Sarge KD, Abravaya K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J Biol Chem. 1992;267:21987–21990. [PubMed] [Google Scholar]
- Mosser DD, Caron AW, Bourget L, Denis-Larose C, Massie B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol Cell Biol. 1997;17:5317–5327. doi: 10.1128/mcb.17.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Martin LH. Induced thermotolerance to apoptosis in a human T lymphocyte cell line. J Cell Physiol. 1992;151:561–570. doi: 10.1002/jcp.1041510316. [DOI] [PubMed] [Google Scholar]
- Park SH, Lee SJ, Chung HY, Kim TH, Cho CK, Yoo SY, Lee YS. Inducible heat-shock protein 70 is involved in the radioadaptive response. Radiat Res. 2000;153:318–326. doi: 10.1667/0033-7587(2000)153[0318:IHSPII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Piano A, Valbonesi P, Fabbri E. Expression of cytoprotective proteins, heat shock protein 70 and metallothioneins, in tissues of Ostrea edulis exposed to heat and heavy metals. Cell Stress Chaperones. 2004;9:134–142. doi: 10.1379/483.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali A, Cotter TG. Heat shock proteins increase resistance to apoptosis. Exp Cell Res. 1996;223:163–170. doi: 10.1006/excr.1996.0070. [DOI] [PubMed] [Google Scholar]
- Shamseldin AA, Clegg JS, Friedman CS, Cherr GN, Pillai MC. Induced thermotolerance in the Pacific oyster, Crassostrea gigas. J Shellfish Res. 1997;16:487–491. [Google Scholar]
- Silar P, Butler G, Thiele DJ. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol Cell Biol. 1991;11:1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MM, Reikerstorfer A, Schwarz A, Krone C, Luger TA, Jaattela M, Schwarz T. Heat shock protein 70 overexpression affects the response to ultraviolet light in murine fibroblasts. Evidence for increased cell viability and suppression of cytokine release. J Clin Invest. 1995;95:926–933. doi: 10.1172/JCI117800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soletchnik P, Lambert C, Costil K. Summer mortality of Crassostrea gigas (Thunberg) in relation to environmental rearing conditions. J Shellfish Res. 2005;24:197–207. [Google Scholar]
- Sorger PK. Heat shock factor and the heat shock response. Cell. 1991;65:363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- Snyder MJ, Girvetz E, Mulder EP. Induction of marine mollusc stress proteins by chemical or physical stress. Arch Environ Contam Toxicol. 2001;41:22–29. doi: 10.1007/s002440010217. [DOI] [PubMed] [Google Scholar]
- Sturzbecher HW, Chumakov P, Welch WJ, Jenkins JR. Mutant p53 proteins bind hsp 72/73 cellular heat shock-related proteins in SV40-transformed monkey cells. Oncogene. 1987;1:201–211. [PubMed] [Google Scholar]
- Tchénio T, Havard M, Martinez LA, Dautry F. Heat shock-independent induction of multidrug resistance by heat shock factor 1. Mol Cell Biol. 2006;26:580–591. doi: 10.1128/MCB.26.2.580-591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H, Müller L, Buchner J. Hsp70 and Hsp90—a relay team for protein folding. Rev Physiol, Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]