Abstract
For many years, there has been uncertainty concerning the reason for Hsp70 translocation to the nucleus and nucleolus. Herein, we propose that Hsp70 translocates to the nucleus and nucleoli in order to participate in pathways related to the protection of the nucleoplasmic DNA or ribosomal DNA from single-strand breaks. The absence of Hsp70 in HeLa cells, via Hsp70 gene silencing (knockdown), indicated the essential role of Hsp70 in DNA integrity. Therefore, HeLa Hsp70 depleted cells were very sensitive in heat treatment and their DNA breaks were multiple compared to that of control HeLa cells. The molecular mechanism with which Hsp70 performs its role at the level of nucleus and nucleolus during stress was examined. Hsp70 co-localizes with PARP1 in the nucleus/nucleoli as was observed in confocal studies and binds to the BCRT domain of PARP1 as was revealed with protein–protein interaction assays. It was also found that Hsp70 binds simultaneously to XRCC1 and PARP-1, indicating that Hsp70 function takes place at the level of DNA repair and possibly at the base excision repair system. Making a hypothetical model, we have suggested that Hsp70 is the molecule that binds and interrelates with PARP1 creating the repair proteins simultaneously, such as XRCC1, at the single-strand DNA breaks. Our data partially clarify a previously unrecognized cellular response to heat stress. Finally, we can speculate that Hsp70 plays a role in the quality and integrity of DNA.
Keywords: Hsp70, PARP-1, XRCC1, DNA damage, DNA repair, DNA breaks, Apoptosis, Heat shock
Introduction
Hsp70 (used herein to denote HSP70A1A) is the main member of the 70-kDa heat shock protein family (Kampinga et al. 2009), and its expression is dramatically induced in elevated environmental temperatures (Lindquist and Craig 1988; Morimoto et al. 1990; Hightower 1991). At the molecular level, Hsp70 is a molecular chaperone guiding the pathways of protein folding (Beckman et al. 1990), protein translocation (Chirico et al. 1988), and protein degradation (Chiang et al. 1989; Saliba et al. 2002). Hsp70 acts in cooperation with a team of co-chaperone proteins to form functional chaperone complexes (Minami et al. 1996; Huang et al. 2001; Bozidis et al. 2002). Evidence concerning the oligomerization state of Hsp70s led to suggestions that oligomerized Hsp70 is inactive and that its chaperoning function is performed by the monomeric form (Benaroudj et al. 1996; Angelidis et al. 1999). At the cellular level, Hsp70 is involved both in cell viability (Angelidis et al. 1991, 1996) and cell death (Samali and Cotter 1996). Defects in Hsp70 function is often associated with various diseases and pathophysiological situations, including ischemic injury (Plumier et al. 1995), neurodegenerative diseases (Cummings et al. 2001; Adachi et al. 2003), and cancer (Scott and Frydman 2003; Mosser and Morimoto 2004). Structural analysis demonstrated that Hsp70s are composed of two domains, a highly conserved N-terminal ATPase domain of 44 kDa and a less conserved C-terminal domain of 25 kDa (Flaherty et al. 1990). A peptide-binding site (PBD) of the 18-kDa portion of the C-terminal domain adjacent to the ATPase domain of the protein was shown to mediate the interaction of Hsp70 with misfolded, unfolded, or partially denatured polypeptide substrates (Wang et al. 1993). In response to heat shock treatment, Hsp70 is induced and translocates from the cytoplasm to the nucleus and nucleoli (Welch and Feramisco 1984), but the mechanism involved remains unknown. We propose that Hsp70 translocates to the nucleus and nucleoli in order to regulate pathways involved in nucleoplasmic or ribosomal DNA (rDNA) quality and, moreover, in nucleolar integrity. Nucleoli are plurifunctional nuclear domains involved in the regulation of several cellular processes such as ribosome biogenesis, the biogenesis of non-ribosomal ribonucleoprotein complexes, cellular aging, and the cell cycle. Each nucleolus contains up to 700 proteins (Coute et al. 2005), and the sites containing rDNA genes (nucleolar organizer regions, NORs) are located on human chromosomes of 13, 14, 15, 21, and 22. Each NOR comprises 30–50 repeated rDNA genes in a tandem array separated by non-transcribed spacer DNA (Henderson et al. 1972).
Parp-1 (poly[ADP-ribose] polymerase 1) has been the most extensively studied molecule among the members of the PARP family. It can covalently ribosylate both itself and a limited number of other acceptor proteins such as histones, HMG proteins, topoisomerases I and II, DNA helicases, various transcription factors, and factors participating in the single-strand break repair (SSBR) and base excision repair (BER) systems. In response to DNA strand breaks, PARP-1 is activated more than 500-fold and binds transiently to the damaged DNA. This binding may also serve as a signal for the initiation of DNA repair reaction by recruiting other proteins, XRCC1 (X-ray repair cross-complementing group) among them (Caldecott et al. 1996; Schreiber et al. 2006). It has been shown that XRCC1, a protein of 633 AA and MW of 70 kDa, constitutes a SSBR/BER scaffold protein in mammalian cells that binds preferentially to PARP-1 (Masson et al. 1998). Its functions have been mainly attributed to its interactions with partner proteins. During BER and SSBR, it interacts with polynucleotide kinase poly[ADP-ribose] polymerase 1 and 2, DNA ligase 3a, AP endonuclease 1 (APE1), and DNA polβ (Caldecott et al. 1996). It is suggested that the combined capacities of XRCC1 to interact with the DNA repair enzymes and their DNA substrates allows a more efficient repair process by holding the damaged DNA together (Rice 1999), creating a transient sealing and therefore ensuring that no nicked DNA is left unrepaired. Subsequently, by initiating interactions at both sites of a single-stranded DNA (ssDNA) break, XRCC1 could recruit just those necessary enzymes (Caldecott et al. 1996).
In this report, we tried to partially decipher one issue concerning the role of Hsp70 translocation to nucleoplasm/nucleoli during heat stress. Extensive review of the literature revealed a limited number of studies suggesting a possible relationship between Hsp70 levels and DNA damage (Calini et al. 2003; Niu et al. 2006). Moreover, few researchers have suggested that Hsp70 may be involved in DNA repair systems. Recent studies showed that Hsp70 enhances deoxyribonucleic acid BER in human leukemic cells after ionizing radiation (Bases 2006; Mendez et al. 2003). Our group has also demonstrated that Hsp70 is implicated in the mechanism of hydrogen-peroxide-induced DNA damage (Doulias et al. 2007).
Having understood that the nucleolus is a key organelle coordinating the synthesis and assembly of ribosomal subunits (Andersen et al. 2005), we were led to question whether this region is under the control of a DNA break protective molecular mechanism similar to that of the nucleoplasm and if by any chance Hsp70 is part of both of these mechanisms. Initially, we modified two preexisting methods, one for nucleoli isolation and a second one for DNA (ssDNA) breaks identification in nucleoli, which we called “nucleolar comet assay” (NCA). Using the NCA method, we studied the differences in rDNA breaks under normal and stress conditions. The NCA experiments revealed that both rDNA and nuclear DNA suffer single-strand DNA breaks during exposure of cells to heat and that cells deficient in Hsp70 were more sensitive to DNA damage (ssDNA breaks). Furthermore, we determined that Hsp70 co-localizes into the nucleoplasm and nucleolus, interacting simultaneously with PARP-1 and XRCC1, two major molecules participating in ssDNA breaks repairing machines (Caldecott et al. 1996; Masson et al. 1998).
Taking all these into consideration, we propose a model in which Hsp70 translocates to the nucleoplasm/nucleoli in order to facilitate the DNA repair molecules, accelerating their ability to compose molecular machines whose functioning leads to high-quality DNA. This result is possibly an outcome of protein–protein interactions between Hsp70, PARP-1, and XRCC1.
Experimental procedures
Cell culture, heat treatment, cell extracts, and nucleoli isolation
Cells were grown in monolayer as described previously (Angelidis et al. 1988).
Sub-confluent cells were heat-treated by immersing the culture dishes in a water bath set at the desired temperature.
Control or heat-treated cells were harvested, washed with NaCl/Pi, resuspended in RIPA buffer, and lysates prepared as described (Bozidis et al. 2002). The lysates were mixed with sodium dodecyl phosphate (SDS) sample buffer to 1× final concentration, boiled for 3 min, and subjected to SDS/polyacrylamide gel electrophoresis (PAGE) and Western blotting analysis using the enhanced chemiluminescence’s method (PIERCE, SuperSignal West Pico, Chemiluminescent Substrate CA 47079).
Nucleoli were purified based on previously published protocols (Muramatsu and Onishi 1978; Ochs 1998) with some modifications. For a typical experiment, 3 × 108 HeLa cells were plated onto 245 × 245-mm Petri dishes and at ∼80% confluence resuspended in 15 volumes of an hypotonic buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, and 1 mM MgCl2) and incubated on ice for 30 min. Cell lysis was performed by addition of a final concentration of 0.3% Nonidet P-40 (Roche Applied Science, Mannheim, Germany), and homogenization was performed using a 0.4-mm clearance Dounce homogenizer (Kimblr/Kontes, Owens, IL, USA). Nuclei were collected by centrifugation at 1,200×g for 5 min and resuspended in 10 volumes of 0.25 M sucrose-containing 10 mM MgCl2. The supernatant—containing the cytoplasmic fraction—was harvested for further analysis. Nuclei were then purified by centrifugation at 1,200×g for 10 min through a 0.88 M sucrose cushion containing 0.05 mM MgCl2. Purified nuclei were resuspended in 10 volumes of 0.34 M sucrose containing 0.05 mM MgCl2 and sonicated on ice for several bursts of 30 s with 5-min intervals between them. Nucleoli were then purified from the resulting homogenate by centrifugation at 2,000×g for 20 min through a 0.88 M sucrose cushion containing 0.05 mM MgCl2. The supernatant containing the “nucleoplasmic fraction” devoid of nucleoli was harvested for further analyses. Purified nucleoli were resuspended in RIPA for protein quantification and for further analyses.
Plasmid construction, cell lines, and cell transfection
The DNA fragments of GST and GST-fused hPARP-1 fragments of GST-DBD1 (1–213), GST-DBD2 (214–337), GST-AUTO (338–523), GST-BRCT (384–460), GST-NAD1 (524–780), and GST-NAD2 (764–1014) were subcloned into pGEX4T-1 (Amersham Pharmacia Biotech, Uppsala, Sweden) by Gwack Y. as previously described (Gwack et al. 2003). In the Apr-SMA-LDH, Apr-NSC-LDH, and Apr-28K-LDH expression vectors (unpublished data) have been respectively subcloned the ATPase domain (Angelidis et al. 1999), the PBD, and the 28-kDa amino-end of the human Hsp70, tagged with the LDH sequences, under the control of the Human β-actin promoter. The pBC-D plasmid that is able to overexpress the BRCT-GST-fused peptide in mammalian cells was constructed as previously described (Dantzer et al. 2004).
HeLa, HeLa–siRNA–Hsp70 (Doulias et al. 2007), HeLa-BRCT-GST (unpublished data), CV1-SMA-LDH, CV1-NSC-LDH (Milarski and Morimoto 1989; Angelidis et al. 1999), and CV1-28K-LDH cells (unpublished data) were grown as described above (see cell culture).
Cells seeded in 5-cm culture dishes were transfected with 2 μg plasmid DNA using the Lipofectamine plus™ reagent (Invitrogen, cat. no. 10964-013). Selection of cells containing stably integrated copies of the transfected plasmid was accomplished by adding geneticin (Geneticin, G-418: BRL-GIBCO 11811-031) to the medium at a concentration of 700–1000 μg/ml.
Flow cytometry studies, Hsp70 expression, and Hsp70 anti-apoptotic activity
Analysis of Hsp70 expression was performed by flow cytometry (Partec CyFlow ML) after cytoplasmic or nuclear and nucleolar staining.
HeLa or HeLa–siRNA–Hsp70 cells were washed twice with cold phosphate-buffered saline (PBS), resuspended in 1% paraformaldehyde/PBS to a concentration of 1 × 106 cells/ml, and incubated for 10 min at room temperature. The samples were prepared for FACScan flow cytometer analysis using 1.5 μg of MAb C92 as previously reported (Dressel et al. 1998).
Once 100% confluence was reached, each cell line was divided in five aliquots of 1.5 × 106 cells. One was analyzed immediately (control) after Hoechst 33342 staining at 20 μg/ml for 60 min. The others were subjected to heat shock at 45°C for time intervals of 15, 30, 60, and 90 min, respectively, followed by recovery for 90 min at 37°C, stained with Hoechst 33342, and then analyzed. Apoptosis was measured by DNA flow cytometric analysis of sub-G1 peaks after Hoechst 33342 staining. The cytometric analysis was performed in a Partec ML flow cytometer and the results were analyzed by Partec FloMax software.
Protein purification
Glutathione S-transferase (GST) and GST-fused proteins were expressed in BL21 (DE3) cells and purified from lysates according to standard procedures (Sambrook et al. 1989).
Recombinant human-Hsp70 was purified using the Bac-to-Bac expression system (Salma et al. 2007) according to the manufacturer’s recommendation (Invitrogen).
Nucleolar comet assay
The alkaline nucleolar comet assay was performed based on previous reported data concerning whole cells (Collins et al. 1995) with some modifications. Cells were fractionated to cytoplasm, nucleoplasm, and nucleoli, as it is subscribed above, and purified nucleoli resuspended to 100 μl of 1.5% L.M.P agarose/PBS pH 7.4 at 37°C. The agarose sample was spread onto frosted slide that was covered with a bed of 1% normal agarose/PBS. On top of this arrangement, a coverglass was laid and the agarose/nucleoli mixture allowed to gel at 4°C. The coverglass was carefully removed and the slide immersed in cold lysis solution (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris, NaOH to pH 10, and 1% Titon-X100) for 1 h at 4°C in the dark. Slide was washed three times for 5 min each time with cold PBS and putted in electrophoresis tank containing fresh solution of 0.3 M NaOH and 1 mM Na2EDTA (pH > 13) for 40 min at 4°C in the dark. Electrophoresis was then carried out at 30 V/300 mA for 30 min at 4°C (temperature < 15°C). Slide was rinsed three times/5 min each with cold PBS pH 7.4, stained with 50 μl DAPI (5 μg/ml PBS), rewashed one time/5 min with cold PBS pH 7.4, and the slide was subjected for observation.
Antibodies
Mouse anti-Hsp70 (StressGen Biotechnologies, Victoria, Canada: SPA 810, a mouse monoclonal antibody that binds to human HSP70 A1A specifically), rabbit anti-PARP-1 (Santa Cruz Biotechnology, sc-7150), rabbit anti-B23 (sc-5564 Santa Cruz Biotechnology, also known as nucleophosmin, numatrin, NO38, NPM), rabbit anti-XRCC1 (H300: sc-11429, Santa Cruz Biotechnology), and rabbit anti-Luciferase (Promega) were used in our experiments.
Immunofluorescence in confocal microscopy
HeLa cells or HeLa–siRNA–Hsp70 stable transfected cell line growing in coverslips were incubated at 37°C or at 43.5°C for time periods indicated in the text and figures legend. Indirect immunofluorescence was performed as described previously (Bozidis et al. 2002). Briefly, cells were washed with NaCl/Pi, fixed with 2% paraformaldehyde for 10 min, and permeabilized for 3–5 min with absolute methanol. Then, the cells were washed with NaCl/Pi, incubated in 3% bovine serum albumin to prevent nonspecific staining, and subjected to incubation with the appropriate antibodies.
Immunoprecipitation and Western blotting
HeLa, HeLa–siRNA–Hsp70, or CV1 cells (2 × 107 cells/sample) were cultured and cell extracts were prepared in RIPA buffer. In the cases of cytoplasm, nuclei, and nucleoli, we immediately used the samples for immunoprecipitations in buffers as indicated in the text. The lysates were incubated overnight at 4°C by end-over-end rocking with 3 μg PARP-1-specific antibody, or 3 μg of Hsp70-specific antibody, or 3 μg XRCC1, or 5 μg LDH-specific antibodies and subjected to immunoprecipitation as previously reported (Sainis et al. 2000). The immunoprecipitates were subjected to SDS/PAGE and Western blotting analysis as previously reported (Bozidis et al. 2002).
To determine the levels of protein expression in the whole cell, cytoplasm, nucleus, or nucleolus, we prepared protein extracts that were analyzed by SDS-PAGE. After electrophoresis, the proteins (20 μg/sample) were transferred to nitrocellulose membranes and the identification of the proteins was carried out with antibodies specific for Hsp70, PARP-1, or for XRCC1 and detected by ECL regent (Santa Cruz Biosciences).
GST pull-down experiments
Glutathione-agarose beads were washed three times in buffer S (150 mM NaCl, 20 mM Tris–HCl, pH 7.5, 250 mM sucrose, 2 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 1 mM PMSF, protease inhibitors). GST fusion proteins (GST-DBD1, GST-DBD2, GST-AUTO, GST-BRCT, GST-NAD1, GST-NAD2, or GST alone; 1 μg) were incubated in buffer ST (buffer S plus 1% Triton X-100) and 30 μl buffer fish skin gelatin 10% (for blocking) for 30 min at room temperature using 30 μl of glutathione-agarose pre-washed beads. After washing three times with the same medium (S), the beads were combined with purified Hsp70 (500 ng) and further incubated for 1 h at room temperature. The beads were washed five times with buffer ST and once with isotonic buffer (150 mM NaCl, 20 mM Tris–HCl, pH 7.5, 250 mM sucrose, 2 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 1 mM PMSF) before eluting the bound proteins with hot SDS sample buffer (Makatsori et al. 2004).
Experiments with GST-fusion purified proteins and cell extracts
RIPA cell extracts from HeLa cells (3 × 106) overexpressing the GST-BRCT domain (HeLa-GST-BRCT) were accompanied with glutathione-agarose beads and incubated for 2 h at 4°C. The samples were washed three times with RIPA before eluting the bound proteins with hot SDS sample buffer.
Statistical analysis
Percentages of apoptotic cells and three types (I, II, III) of nucleolus were measured after every treatment. Statistical analysis for differences in our values was done using Student’s test for comparing two sample sets and analysis of variance for multiple sample sets. Values of P less than 0.01 were considered statistically significant
Results
Hsp70 down-regulation induces apoptosis in response to heat treatment
To investigate the in vivo function of Hsp70 in response to heat shock, we used a previously described HeLa cell line stably depleted of Hsp70 (Doulias et al. 2007). In a preliminary experiment, the expression of Hsp70 in one of the G-418 isolated clones, c-25, was assayed by Western blotting before and after heat shock at 43.5°C for 90 min and recovery at 37°C for 0 or 3 h. As shown in Fig. 1a, no detectable levels of Hsp70 were observed in both untreated and in heat-shock-treated HeLa siRNA-Hsp70. The depletion of Hsp70 was also monitored by flow cytometry analysis using a mouse monoclonal HSP70A1A specific antibody both in control and heat-shocked cells (Fig. 1b). The heat-shock-dependent induction of Hsp70 was observed in control HeLa (Fig. 1b, lower panel) cells only, not in HeLa–siRNA–Hsp70 (Hsp70-depleted cells; Fig. 1b, upper panel). Together, these results show an almost complete Hsp70 down-regulation in HeLa–siRNA–Hsp70 cells. We next analyzed the stress-induced apoptotic response in HeLa–siRNA–Hsp70 compared to the control cells by following PARP-1 cleavage and flow cytometry analysis using Hoechst staining. In the absence of Hsp70, PARP-1 was activated earlier (15 min at 45°C and 90 min at 37°C) and strongly 45 min later (60 min at 45°C and 90 min at 37°C) than in those cells expressing Hsp70 in high levels (HeLa; Fig. 1c). Therefore, we can suggest that the Hsp70 depletion increases the apoptosis of HeLa cells. Similar results were obtained, under the same conditions, when we compared the same cells using a DNA fluorochrome and flow cytometry analysis. HeLa and HeLa–siRNA–Hsp70 cells were grown under standard conditions and were analyzed for apoptosis before and after heat shock (Fig. 1d). Comparing their histogram graphs showing the percentage of apoptotic cells, we can ascertain that cells depleted of Hsp70 showed increased apoptotic activity (Fig. 1d). All the above results suggest, as in previous studies (Samali and Cotter 1996), that Hsp70 constitutes one of the main anti-apoptotic members, and cells lacking Hsp70 are led to apoptotic death triggered by heat. However, little is known about the mechanism of heat action and the Hsp70 translocation to nucleus and nucleoli.
Fig. 1.
The down-regulation of Hsp70 is followed by increase of apoptotic activity in HeLa–siRNA–Hsp70 cells. a HeLa and HeLa–siRNA–Hsp70 cells were heat-treated (43.5 The down-regulation of Hsp70 is followed by increase of apoptotic activity in HeLa–siRNA–Hsp70 cells. a HeLa and HeLa–siRNA–Hsp70 cells were heat-treated (43.5°C for 90 min), or not, as indicated. Western blotting analysis using a MAb specific for the inducible Hsp70 (C92) revealed that Hsp70 is absent in HeLa–siRNA–Hsp70 even after 3 h of recovery at 37°C. b The same cells were heat-shocked at 43.5°C with 90 min recovery at 37°C or kept at 37°C. Flow cytometric analysis of intracellular Hsp70, using the same anti-Hsp70 antibody as in a, demonstrated a constitutive Hsp70 expression in control cells and an increased amount in heat treated cells. The fluorescence of HeLa–siRNA–Hsp70 was identical to the isotype control (mouse IgG-FITC) and was the same before and after heat shock. c HeLa and HeLa–siRNA–Hsp70 cells were heat-treated or kept at 37°C as indicated. RIPA cell extracts were selected and analyzed by SDS-PAGE and Western blotting using specific antibodies for PARP-1 and α-tubulin (as internal standard). The clone of HeLa–siRNA–Hsp70 possesses increased apoptotic activity. d The same cells were exposed to heat treatment as indicated and analyzed for Hoechst fluorescence by flow cytometry. Each point represents the mean value ± SD of three measurements in three separate experiments
Increased single-strand breaks in the nucleoli of Hsp70-depleted HeLa cells
It was previously reported that heat induces oxygen-derived radicals (Reddy and Gangadharam 1992; Bruskov et al. 2002), leading to oxidative damage to DNA. In our previous work, we have shown that Hsp70 is involved in the mechanism of hydrogen-peroxide-induced DNA damage (Doulias et al. 2007). To further characterize the role of Hsp70 in cellular response to DNA strand breaks, we first monitored the level of heat-shock-induced double-strand breaks in HeLa–siRNA–Hsp70 cells compared to the control cells using nucleoplasmic DNA or nucleolar rDNA and DNA fragmentation assays. HeLa and HeLa–siRNA–Hsp70 cells were either exposed or not to heat (43.5°C for 90 min and 8 h at 37°C), and samples of isolated DNAs were analyzed in an agarose gel. Under these conditions, no differences in DNA laddering were observed in either the presence or the absence of Hsp70. This means that the nucleoplasmic DNA (chromosomes: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 16, 17, 18, 19, 20, 22, ΧΧ) or rDNA (the rest of chromosomes: 13, 14, 15, and 21) double-strand breaks are not in any way dependent upon the Hsp70 accumulation in the nucleus/nucleoli (data not shown). This suggestion was supported by previously reported data (Doulias et al. 2003) which claimed that the exposure of HeLa cells to oxidative stress induced a dose- and time-dependent increase of nuclear DNA single-strand breaks, and these breaks were regulated by Hsp70 expression in the cells (Doulias et al. 2007). We next monitored the role of Hsp70 in the cellular response to single-strand breaks. To do so, we developed a new approach to evaluate the level of single-strand breaks in rDNA using a modified NCA (Collins et al. 1995). Using the modified technique (see “Experimental procedures”), nuclei (Fig. 2a, A1, left panel) and nucleoli (Fig. 2a, A1, middle and right panel) were isolated and analyzed with NCA (Fig. 2a, A2, A3). In order to confirm the purity of these subcellular cell extracts or cellular extracts from organelles, we used antibodies specific for B23 protein, a specific indicator of nucleoli (Fig. 2, A4). Combining all insets of Fig. 2a, the excellent quality of the purified nucleoli is obvious. Inset A2 (Fig. 2) shows the different sizes of nuclei and nucleoli and their morphology as observed during an alkaline comet assay. Inset A3 (Fig. 2) shows randomly selected nuclei, during NCA analysis, from HeLa or HeLa–siRNA–Hsp70 that constitute a comparable substrate for the nucleoli, meaning their damaged DNA as it is represented in an alkaline comet assay. Subsequently, nucleoli were arbitrary classified into three groups: type I which are characterized by intact nucleoli, type II with mild disruption, and type III with complete disruption (Fig. 2b, B1). Types IΙ and IIΙ represent an increasing extent of DNA damage, visualized as an extending tail and increased nucleoli disruption. Thus, upon heat shock, a significant number of nucleoli appeared with extended tails and disruptions in their central compartment compared to those in controls (Fig. 2b, Β2). In the absence of heat treatment, a weak but significant increase of type II and type III (20% and 5% corresponding) in untreated HeLa–siRNA–Hsp70 cells compared to untreated control cells was observed, thus indicating a higher level of endogenous single strand breaks (Fig. 2b, B2). After heat shock, there was a very significant increase in the percentage of type II and, specifically, type III. Together, these results provide evidence of a role of Hsp70 in cellular response to single-strand breaks.
Fig. 2.
Hsp70 protects HeLa cells from apoptosis, intervening only at the ssDNA repairing system. a Isolation of HeLa nuclei and nucleoli (A1). Size comparison among nuclei and nucleoli during comet assay (A2) and comet assay of randomly isolated nuclei from HeLa and HeLa–siRNA–Hsp70 under control or heat shock (90 min at 43.5 Hsp70 protects HeLa cells from apoptosis, intervening only at the ssDNA repairing system. a Isolation of HeLa nuclei and nucleoli (A1). Size comparison among nuclei and nucleoli during comet assay (A2) and comet assay of randomly isolated nuclei from HeLa and HeLa–siRNA–Hsp70 under control or heat shock (90 min at 43.5°C and 90 min recovery at 37°C) (A3). Ten micrograms of each HeLa cellular fraction was analyzed by Western blot analysis with anti-B23 antibody as indicated (A4). b Nucleolar comet assay (NCA) of HeLa and HeLa–siRNA–Hsp70 cells under control or heat treatment (90 min at 43.5°C and 90 min recovery at 37°C). Type I, type II, and type III are the three different types of nucleolar disruptions (B1). The same cells were treated as above and measured for nucleoli disruption. As indicated, the type I (intact nucleolus) is eliminated and changed to type II (moderately disrupted nucleolus) and type III (completely disrupted nucleolus) (B2). Columns represent percentage of nucleoli type I, II, or III as the mean value of four independent experiments with four microscope slides per sample with standard errors indicated in bars. Each point represents the mean value ± SD of triplicate measurements from five separate experiments. Bar, 10, 2 or 1.25 μm
Hsp70 co-localizes with PARP1 and XRCC1 in the nucleus and nucleolus after heat shock treatment
We next analyzed the subcellular localization of Hsp70, PARP-1, and XRCC1 in response to heat shock. Control HeLa and HeLa–siRNA–Hsp70 cells were heat shocked and the localization of each partner was monitored by IF using anti-Hsp70 and anti-XRCC1 specific antibodies followed by green-labeled and red-labeled secondary antibodies, respectively.
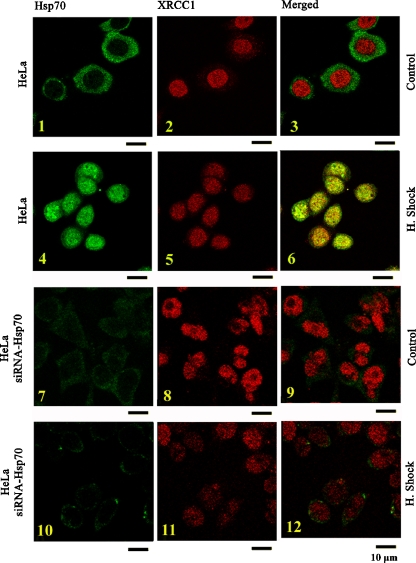
Concretely, under heat stress at 43.5°C for 90 min and 90 min recovery at 37°C, a large population of Hsp70 molecules translocates to the nucleus/nucleolus (Fig. 3, insets 4, 6) and is co-localized to PARP-1 (Fig. 3, inset 6). In the absence of Hsp70 (Fig. 3, insets 7-12), PARP-1 continues to participate in nuclei/nucleoli during heat shock (Fig. 3, inset 11). This means that PARP-1 translocates to the nucleolus, independently of Hsp70, as is shown below (Fig. 3, insets 8 and 11, 9, and 12). Secondly, Hsp70 co-localizes to XRCC1 (Fig. 4, inset 6), a molecule that coordinates and facilitates SSB and BER in mammalian cells (Nazarkina et al. 2007). This suggests that Hsp70 translocates to the nucleus/nucleolar during stress owing to the DNA breaks and coordinates known molecules responsible for DNA damage such as XRCC1.
Fig. 3.
Hsp70 and PARP-1 accumulate in the nucleoplasm and nucleolus during heat shock and mainly co-localize at the same sites. Confocal sections showing the simultaneous immunodetection of Hsp70 and PARP-1 in HeLa (insets 1–6) or in HeLa–siRNA–Hsp70 cells (insets 7–12). Cells were exposed, or not, to heat shock (90 min at 43.5 Hsp70 and PARP-1 accumulate in the nucleoplasm and nucleolus during heat shock and mainly co-localize at the same sites. Confocal sections showing the simultaneous immunodetection of Hsp70 and PARP-1 in HeLa (insets 1–6) or in HeLa–siRNA–Hsp70 cells (insets 7–12). Cells were exposed, or not, to heat shock (90 min at 43.5°C and 90 min recovery at 37°C), fixed with 2% formaldehyde, and double immunofluorescence were applied using antibodies specific for Hsp70 and PARP-1. Bar, 10 μm
Fig. 4.
Subcellular distribution of endogenous Hsp70 and XRCC1 during control or heat shock. Confocal sections showing the simultaneous immunodetection of Hsp70 and XRCC1 in HeLa cells (insets 1–6) or in HeLa–siRNA–Hsp70 cells (insets 7–12). Cells were exposed, or not, to heat shock (90 min at 43.5 Subcellular distribution of endogenous Hsp70 and XRCC1 during control or heat shock. Confocal sections showing the simultaneous immunodetection of Hsp70 and XRCC1 in HeLa cells (insets 1–6) or in HeLa–siRNA–Hsp70 cells (insets 7–12). Cells were exposed, or not, to heat shock (90 min at 43.5°C and 90 min recovery at 37°C), fixed with 2% formaldehyde, and double immunofluorescence were applied using antibodies specific for Hsp70 and XRCC1. Bar, 10 μm
Hsp70 is associated with PARP-1 and XRCC1
To explore further the role of Hsp70 in DNA damage pathways, we examined whether it could biochemically associate with the SSBR/BER proteins: PARP-1 or XRCC1. Cell lysates from HeLa cells were immunoprecipitated with either an anti-Hsp70, either anti-PARP-1, or anti-XRCC1, and the immunoprecipitates were analyzed by immunoblotting for the presence of PARP-1, Hsp70, and PARP-1/Hsp70, respectively. As we expected, Hsp70 antibody co-precipitated both Hsp70 and PARP-1 and reversely, PARP-1 antibody co-precipitated both PARP-1 and Hsp70 (Fig. 5a, A1, A2) in both untreated and treated cells. Also, as shown in Fig. 5b, PARP-1 (Fig. 5b, B1) and Hsp70 (Fig. 5b, B2) were co-immunoprecipitated using an anti-XRCC1 antibody in both untreated and treated cells. As an additional immunoprecipitation (IP) control, we used cell lysates from HeLa–siRNA–Hsp70 cells and antibodies specific for Hsp70 (Fig. 5c). Western blotting analysis of the received IPs with anti-XRCC1 and anti-PARP-1 confirmed the expected results, namely, in the absence of Hsp70 (Fig. 5c, C3), PARP1, or XRCC1 do not precipitate (Fig. 5c, C1, C2)
Fig. 5.
Hsp70 associates with the DNA repairing molecules, PARP-1 and XRCC1. HeLa or HeLa–siRNA–Hsp70 whole cell extracts, from control or heat-shocked (90 min at 43.5 Hsp70 associates with the DNA repairing molecules, PARP-1 and XRCC1. HeLa or HeLa–siRNA–Hsp70 whole cell extracts, from control or heat-shocked (90 min at 43.5°C and 90 min recovery at 37°C) cells (2 × 107 cells/sample), were prepared in RIPA buffer. Subsequently, control or heat-shocked (90 min at 43.5°C and 90 min recovery at 37°C) cell extracts were subjected to immunoprecipitation with anti-Hsp70 (a, c), anti-PARP-1 (a), or anti-XRCC1 (b). The immunoprecipitations were analyzed with SDS-PAGE and Western blotting using specific antibodies against PARP-1, Hsp70, or XRCC1 to track bindings between PARP-1 (A2, B1, C1), Hsp70 (A1, B2, C3), and XRCC1 (B3, C2) proteins. CE cell extract, IP immunoprecipitation, B beads
Together, these results indicate either an indirect or a direct interaction of Hsp70 with PARP-1 and XRCC1 and further implicate the role of Hsp70 in DNA repairing systems.
Hsp70 binds to the PARP-1 BRCT domain
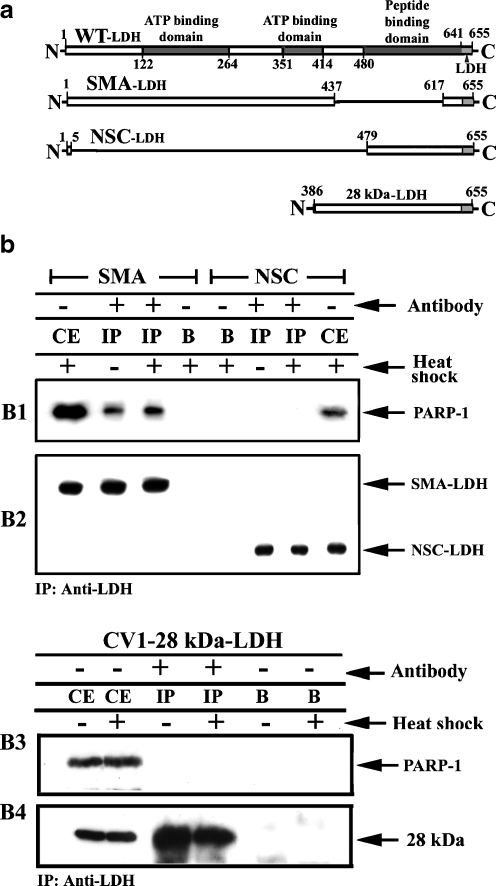
PARP-1 contains multiple domains, such as DNA binding domain 1 (DBD1; AA 1 to 213), DNA binding domain 2 (DBD2; AA 214 to 337), automodification domain (AUTO; AA 338 to 523), BRCA1 C-terminal domain (BRCT; AA 384 to 460), NAD binding domain 1 (NAD1; AA 524 to 780), and NAD binding domain 2 (NAD2; AA 764 to 1014). To map the Hsp70 binding region within PARP-1, we generated truncated versions of PARP-1 fused to GST: (DBD1; aa 1 to 213), DNA binding domain 2 (DBD2; aa 214 to 337), automodification domain (AUTO; aa 338 to 523), BRCA1 C-terminal domain (BRCT; aa 384 to 460), NAD binding domain 1 (NAD1; aa 524 to 780), and NAD binding domain 2 (NAD2; aa 764 to 1014). GST alone or GST-fused domains of PARP-1 were mixed with purified recombinant human Hsp70 (Fig. 6b, B1) or firefly luciferase (Fig. 6b, B2; Angelidis et al. 1999) and analyzed by GST pull down (Fig. 6b, B3; Salma et al. 2007).
Fig. 6.
Human purified Hsp70 binds to BRCT domain of PARP-1. a Schematic presentation of PARP-1 domains based on Jung J.U., 2003. b 1 μg of GST-DBD1, GST-DBD2, GST-AUTO, GST-BRCT, GST-NAD1, GST-NAD2, or GST alone (B3) was each mixed with 0.5 μg of human Hsp70 (B1) or 0.5 μg of purified firefly luciferase (B2) (refolded according to Qiagen methods) and the mixtures subjected to pull-down experiments. The resulted precipitates were analyzed by SDS-PAGE and Western blotting using antibodies specific to Hsp70 (B1), luciferase (B2), or GST (B3)
As shown in Fig. 6, Hsp70 binds to the automodification domain of PARP-1 containing the BRCT domain (Fig. 6, B1). In contrast, no binding was revealed between luciferase and PARP-1 domains (Fig. 6, B2). This finding was very important given that similar domains possess some proteins participating in ssDNA repairing systems, among them, XRCC1, DNA ligase III, and BRCA-1 (Rodriguez et al. 2003). Alternatively, we tried to repeat the in vitro binding of the BRCT domain to Hsp70 in a semi-in vivo system. HeLa cells were stably transfected with pBC-D plasmid and cell clones, termed HeLa-GST-BRCT and overexpressing the GST-BRCT domain, were selected (Fig. 7a).
Fig. 7.
The BRCT domain of PARP-1 associates with Hsp70 and XRCC1 in HeLa cells overexpressing the GST-BRCT fused protein. a HeLa cells were stable transfected with the pBC-D plasmid and cell clones, called HeLa-GST-BRCT cells, were isolated using G-418. The expression of GST-BRCT fused protein was monitored using a monoclonal anti-GST antibody and Western blotting analysis. In all experiments, the HeLa-GST-BRCT (clone 26) was used (indicated by the asterisk). b HeLa-GST-BRCT (clone 26) was used in order to prepare whole cell extracts from control or heat-shocked (90 min at 43.5°C and 90 min recovery at 37°C) cells. Pull-down experiment using these extracts indicated the binding between Hsp70 and BRCT domain. c The same cells were fractionated to cytoplasm (Cyt), nucleus (Nu), nucleoplasm (Np), nucleolus (Nli), and each fraction was analyzed for Hsp70 and BRCT-GST accumulation under control or heat shock conditions by Western blotting. d The same fractions, as in c, were used for pull-down experiments. As shown, Hsp70 binds to BRCT-GST fused protein only in nucleus, nucleoplasm, and nucleolus independently from the heat treatment
HeLa-GST-BRCT cells (clone 26) were used for cell extract selection, as indicated in the Fig. 7, and the lysates were subjected to Western blotting (a, c) or pull-down experiments (b, d). The same finding as with that of purified proteins was obtained, indicating that the BRCT domain is indeed recognized, directly or indirectly, by Hsp70 and their binding takes place independent of heat shock (Fig. 7b).
Next, to verify whether Hsp70-PARP-1/BRCT interaction occurs within all subcellular compartments, HeLa-GST-BRCT cells were divided in cytosolic, nuclear, nucleoplasmic, and nucleolar fractions and each fraction was subjected to GST pull down and analyzed for the co-immunoprecipitation of Hsp70 and XRCC1. As shown in Fig. 7d, Hsp70 binds to the BRCT domain of PARP-1 in the nucleus, nucleoplasm, and nucleoli. XRCC1 was also co-immunopurified in the same subcellular lysates (Fig. 7d). No interaction between Hsp70 and GST-BRCT could be detected in the cytoplasm, although both proteins are present (Fig. 7c). Thus, the failed binding among Hsp70/GST-BRCT in the cytoplasm implies a competition for Hsp70 binding with another cytoplasmic protein or different Hsp70 chemical modifications between cytoplasm and nucleus. The significance of this observation remains to be determined.
PARP-1 associates with the ATPase domain of Hsp70
We next mapped the interaction domain of PARP-1 within Hsp70. CV1 cell lines overexpressing either the Hsp70 ATPase domain in fusion with testis lactate dehydrogenase (SMA-LDH) or the Hsp70 peptide binding domain PBD (Fig. 8a) in fusion with LDH (NSC-LDH; Milarski and Morimoto 1989; Angelidis et al. 1999) were immunoprecipitated using an anti-LDH antibody (Fig. 8b, B2) and analyzed for the presence of PARP-1. PARP-1 was co-immunopurified with the ATPase domain of Hsp70 (SMA-LDH) in both untreated and heat treated cells, but not with the peptide binding domain NSC-LDH (Fig. 8b, B1 and B2). As expected, similar experiments using lysates from CV1 cells overexpressing the 28-kDa LDH COOH terminus of the Hsp70 confirmed that there is no PARP-1 binding to this Hsp70 fragment (Fig. 8b, B3, B4) and only the Hsp70-ATPase domain binds to BRCT domain of PARP-1 (Fig. 8b, B1). Note that in SMA-LDH mutant, the epitope of c-92 (Anti-Hsp70 antibody) has been deleted, in contrast to that of 28-kDa LDH.
Fig. 8.
Hsp70 forms a complex containing PARP-1 via its ATPase domain. a Representation of the Hsp70 mutated forms were used in this work based partially on previous schema (Milarski, K.L. & Morimoto, R.I., 1989, Angelidis et al., unpublished data). b CV1 stable transfected cells overexpressing the ATP binding domain (SMA-LDH), the peptide binding domain (NSC-LDH), or the carboxy terminus 28-kDa fragment were exposed/or not to heat treatment (43.5°C) for 90 min and 90 min recovery at 37°C. Immunoprecipitation assays using antibodies against LDH (B1/B2, B3/B4) revealed that only the ATPase domain of Hsp70 interacts with PARP-1 (B1). This result was confirmed by the failed bindings among the Hsp70 28-kDa fragment containing the entire PBD- and PARP-1 (B3/B4). CE cell extract, IP immunoprecipitation, B beads
Overall, we can conclude that the PARP-1 BRCT domain associates either directly or indirectly with the Hsp70 ATPase domain, thus creating an initiator signal for the recruit of molecules participating to ssDNA repairing system.
Discussion
For many years, the purpose of Hsp70 translocation and accumulation in the nucleus and nucleolus during heat stress has remained nearly unknown. Some years ago, it was proposed and published that Hsp70 mainly accumulates along with denatured substrates in the nucleoli and that these subcellular compartments are places where Hsp70 chaperoning activity is performed during recovery of stressed cells in their normal growth temperature (Nollen et al. 2001). Furthermore, some previous observations in yeast have revealed that Rad9 complexes, Rad9 being the prototype DNA damage checkpoint gene, contain Ssa1 and or Ssa2 chaperone proteins, both of which carry out the functions of the corresponding Hsp70 in mammalian cells (Gilbert et al. 2003).
In order to examine whether this rapid translocation is associated with other basic cellular pathways, we tried to determine the target of the Ηsp70 in the nucleoplasm and nucleolus. We first examined the role of Hsp70 in cell apoptosis using HeLa cell line which was Hsp70-depleted using siRNA technology. These cells were sensitive to heat- and possess-enhanced apoptotic activity. Subsequently, we modified an existing technique in order to further study nuclear apoptosis, and we provided evidence for the protective effect of Hsp70 in the integrity of ribosomal DNA during exposure of cells to stresses such as heat shock. The NCA provides a powerful technique for nuclear apoptosis studies, making the ssDNA breaks in heat-treated nucleoli visible. Using this methodology and flow cytometry analysis, it was shown that the Hsp70 depletion in HeLa cells produces an increase in apoptosis. However, for many years, the apoptotic pathway of heat was unknown. Previous studies with in vitro experiments demonstrated that heat possibly acts via ROS attack, leading to oxidative damage to DNA (Reddy and Gangadharam 1992; Bruskov et al. 2002). Regarding all the above to be valid, we attempted to search for Hsp70 partners in sites where Hsp70 translocates during stress, that is, nucleoplasm and nucleolus. Note that in the same areas, it is mainly the ROS targets such as nucleoplasmic or ribosomic DNA.
At the beginning, we followed a paradoxical way of thinking, considering the fact that the Hsp70 was not a DNA binding protein; therefore, it could only be an indirect participant of a DNA repairing system. Thus, using confocal microscopy, we found that Hsp70 co-localizes to PARP-1 and XRCC1. Our interest was focused on PARP-1 and XRCC1 because of their participation, along with DNA ligaseIII (Lig3), in the single-strand DNA repairing system (Masson et al. 1998; Caldecott et al. 1996) which is distinct from that which deals with double-strand DNA breaks (DSBs), which include DNA ligase IV (Lig4) and its partner protein, XRCC4 (Grawunder et al. 1998). Given that PARP-1 binds both SSBs and DSBs (Ikejima et al. 1990; Caldecott 2003), some functional overlaps may exist between the SSBR and DSBR mechanism. Later, we determined these possible interactions with immunoprecipitations and pull-down experiments using GST-purified proteins or cell extracts. We determined that Hsp70 binds to PARP1 both in nucleus and nucleoli. Subsequently, the identification of XRCC1 in Hsp70 bindings/co-localization revealed the participation and the possible function of Hsp70 in the ssDNA breaks repairing system.
This study provides the first biochemical evidence that HSP70 plays a protective role against nuclear and nucleolar DNA damages induced by heat stress. Given that Hsp70 is massively induced and translocated to the nucleus/nucleoli only under stress conditions or during the S-phase (Milarski and Morimoto 1986), we could not exclude the potential role of Hsp70 in DNA replication, a major cellular S-phase pathway where ssDNA breaks are produced (Hang and Fox 1995). Overall, these observations indicate that Hsp70 translocates to the nucleus and nucleolus, during S-phase or during a heat shock, in order to participate in pathways involving the integrity and the accuracy of the DNA molecules. Furthermore, this protective role seems to be involved in the regulation of apoptotic pathways containing nucleoplasmic/nucleolic ssDNA breaks. Possibly, the ssDNA breaks are one of the last stages in apoptotic pathways where Hsp70 plays an anti-apoptotic role. Analyzing our results, we are unable to propose a mechanism through which Hsp70 and PARP-1 bindings cause any functional differences between non-heated and heated HeLa cells. Single-strand DNA breaks are produced all the time and their number increases significantly during the S-phase and heat stress. In other words, the complex of Hsp70 and PARP-1 could exist all the time.
Given that Hsp70 is a protein quality regulator molecule, we are here proposing a new Hsp70 role having to do with ssDNA breaks protection. Both functions render Hsp70 as the major chaperone that plays a dominant role in what we call large biomolecule protection or protein and DNA quality maintenance. Also, it is among the first data showing what the role of Hsp70 in the nucleoplasm and nucleolus may be, suggesting ssDNA breaks protection in both subnuclear compartments. Additionally, this is one of the first observations functionally connecting the Hsp70 with different substrates such as proteins and DNAs.
Our further efforts were aimed at identifying the partners for Hsp70 function in nucleus and nucleolus in order to clarify the DNA protective mechanism. Initially, we can propose a more complete single-strand DNA breaks repairing system that includes one more very significant molecule, the Hsp70. Consistent with the previous and present data, the Hsp70 exists in a monomer or oligomer state, directing all the repairing molecules to the ssDNA breaks, especially those that have BRCT motifs. The 95 amino acid BRCT domains (BRCA1 C-Terminus) possess protein–protein interaction capacities and can either bind to different BRCT domains or interact with other unknown protein folds (Zhang et al. 1998; Huyton et al. 2000). These BRCT capacities enable the Hsp70 molecule to come in between molecules and bind to them via BRCT domain, thus inactivating their function when a new ssDNA break is created. This is supported by previous studies that detected bindings via BRCT domain among DNA repair enzymes such as PARP-1, XRCC1, and DNA-ligase III (Caldecott et al. 1996). Thus, it would be a clever way for the cell to regulate their DNA repairing system utilizing the BRCT domain of their partners. As to how Hsp70 binds simultaneously to both proteins (PARP-1 and XRCC1) in immunoprecipitation experiments (Fig. 5, a, b), the answer contains more than one explanation. Firstly, PARP1 and XRCC1 compete for Hsp70 binding, secondly Hsp70 binds to the BRCTb domain and PARP-1 to the BRCTa domain of XRCC1, and thirdly Hsp70 binds directly to the BRCT domain of one PARP-1 molecule and XRCC1 in the other PARP-1 molecule. The failed bindings among Hsp70 and GST-BRCT in the cytoplasm of HeLa-GST-BRCT (Fig. 7, d) suggest that Hsp70 might be modified during translocation to the nucleus/nucleolus to facilitate its interaction with PARP-1.
Finally, an unexpected and new finding is hereby reported. In general, we can assume that Hsp70 simultaneously protects the quality of both proteins (Bukau et al. 2006) and DNAs. It is accepted today that Hsp70 directly participates and regulates the molecular nanomachines for folding and, as we are proposing, indirectly affects the regulation of the ssDNA breaks in nucleoplasmic DNA and rDNA. This suggestion is supported by our observations concerning the cooperation between Hsp70, XRCC1, and PARP-1 and the fact that the last two proteins are partners for the ssDNA breaks repairing system (Masson et al. 1998). All the results presented here have prompted us to consider a new model of Hsp70 function in the repairing systems. According to this model, Hsp70, via ATPase domain, binds to BRCT domain of PARP-1 and recruits the molecules of the repairing partners (Fig. 9). PARP-1 is dimerized, possibly via Hsp70, and labels the histone and non-histone proteins, as well as itself, while XRCC1 holds both ends in the break stable until the formation of the repairing complex is completed (Fig. 9). Finally, the ssDNA breaks molecular machine undertakes the repairing function. Hsp70 may provide a bridge with a lot of arms, accelerating the creation and moving of the DNA repairing molecules between the ssDNA breaks and the DNA-repairing enzymes (Fig. 9).
Fig. 9.
Hypothetical model of Hsp70 function on DNA damage response in nucleoplasm and nucleoli. Hsp70 facilitates the ssDNA breaks repairing machines by creating and organizing their molecules and simultaneously accelerating their function. Hsp70 exists as monomer or oligomer and creates repairing molecules at the side of an ssDNA-break (1). There is one molecule of PARP-1 for every 1,000 bp of DNA (2). PARP-1 is dimerized, possibly via Hsp70 folding function, and catalyzes poly(ADP-ribosyl)ation reactions (3). XRCC1 binds to the DNA ends of a ssDNA break and holds them in a position easy to be repaired (4). DNA ligase III and DNA polβ complete the repairing of the DNA damage (5)
Acknowledgments
We are very thankful to Professor of Biochemistry, Molecular Biology, and Cell Biology, R. Morimoto., Professor of Biological Chemistry D. Galaris and Emeritus Professor Pagoulatos Gerassimos for their support with reagents or useful discussions. We also thank Dr. Doulias P-T., Dr. Skyrlas A., Dr. Markopoulos G., Dr. Noutsopoulos D., and Dr. Zerikiotis S. for their technical and scientific support. This research was partially co-funded by the European Union and the Hellenic Ministry of Education (program “Herakleitos”) within the “Operational Program for Education and Initial Vocational Training. It was also partially supported by a grant to P. Vezyraki from the Empeirikio Institution, Athens.
Abbreviations
- NCA
Nucleolar comet assay
Footnotes
Outlining prior scientific knowledge on the subject and novel information: The role of Hsp70 translocation to the nucleus and nucleolus during heat stress has been nearly unknown. It has been proposed that this biological phenomenon is correlated to Hsp70-chaperoning activity. Furthermore, some previous observations in yeast have revealed that Rad9 complexes—Rad9 being the prototype DNA-damage checkpoint gene—contain Ssa1 and or Ssa2 chaperone proteins, both reconstituting the functions of the corresponding Hsp70 in mammalian cells. Here, we propose that Hsp70 translocates to the nuclei/nucleoli during heat stress, binds to PARP-1 and/or XRCC1, and protects HeLa cells from increased single-strand DNA breaks.
References
- Adachi H, Katsuno M, Minamiyama M, Sang C, Pagoulatos G, Angelidis C, Kusakabe M, Yoshiki A, Kobayashi Y, Doyu M, Sobue G. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Angelidis Ch, Lazaridis I, Pagoulatos G. Specific inhibition of simian virus 40 protein synthesis by heat and arsenite treatment. Eur J Biochem. 1988;172:27–34. doi: 10.1111/j.1432-1033.1988.tb13851.x. [DOI] [PubMed] [Google Scholar]
- Angelidis Ch, Lazaridis I, Pagoulatos G. Constitutive expression of heat-shock protein 70 in mammalian cells confers thermoresistance. Eur J Biochem. 1991;199:35–39. doi: 10.1111/j.1432-1033.1991.tb16088.x. [DOI] [PubMed] [Google Scholar]
- Angelidis CE, Nova C, Lazaridis I, Kontoyiannis D, Kollias G, Pagoulatos GN. Overexpression of HSP70 in transgenic mice results in increased cell thermotolerance. Transgenics. 1996;2:111–117. [Google Scholar]
- Angelidis CE, Lazaridis I, Pagoulatos GN. Aggregation of hsp70 and hsc70 in vivo is distinct and temperature-dependent and their chaperone function is directly related to non-aggregated forms. Eur J Biochem. 1999;259:505–512. doi: 10.1046/j.1432-1327.1999.00078.x. [DOI] [PubMed] [Google Scholar]
- Bases R. Heat shock protein 70 enhanced deoxyribonucleic acid base excision repair in human leukemic cells after ionizing radiation. Cell Stress Chaperones. 2006;11:240–249. doi: 10.1379/CSC-185R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman RP, Mizzen LA, Welch WJ. Interaction of hsp70 with newly synthesized proteins: Implication s for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Triniolles F, Ladjimi MM. Effect of nucleotides, peptides and unfolded proteins on the self-association of the molecular chaperone hsc70. J Biol Chem. 1996;271:18471–18476. doi: 10.1074/jbc.271.31.18471. [DOI] [PubMed] [Google Scholar]
- Bozidis P, Lazaridis I, Pagoulatos G, Angelidis C. Mydj2 as a potent partner of hsc70 in mammalian cells. Eur J Biochem. 2002;269:1553–1560. doi: 10.1046/j.1432-1033.2002.02807.x. [DOI] [PubMed] [Google Scholar]
- Bruskov VI, Malakhova LV, Masalimov ZK, Chernikov AV. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acid Res. 2002;30:1354–1363. doi: 10.1093/nar/30.6.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/S1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly(ADP-ribose) polymerase, and DNA ligase III is a novel molecular “nick sensor” in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calini V, Urani C, Camatini M. Overexpression of Hsp70 is induced by ionizing radiation in C3H 10T1/2 cells and protects from DNA damage. Toxicol In Vitro. 2003;17:561–566. doi: 10.1016/S0887-2333(03)00116-4. [DOI] [PubMed] [Google Scholar]
- Chiang H-L, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degredation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Chirico W, Waters MG, Blobel G. 70 K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Collins AR, Ma AG, Duthie SJ. The kinetics of repair of oxidative DNA damage (strand breaks and oxidised pyrimidines) in human cells. Mutat Res. 1995;336:69–77. doi: 10.1016/0921-8777(94)00043-6. [DOI] [PubMed] [Google Scholar]
- Coute Y, Burgess JA, Diaz J-J, Chichester C, Lisacek F, Greco A, Sanchez J-C. Deciphering the human nucleolar proteome. Mass Spectrom Rev. 2005;25:215–234. doi: 10.1002/mas.20067. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Sun Y, Opal P, Antalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Giraud-Panis M-J, Jaco I, Amé J-C, Schultz I, Blasco M, Koering C-El, Gilson E, Murcia JM, Murcia G, Schreiber V. Functional Interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias P-T, Christoforidis S, Brunk UT, Galaris D. Endosomal and lysosomal effects of desferrioxamine: protection of HeLa cells from hydrogen peroxide-induced DNA damage and induction of cell-cycle arrest. Free Radic Biol Med. 2003;35:719–728. doi: 10.1016/S0891-5849(03)00396-4. [DOI] [PubMed] [Google Scholar]
- Doulias P-T, Kotoglou P, Tenopoulou M, Keramisanou D, Tzavaras T, Brunk U, Galaris D, Angelidis C. Involvement of heat shock protein-70 in the mechanism of hydrogen peroxide-induced DNA damage: the role of lysosomes and iron. Free Radic Biol Med. 2007;42:567–577. doi: 10.1016/j.freeradbiomed.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Dressel R, Johnson JP, Gunther E. Heterogeneous patterns of constitutive and heat shock induced expression of HLA-linked HSP70-1 and HSP70-2 heat shock genes in human melanoma cell lines. Melanoma Res. 1998;8:482–492. doi: 10.1097/00008390-199812000-00002. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three dimensional structure of the ATPase fragment of a 70 K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Gilbert CS, Bosch M, Green MC, Vialard JE, Grenon M, Erdjument-Bromage H, Tempst P, Lowndes NF. The budding yeast Rad9 checkpoint complex: chaperone proteins are required for its function. EMBO Rep. 2003;4:953–958. doi: 10.1038/sj.embor.embor935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Lieber RM. DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains. Curr Biol. 1998;8:873–879. doi: 10.1016/S0960-9822(07)00349-1. [DOI] [PubMed] [Google Scholar]
- Gwack Y, Nakamura H, Lee SH, Souvlis J, Yustein JT, Gygi S, Kung HJ, Jung JU. Poly(ADP-Ribose)polymerase I and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol Cell Biol. 2003;23:8282–8294. doi: 10.1128/MCB.23.22.8282-8294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang H, Fox MH. Expression of hsp70 induced in CHO cells by 45°C hyperthermia is cell cycle associated and DNA synthesis dependent. Cytometry. 1995;19:119. doi: 10.1002/cyto.990190206. [DOI] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- Huang H-C, Sherman My, Kandror O, Goldberg AL. The molecular chaperone DnaJ is required for the degradation of a soluble abnormal protein in E. coli. J Biol Chem. 2001;276:3920–3926. doi: 10.1074/jbc.M002937200. [DOI] [PubMed] [Google Scholar]
- Huyton T, Bates PA, Zhang X, Sternberg MJE, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat Res. 2000;460:319–332. doi: 10.1016/s0921-8777(00)00034-3. [DOI] [PubMed] [Google Scholar]
- Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill DM, Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J Biol Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, Theodoropoulos PA, Singh PB, Georgatos SD. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem. 2004;279:25567–25573. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, deMurcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez F, Kozin E, Bases R. Heat shock protein 70 stimulation of the deoxyribonucleic acid base excision repair enzyme polymerase beta. Cell Stress Chaperones. 2003;8:153–161. doi: 10.1379/1466-1268(2003)008<0153:HSPSOT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milarski KL, Morimoto RI. Expression of human Hsp70 during the synthetic phase of the cell cycle. Proc Natl Acad Sci U S A. 1986;83:9517–9521. doi: 10.1073/pnas.83.24.9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milarski KL, Morimoto RI. Mutational analysis of the human hsp70 protein: distinct domains for nucleolar localization and ATP-binding. J Cell Biol. 1989;109:1947–1962. doi: 10.1083/jcb.109.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Hohfeld J, Ohtsuka K, Hartl F-U. Regulation of the heat shock protein 70 reaction cycle by the mammalian DnaJ homolog, hsp40. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.5.2641. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Tissières A, Georgopoulos C. The stress response, functions of the proteins, and perspectives. In: Morimoto RI, Tissières A, Georgopoulus C, editors. Stress proteins in biology and medicine. New York: Cold Spring Harbor Laboratory Press; 1990. pp. 1–36. [Google Scholar]
- Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–2918. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Onishi T. Isolation and purification of nucleoli and nucleolar chromatin from mammalian cells. Methods Cell Biol. 1978;17:141–161. doi: 10.1016/S0091-679X(08)61142-5. [DOI] [PubMed] [Google Scholar]
- Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair. 2007;6:254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Niu P, Liu L, Gong Z, Tan H, Wang F, Yuan J, Feng Y, Wei Q, Tangway RM, Wu T. Overexpressed heat shock protein 70 protects cells against DNA damage caused by ultraviolet C in a dose-dependent manner. Cell Stress Chaperones. 2006;11:162–169. doi: 10.1379/CSC-175R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Salomons FA, Brunsting JF, Want JJ, Sibon OC, Kampinga HH. Dynamic changes in the localization of thermally unfolded nuclear proteins associated with chaperone-dependent protection. Proc Natl Acad Sci U S A. 2001;98:12038–12043. doi: 10.1073/pnas.201112398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL. Methods used to study structure and function of the nucleolus. Methods Cell Biol. 1998;53:303–321. doi: 10.1016/S0091-679X(08)60884-5. [DOI] [PubMed] [Google Scholar]
- Plumier J-CL, Ross BM, Currie RW, Angelidis CH, Kazlaris H, Kollias G, Pagoulatos GN. Transgenic mice expressing the human hsp70 have improved post-ischemic myocardial recovery. J Clin Invest. 1995;95:1854–1860. doi: 10.1172/JCI117865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MV, Gangadharam PRJ. Heat shock treatment of macrophages causes increased release of superoxide anion. Infect Immun. 1992;60:2386–2390. doi: 10.1128/iai.60.6.2386-2390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA. Holding damaged DNA together. Nat Struct Biol. 1999;6:805–806. doi: 10.1038/12257. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- Sainis I, Angelidis C, Pagoulatos GN, Lazardis I. Hsc70 interactions with SV40 viral proteins differ between per missive and non-permissive mammalian cells. Cell Stress Chaperones. 2000;5:132–138. doi: 10.1379/1466-1268(2000)005<0132:HIWSVP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- Salma A, Tsiapos A, Lazaridis I. The viral SV40 T antigen cooperates with dj2 to enhance hsc70 chaperone function. FEBS J. 2007;274:5021–5027. doi: 10.1111/j.1742-4658.2007.06019.x. [DOI] [PubMed] [Google Scholar]
- Samali A, Cotter TG. Heat shock proteins increase resistance to apoptosis. Exp Cell Res. 1996;223:163–170. doi: 10.1006/excr.1996.0070. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TM. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schreiber V, Dantzer F, Ame J-C, Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Scott MD, Frydman J. Aberrant protein folding as the molecular basis of cancer. Methods Mol Biol. 2003;232:67–76. doi: 10.1385/1-59259-394-1:67. [DOI] [PubMed] [Google Scholar]
- Wang TF, Chang JH, Wang C. Identification of the peptide binding domain of hsc70. 18-Kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J Biol Chem. 1993;268:26049–26051. [PubMed] [Google Scholar]
- Welch WJ, Feramisco JR. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. J Biol Chem. 1984;259:4501–4513. [PubMed] [Google Scholar]
- Zhang X, Morena S, Bates PA, Whitehead C, Coffer AI, Hainbucher K, Nash R, Sternbera JE, Lindahk T, Freemont PS. Structure of an XRCC1 BRCT domain: a new protein–protein interaction module. EMBO J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]