Abstract
MicroRNAs comprise a novel class of endogenous, small, noncoding RNAs that negatively regulate gene expression. Functionally, an individual miRNA is as important as a transcription factor because it is able to regulate the expression of its multiple target genes. Recently, miR-221 and miR-222 have been found to play a critical role in cancer cell proliferation. However, their roles in vascular smooth muscle cell (VSMC) biology are currently unknown. In the current study, the time course changes and cellular distribution of miR-221 and miR-222 expression were identified in rat carotid arteries after angioplasty, in which their expression was upregulated and localized in VSMCs in the injured vascular walls. In cultured VSMCs, miR-221 and miR-222 expression was increased by growth stimulators. Knockdown of miR-221 and miR-222 resulted in decreased VSMC proliferation in vitro. Using both gain-of-function and loss-of-function approaches, we found that p27(Kip1) and p57(Kip2) were two target genes that were involved in miR-221 and miR-222-mediated effect on VSMC growth. Finally, knockdown of miR-221 and miR-222 in rat carotid arteries suppressed VSMC proliferation in vivo and neointimal lesion formation after angioplasty. The results indicate that miR-221 and miR-222 are novel regulators for VSMC proliferation and neointimal hyperplasia. These findings may also represent promising therapeutic targets in proliferative vascular diseases.
Keywords: MicroRNAs, vascular smooth muscle cells, gene regulation, proliferation, vascular disease
Introduction
Recently, the most significant breakthrough regarding gene expression regulation has been the discovery of microRNAs (miRNAs) (1). miRNAs comprise a novel class of endogenous, small, non-coding RNAs that negatively regulate gene expression via degradation or translational inhibition of their target mRNAs (1–5). More importantly, one miRNA is able to regulate the expression of multiple genes because it can bind to its mRNA targets as either an imperfect or perfect complementarity. Thus, one miRNA is as functionally important as a transcription factor (6). Over 700 miRNAs have been identified and sequenced in humans (7), and the estimated number of miRNA genes is as high as 1000 in the human genome (8). As a group, miRNAs may directly regulate at least 30% of the genes in the human genome (9). It is not surprising that miRNAs are involved in the regulation of almost all major cellular functions, such as cell differentiation, proliferation/growth, mobility and apoptosis (1). For that reason, miRNAs could be the pivotal regulators in normal development, physiology, as well as disease development including cancer and cardiovascular disease (10–13).
The biological function of an individual miRNA is cell specific. One miRNA may have different cellular effects on different cells. For example, miR-21 has an anti-apoptotic effect on glioblastoma cells, but increases Hela cell apoptosis (14, 15). Recent studies have revealed that miR-221 and miR-222 are upregulated in cancer cells (16–21). Both miR-221 and miR-222 have a pro-proliferative effect on cancer cells (16–21). It is well established that aberrant proliferation of vascular smooth muscle cells (VSMCs) is a key cellular event in the pathogenesis of a variety of proliferative vascular diseases (22). In our recent study, we have demonstrated via microarray analysis that miR-221 and miR-222 are upregulated in vascular walls with neointimal lesion formation (23). However, their roles in VSMC biology are currently unknown. The objective of the current study is to establish the roles miR-221 and miR-222 in VSMC growth and neointimal formation and their molecular mechanisms using both cultured VSMCs in vitro and balloon-injured rat carotid arteries in vivo.
Materials and Methods
An expanded Materials and Methods section is provided in the online data supplement at http://circres.ahajournals.org.
Rat carotid artery balloon injury model
Carotid artery balloon injury was induced in male Sprague-Dawley rats (250 to 300 g) as described (23).
Local oligo delivery into the injured vascular walls
miR-221 and miR-222 were downregulated in the balloon-injured rat carotid artery via miR-222 and miR-221 inhibitor (2'OMe-miR-222) using an established local oligo delivery model with little modification (23).
Morphometric analysis for neointimal lesion formation
Morphometric analysis was performed in carotid artery sections stained with Masson's trichrome staining as described (23).
Immunofluorescence of proliferating cell nuclear antigen (PCAN) and smooth muscle (SM) α-actin
Proliferating cells were evaluated in vessel sections by using the immunofluorescence of PCAN. SM α-actin was detected by immunofluorescence.
Cell culture
VSMCs and endothelial cells were obtained from rat aortas (23).
Oligo transfection, miR-221 and miR-222 knockdown, miR-221 and miR-222 overexpression, and p27(Kip1) and p57(Kip2) gene knockdown in cultured VSMCs
Oligo transfection, miR-221 and miR-222 knockdown, miR-221 and miR-222 overexpression, and p27(Kip1) and p57(Kip2) gene knockdown in cultured VSMCs were performed as described in the expanded Materials and Methods.
VSMC Proliferation in vitro
VSMC proliferation in vitro was determined by cell counting and bromodeoxyuridine (BrdU) incorporation assay (23).
RNA levels were determined by qRT-PCR
miRNAs and mRNAs were determined by qRT-PCR as described (23). The sequences of the primers used are shown in online Table I.
Detection of miR-221 and miR-222 in vessel sections by fluorescence in situ hybridization
In situ hybridizations (ISH) of miR-221 and miR-222 and Co-immunofluorescence with the smooth muscle marker SM α-actin were performed in 10-µm vessel sections (24, 25).
Western blot analysis
Proteins were isolated from cultured VSMCs and carotid arteries and protein levels were determined by western blot analysis.
Construction of the adenoviruses
The adenoviruses expressing miR-221 (Ad-miR-221) or miR-222 (Ad-miR-222), and control viruses expressing GFP (Ad-GFP), miR-31 (Ad-miR-31) or miR-145 (Ad-miR-145) were generated using the Adeno-X™ Expression Systems 2 kit (Clontech, CA) according to the manufacturer’s protocols.
Luciferase assay
A construct in which a fragment of the 3’-UTR of p27(kip1) or p57(kip2) mRNA containing the putative or mutated miR-221 and miR-222 binding sequence was used.
Results
The expression of miR-221 and miR-222 are increased in vascular walls with neointimal lesion formation after angioplasty
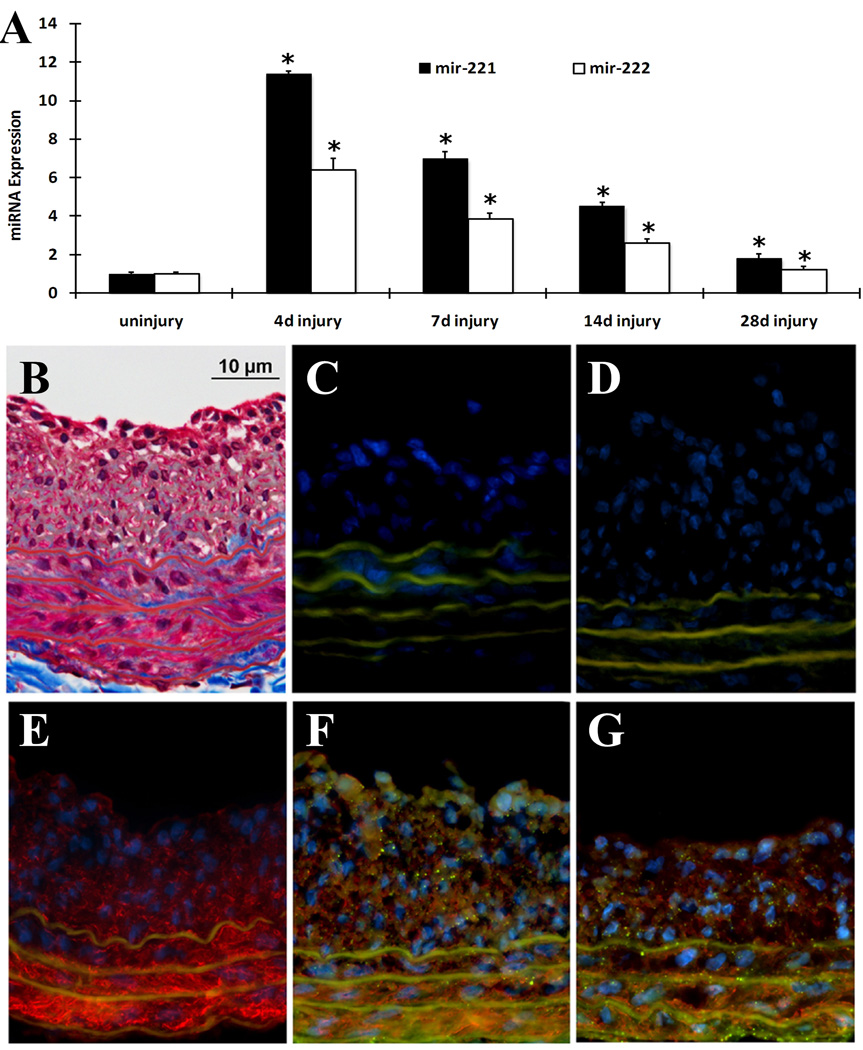
In the current study, the upregulation of miR-221 and miR-222 was verified by qRT-PCR and the time course expression changes of both miRNAs in carotid arteries after angioplasty were displayed in Fig. 1A. Undoubtedly, both miR-221 and miR-222 were upregulated in the balloon-injured arteries. To further confirm the upregulation of miR-221 and miR-222 was unique for injured arteries, we also measured the expression of several miRNAs that did not change based on the microarray data. We found via qRT-PCR that the expression some miRNAs such as miR-152, miR-181C and miR-142 did not change after balloon-injury (Online Figure I).
Fig. 1. Expression and distribution of miR-221 and miR-222 in balloon-injured rat carotid arteries.
(A). The time course changes of miR-221 and miR-222 expression determined by qRT-PCR. Note: n=6; *P<0.05 compared with uninjured control. (B). Representative Masson's trichrome staining. (C) Negative control (no SM α-actin antibody, no miRNA probe) for In situ hybridization and immunofluorescence. (D) Scrambled probe control 1 (no SM α-actin antibody, but had scrambled miRNA probe). (E) Scrambled probe control 2 (had SM α-actin antibody and scrambled miRNA probe). (F) In situ hybridization of miR-221 (dot green color), immunofluorescence of smooth muscle cell marker SM α–actin (red color) and cell nuclear staining by DAPI (blue color). (G) In situ hybridization of miR-222 (dot green color), immunofluorescence of smooth muscle cell marker SM α–actin (red color) and cell nuclear staining by DAPI (blue color). Note: Autofluorescence in the elastic laminae is demonstrated as green color, but is not dot green (C–G).
Both miR-221 and miR-222 are localized in VSMCs of the vascular wall
To determine the distribution of miR-221 and miR-222 expression in the vascular wall, in situ hybridization was performed on the rat carotid arteries. Vessel structure was demonstrated via Masson's trichrome staining in frozen sections of balloon-injured rat carotid arteries as shown in Fig. 1B. In situ hybridization of miR-221 (Fig. 1F) and miR-222 (Fig. 1G) showed that it was expressed in the vessel media and neointima where VSMCs were localized. To confirm that VSMC contained miR-221 and miR-222, co-immunofluorescence with the smooth muscle marker SM α-actin was performed. As expected, SM α-actin was observed in VSMCs that were located in the media and neointima (Fig. 1E–G). In addition, both miR-221 (Fig. 1F) and miR-222 (Fig. 1G) were clearly co-localized with VSMCs. In contrast, there was no miR-221 and miR-222 expression in 3 control sections: negative control (Fig. 1C, no SM α-actin antibody, no miRNA probe), scrambled probe control 1 (Fig. 1D, no SM α-actin antibody, but had scrambled miRNA probe), and scrambled probe control 2 (Fig. 1E, had SM α-actin antibody and scrambled miRNA probe).
It should be noted that we performed in situ staining in balloon-injured vessel sections, in which endothelial cells were removed by balloon-catheter during the injury. To further confirm the cellular distribution, we had also determined the levels of miR-221 and miR-222 in isolated rat aortic VSMCs and endothelial cells using qRT-PCT. As shown in online Figure II, miR-221 and miR-222 were also expressed in endothelial cells, even though the expression levels were a little lower than those in VSMCs.
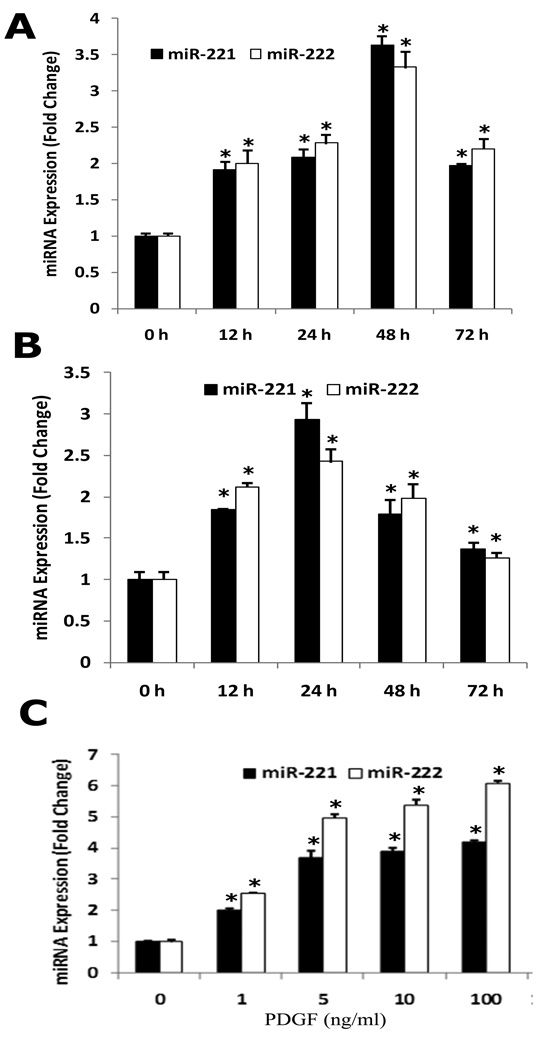
The expression of miR-221 and miR-222 are increased in proliferative VSMCs stimulated by either platelet-derived growth factor (PDGF) or serum
To investigate the potential link between miR-221, miR-222 and the VSMC proliferation, the expression of miR-221 and miR-222 was determined in non-proliferative VSMCs and in proliferative VSMCs stimulated by either PDGF-BB ( 20 ng/ml) or 10% serum. As shown in Fig. 2A & 2B, the expression of miR-221 and miR-222 in both PDGF-BB and serum-treated VSMCs was significantly higher than that in vehicle-treated VSMCs, which was time dependent. The dose-dependent response in miR-221 and miR-222 expression to PDGF was measured in cultured VSMCs after 48h treatment with different concentrations of PDGF and the result was shown in Fig. 2C. These results demonstrated that the expression of miR-221 and miR-222 was upregulated in proliferating VSMCs.
Fig. 2. The effect of platelet-derived growth factor (PDGF) and serum on the expression of miR-221 and miR-222 in cultured rat VSMCs.
(A) PDGF-BB (20 ng/ml) caused a time-dependent increase in miR-221 and miR-222 expression as demonstrated by qRT-PCR. (B) Serum (10%) caused a time-dependent increase in miR-221 and miR-222 expression as demonstrated by qRT-PCR. (C) PDGF-BB caused a dose-dependent increase in miR-221 and miR-222 expression in cultured VSMCs at 48 h after treatment. Note: n=6; *P<0.05 compared with 0 h groups in Fig. 2A & B, and with vehicle control (0) in Fig. 2C.
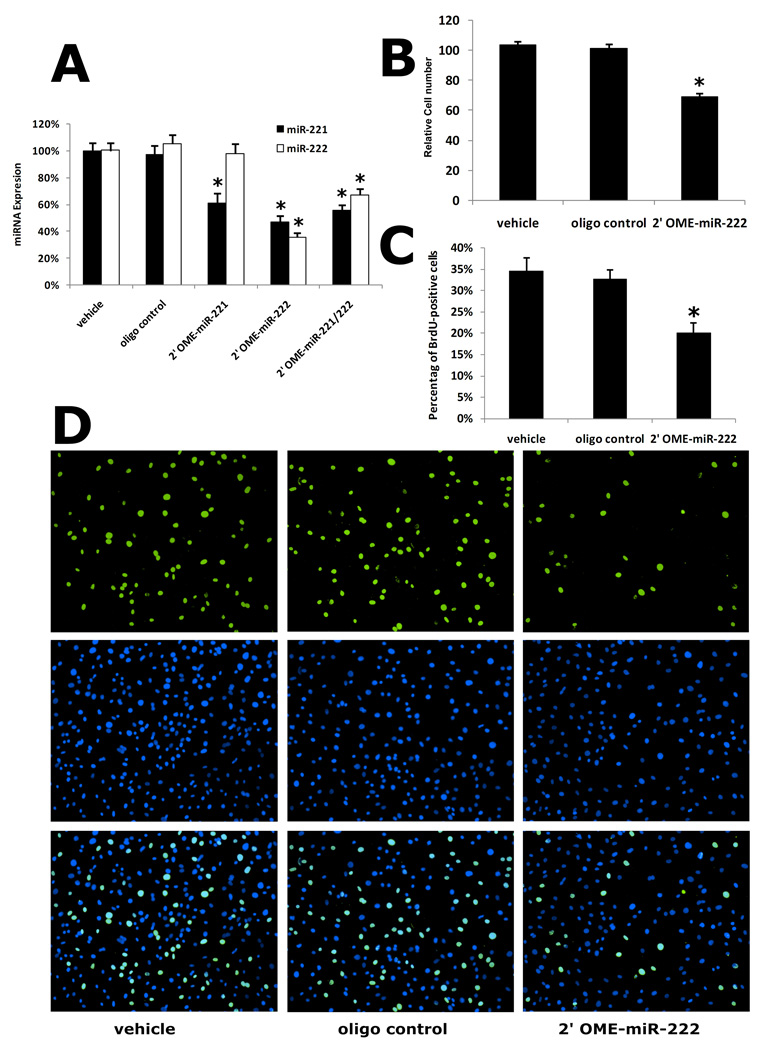
Knocking down of miR-221 and miR-222 inhibits proliferation of cultured VSMCs
To further determine the potential roles of miR-221 and miR-222 in VSMC proliferation, we applied antisense oligonucleotide-mediated miRNA depletion to knock down the miR-221 and miR-222. After 48 h treatment, miR-221 inhibitor (2'OMe-miR-221) inhibited miR-221 expression but had no effect on miR-222 expression (Fig. 3A). However, miR-222 inhibitor (2'OMe-miR-222) inhibited the expression of both miR-221 and miR-222 (Fig. 3A). Addition of both miR-221 and miR-222 inhibitors could not give additional inhibition on the expression of miR-221 and miR-222 compared with 2'OMe-miR-222 only, although the reason was unclear (Fig. 3A). We therefore used 2'OMe-miR-222 as the inhibitor for both miR-221 and miR-222.
Fig. 3. The effect of miR-221 and miR-222 inhibitor on VSMC proliferation in vitro.
(A) Knocking down of miR-221 and miR-222 expression by their inhibitors, 2'OMe-miR-221 (100 nM), 2'OMe-miR-222 (100 nM), and 2'OMe-miR-221 plus 2'OMe-miR-222 (100 nM). (B) 2'OMe-miR-222 (100 nM) decreased cell numbers and (C) BrdU incorporation at 48 h after culture with DMEM containing 10% FBS. (D) Representative BrdU-stained cell photomicrographs (top panel), their corresponding total cell photomicrographs stained by DAPI (medial panel) and merged photomicrographs (bottom panel). Note: n=8; *P<0.05 compared with vehicle control.
In subsequent experiments, we determined the effect of 2'OMe-miR-222 on VSMC proliferation by using two different methods: cell counting and BrdU incorporation assay as described (23). Consistent with the levels of miR-221 and 222 in Fig. 3A, 2'OMe-miR-222 significantly decreased cell numbers and BrdU incorporation at 48 h after culture with DMEM containing 10% FBS (Fig. 3B and 3C). Representative BrdU-stained cell photomicrographs (top panel), their corresponding total cell photomicrographs stained by DAPI (medial panel) and merged photomicrographs (bottom panel) were shown in Fig. 3D. As we found that miR-221 inhibitor, 2'OMe-miR-221 could only inhibit miR-221, we determined the effect of miR-221 inhibition on VSMC proliferation. We found that miR-221 inhibition also inhibited VSMC proliferation. However, the inhibitory effect on VSMC proliferation is smaller than that in both miR-221 and miR-222-inhibited cells via 2'OMe-miR-222 (online Figure III). The results indicated that both miR-2221 and miR-222 had a pro-proliferative effect on cultured VSMCs. In contrast, control oligo had no effect on VSMC proliferation.
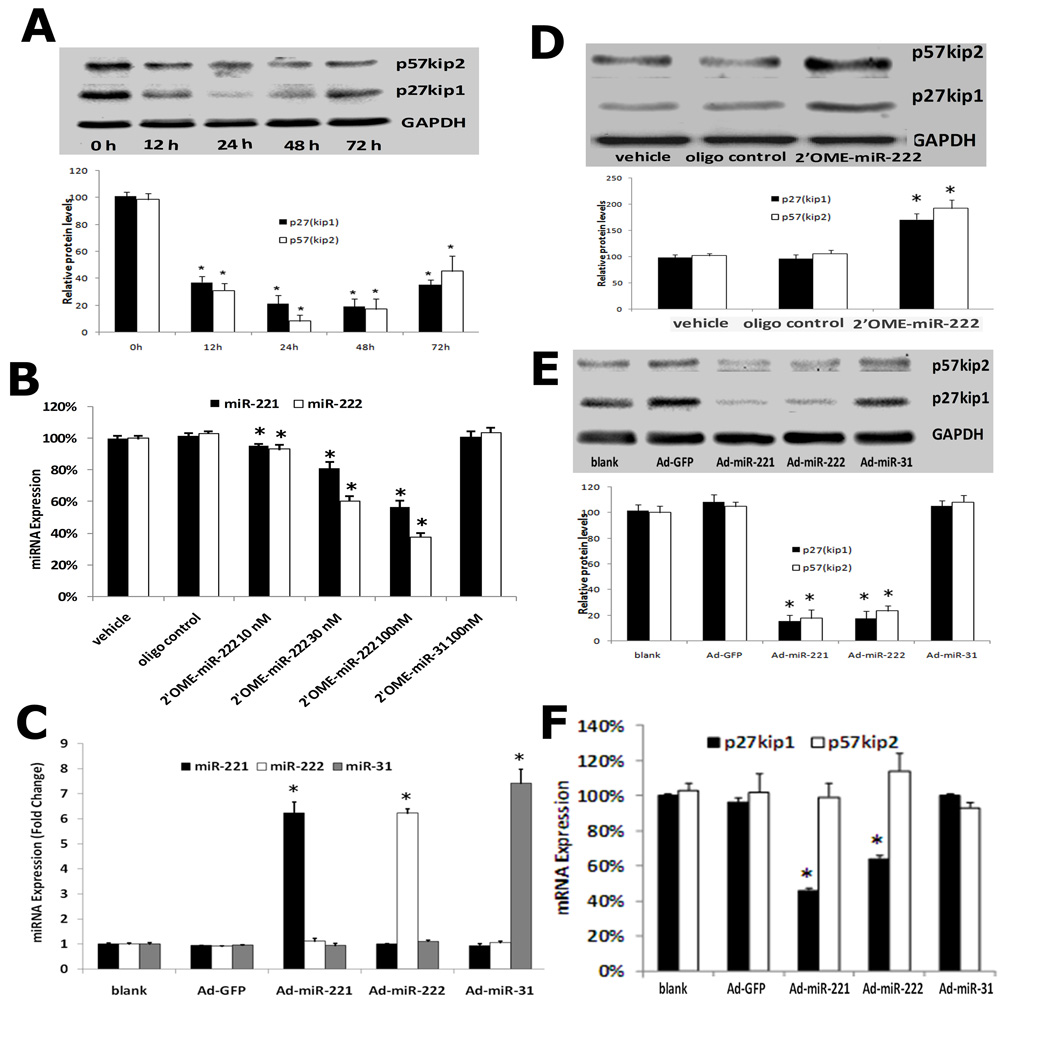
p27(Kip1) and p57(Kip2) are target genes of miR-221 and miR-222 in VSMCs
Computational analysis from previous studies have suggested that p27(Kip1) and p57(Kip2) are two potential target genes of miR-221 and miR-222 (16–21). Online Figure IV shows that both p27(Kip1) and p57(Kip2) have miR-221 and miR-222 binding sites in their 3'-untranslated region (3'-UTR).
If p27(Kip1) and p57(Kip2) are the target genes of miR-221 and miR-222, their expression should be downregulated in proliferative VSMCs because both miR-221 and miR-222 are upregulated in these cells. Indeed, as shown in Fig. 4A (top panel), both p27(Kip1) and p57(Kip2) were downregulated in proliferative VSMCs stimulated by PDGF-BB (20 ng/ml). Quantification of p27(Kip1) and p57(Kip2) protein levels was shown in Fig. 4A (bottom panel).
Fig. 4. p27(Kip1) and p57(Kip2) are target genes of miR-221 and miR-222 in cultured VSMCs.
(A) p27(Kip1) and p57(Kip2) were downregulated in proliferative VSMCs stimulated with PDGF-BB (20 ng/ml). Note: Top panel was the representative western blot and bottom panel was the quantification of p27(Kip1) and p57(Kip2) protein. n=3; *P<0.05 compared with 0 h group. (B) Treatment with 2'OMe-miR-222 for 48 h decreased the expression of miR-221 and miR-222 in cultured VSMCs. In contrast, control oligo or unrelated miRNA inhibitor had no effect on miR-221 and miR-222 expression. Note: n=3; *P<0.05 compared with vehicle group. (C) Ad-miR-221 and Ad-miR-222 increased the expression of miR-221 and miR-222 in cultured VSMCs. In contrast, control adenovirus, Ad-GFP or unrelated adenovirus control, Ad-miR-31 had no effect on the expression of miR-221 and miR-222. Note: n=6; *P<0.05 compared with vehicle (blank) group. (D) p27(Kip1) and p57(Kip2) protein levels were upregulated by 2'OMe-miR-222. Note: Top panel was the representative western blot and bottom panel was the quantification of p27(Kip1) and p57(Kip2) protein. n=3; *P<0.05 compared with vehicle group. (E) p27(Kip1) and p57(Kip2) protein levels were down regulated by Ad-miR-221 or Ad-miR-222. Note: Top panel was the representative western blot and bottom panel was the quantification of p27(Kip1) and p57(Kip2) protein. Note: n=3; *P<0.05 compared with vehicle (blank) group. (F). The effects of miR-221 or miR-222 overexpression by Ad-miR-221 or Ad-miR-222 on mRNA expression of p27(Kip1) and p57(Kip2). Note: n=3; *P<0.05 compared with vehicle (blank) group.
To verify that p27(Kip1) and p57(Kip2) are the target genes of miR-221 and miR-222, both gain-of-function and loss-of-function approaches were applied. As shown in Fig. 4, 2'OMe-miR-222 decreased (Fig. 4B), whereas Ad-miR-221 and Ad-miR-222 increased (Fig. 4C) the expression of miR-221 and miR-222 in cultured VSMCs. In contrast, unrelated adenovirus control, Ad-miR-31 had no effect on the expression of miR-221 and miR-222 (Fig. 4C). As expected, p27(Kip1) and p57(Kip2) were upregulated by 2'OMe-miR-222 (Fig. 4D) and were downregulated by Ad-miR-221 or Ad-miR-222, but not by unrelated adenovirus, Ad-miR-31 (Fig. 4E) at protein levels. Furthermore, overexpression of miR-221 or miR-222 by Ad-miR-221 or Ad-miR-222 decreased p27(Kip1) mRNA level, but had no effect on p57(Kip2) mRNA level (Fig. 4F).
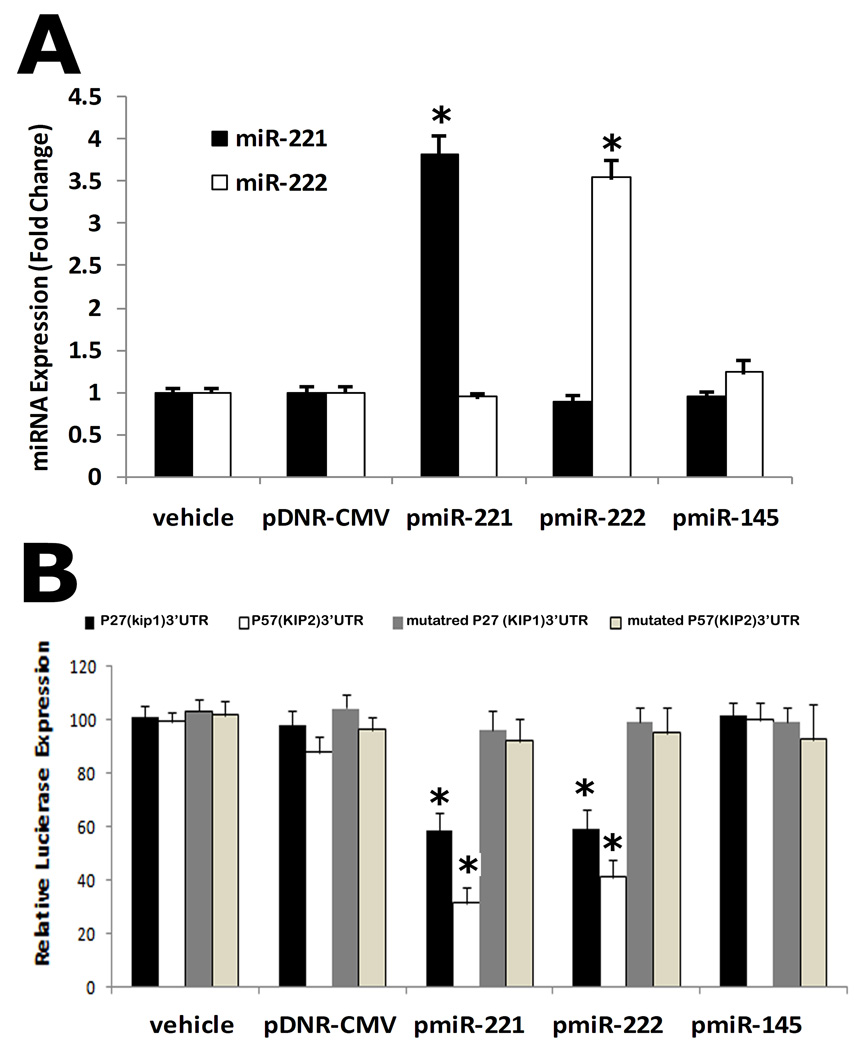
To further confirm that miR-221 and miR-222 are able to directly bind to p27(Kip1) and p57(Kip2) and inhibit their expression, a construct in which a fragment of the 3’-UTR of either p27(Kip1) or p57(Kip2) mRNA containing the putative miR-221 and miR-222 binding sequences was cloned into a firefly luciferase reporter construct and transfected into HEK 293 cells with either vehicle, an empty plasmid (pDNR-CMV), a plasmid expressing miR-221 (pmiR-221), miR-222 (pmiR-222), or a control plasmid expressing an unrelated miRNA, miR-145 (pmiR-145), following the transfection procedures provided by Invitrogen. The constructs with mutated fragment of the 3’-UTR of either p27(Kip1) or p57(Kip2) mRNA without the putative miR-221 and miR-222 binding sequences were used as mutated controls. As expected, pmiR-221 and miR-222, but not pmiR-145 and pDNR-CMV, increased miR-221 or miR-222 expression in HEK 293 cells (Fig. 5A). Consequently, pmiR-221 and pmiR-222, but not pDNR-CMV or pmiR-145 inhibited luciferase activity (Fig. 5B). In the mutated control groups, the inhibitory effect of pmiR-221 and miR-222 was disappear (Fig. 5B). The results suggested that miR-221 and miR-222 were able to bind to p27(Kip1) and p57(Kip2) directly and inhibit their expression.
Fig. 5. miR-221 and miR-222 are able to directly bind to p27(Kip1) and p57(Kip2) and inhibit their expression in HEK 293 cells.
A construct in which a fragment of the 3’-UTR of either p27(Kip1) or p57(Kip2) mRNA containing the putative miR-221 and miR-222 binding sequences was cloned into a firefly luciferase reporter construct and transfected into HEK 293 cells with either vehicle (vehicle), an empty plasmid (pDNR-CMV), a plasmid expressing miR-221 (pmiR-221), miR-222 (pmiR-222), or a control plasmid expressing an unrelated miRNA, miR-145 (pmiR-145). The constructs with mutated fragment of the 3’-UTR of either p27(Kip1) or p57(Kip2) mRNA without the putative miR-221 and miR-222 binding sequences were used as mutated controls. (A) pmiR-221 and pmiR-222, but not pmiR-145 and pDNR-CMV, increased miR-221 or miR-222 expression in HEK 293 cells. (B) pmiR-221 and pmiR-222, but not pDNR-CMV or pmiR-145 inhibited luciferase activity. In the mutated control groups, the inhibitory effect of pmiR-221 and pmiR-222 was disappear. Note; N=5; *P<0.05 compared with vehicle control.
p27(Kip1) and p57(Kip2) are involved in miR-221 and miR-222-mediated effect on VSMC proliferation
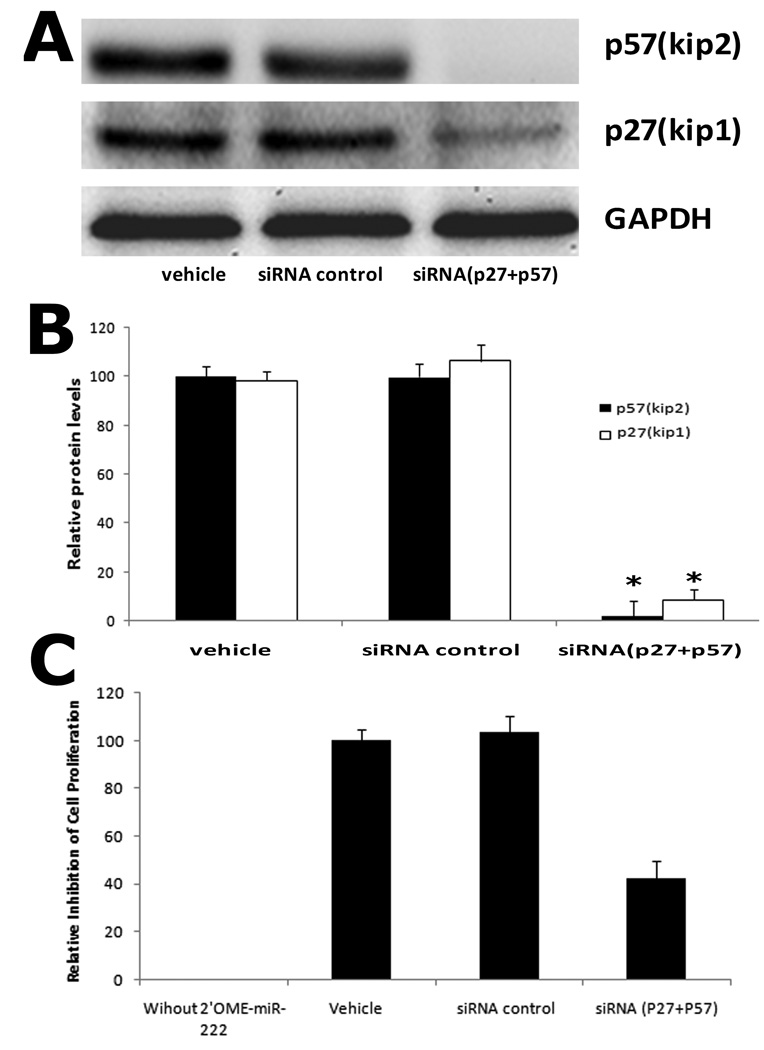
As shown in Fig. 6A and 6B, both p27(Kip1) and p57(Kip2) were depleted by their siRNAs. We next determined the involvement of p27(Kip1) and p57(Kip2) in 2'OMe-miR-222 induced inhibitory effect on proliferation. In this experiment, We defined that the relative inhibition of cells without 2'OMe-miR-222 as 0%, while in 2'OMe-miR-222 plus vehicle-treated cells as 100% inhibition. As shown in Fig. 6C, in the target gene-depleted VSMCs, 2'OMe-miR-222-induced inhibitory effect on proliferation was decreased compared with that in VSMCs without target gene depletion. The results suggested that p27(Kip1) and p57(Kip2) were functional target genes of miR-221 and miR-222 that were involved in miR-221 and miR-222-mediated effect on VSMC proliferation.
Fig. 6. miR-221 and miR-222-mediated effect on VSMC proliferation is decreased in p27(Kip1) and p57(Kip2) deficient cells.
(A) Representative western blot of p27(Kip1) and p57(Kip2) protein. (B) p27(Kip1) and p57(Kip2) expression at protein level was depleted by their siRNAs (25 nM). Note: n=6; *P<0.05 compared with vehicle control. (C) miR-221 and miR-222 inhibitor, 2'OMe-miR-222-mediated inhibitory effect on VSMC proliferation was decreased in p27(Kip1) and p57(Kip2) depleted cells. Note: The relative inhibition of cells without 2'OMe-miR-222 was defined as 0%, while in 2'OMe-miR-222 plus vehicle-treated cells was defined as 100% inhibition. N=8; *P<0.05 compared with vehicle control.
p27(Kip1) and p57(Kip2) are target genes of miR-221 and miR-222 in vascular walls after angioplasty
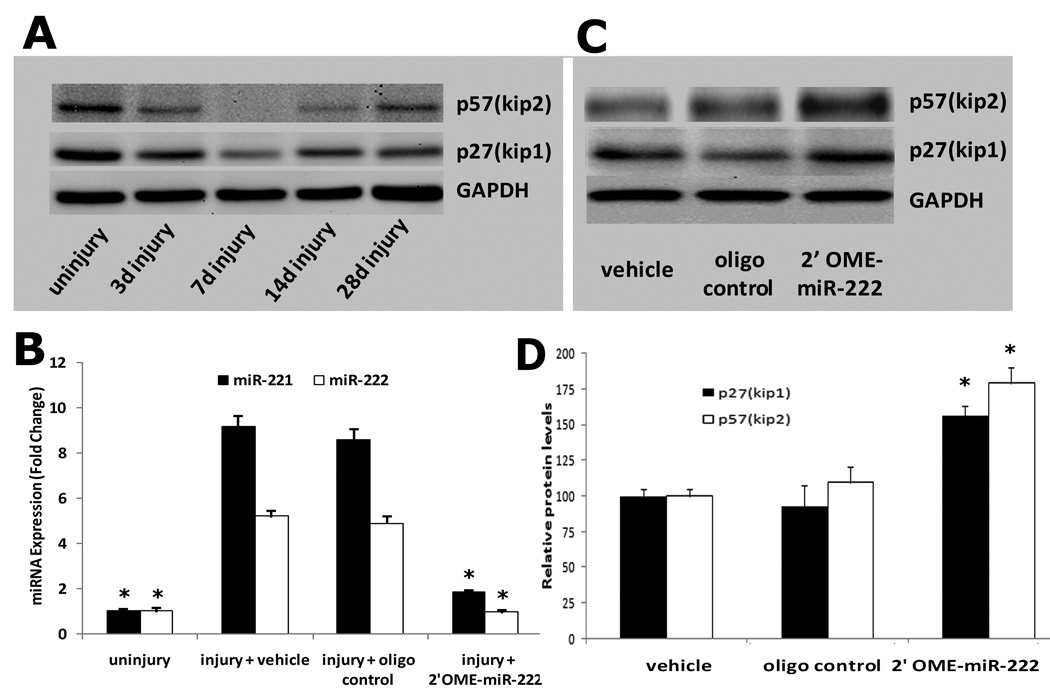
As shown in Fig. 7A, both p27(Kip1) and p57(Kip2) were down regulated in balloon-injured rat carotid arteries. The negative relationship with the expression of miR-221 and miR-222 indicated that p27(Kip1) and p57(Kip2) could be miR-221 and miR-222 target genes in vivo. To further verify this, the expression of miR-221 and miR-222 in balloon-injured rat carotid arteries was modulated by their inhibitor, 2'OMe-miR-222. As shown in Fig. 7B, miR-221 and miR-222 were downregulated by 2'OMe-miR-222. Interestingly, in 2'OMe-miR-222-treated vessels, expression of p27(Kip1) and p57(Kip2) was upregulated (Fig. 7C). The results demonstrated that p27(Kip1) and p57(Kip2) were target genes of miR-221 and miR-222 in vivo.
Fig. 7. p27(Kip1) and p57(Kip2) are target genes of miR-221 and miR-222 in the vascular walls after angioplasty.
A. Both p27(Kip1) and p57(Kip2) proteins were downregulated in balloon-injured rat carotid arteries. (B) miR-221 and miR-222 were downregulated by 2'OMe-miR-222 in balloon-injured vascular walls. (C) Representative western blot of p27(Kip1) and p57(Kip2) protein in the injured vascular walls treated with vehicle, control oligo or 2'OMe-miR-222. (D) In 2'OMe-miR-222-treated vessels, expression of p27(Kip1) and p57(Kip2) was upregulated. Note; N=6; *P<0.05 compared with vehicle control.
Downregulation of miR-221 and miR-222 decreases cell proliferation and neointima formation in rat carotid artery after angioplasty
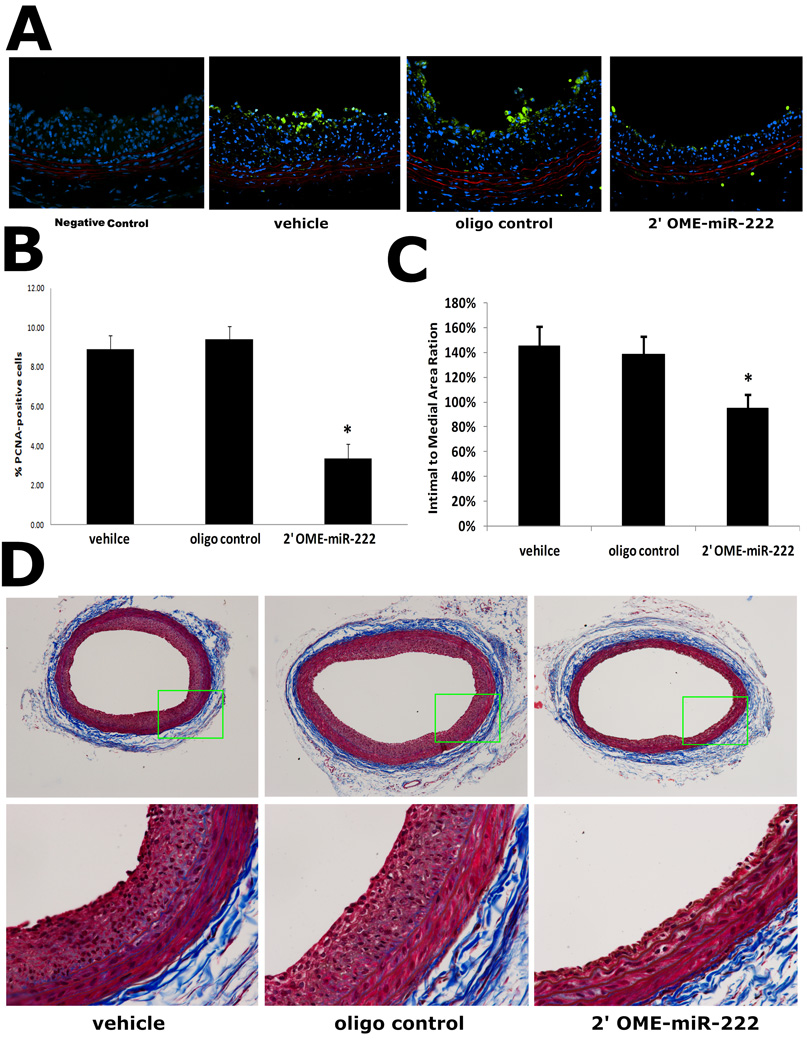
To determine the effects of miR-221 and miR-222 on VSMC proliferation and neointimal growth in vivo, the carotid arteries were isolated 14 days after balloon-injury. Representative immunofluorescence of PCAN and its negative control was shown in Fig. 8A. Compared with vehicle-treated vessel, fewer cells were proliferating in the injured vascular walls treated with 2'OMe-miR-222 (Fig. 8B). Furthermore, downregulation of the miR-221 and miR-222 resulted in nearly 40% decrease in neointima formation after angioplasty (Fig.8C). Representative Masson's trichrome stained photomicrographs of rat carotid arteries from different groups were shown in Fig. 8D.
Fig. 8. Downregulation of miR-221 and miR-222 decreases cell proliferation and neointima formation in rat carotid artery after angioplasty.
(A) Representative immunofluorescence of PCNA in rat carotid arteries at 14 days after balloon-injury. Note: Green color was the immunofluorescence of PCNA that represented proliferating cells. Blue was cell nuclear staining by DAPI that reflects total cells. Red color was the autofluorescence in elastic laminae. (B) Quantification of the proliferative cells showed that, compared with vehicle-treated and control oligo-treated vessels, fewer cells were proliferating in the injured vascular walls treated with 2'OMe-miR-222. (C) Downregulation of the miR-221 and miR-222 by 2'OMe-miR-222 decreased neintimal formation. (D) Representative Masson's trichrome stained photomicrographs of rat carotid arteries from different groups. Note; N=8; *P<0.05 compared with vehicle control.
Discussion
miR-221 and miR-222 are encoded by a gene cluster on the X chromosome. They share the same seed and appear to have identical target genes and similar functions. Recent studies have revealed that miR-221 and miR-222 are overexpressed in several types of cancers, in which they are related to cancer cell proliferation (16–21). VSMC proliferation-mediated expansion of the neointima is a unifying facet of a variety of proliferative vascular diseases such as atherosclerosis, restenosis after angioplasty or bypass, diabetic vascular complication and transplantation arteriopathy. In our recent report, we have identified that miR-221 and miR-222 are two abundant miRNAs in the vascular wall, in which miR-221 is ranked at top 39 and miR-222 is ranked at top 50 among the 140 detected mature miRNAs (23). Interestingly, miR-221 and miR-222 are significantly upregulated in the vascular walls with neointimal lesion formation as determined by microarray analysis (23). However, the roles of miR-221 and miR-222 in VSMC proliferation and proliferative vascular disease are currently unknown.
In the current study, the upregulation of miR-221 and miR-222 is further verified by qRT-PCR. We have found that the increase in their expression is larger than that determined by microarray analysis. We think the different method of miRNA quantification could be the major reason for it. This opinion is also supported by a recent report from Miller et al (26). They have found that there is about a 2-fold increase in miR-221 and miR-222 expression in tamoxifen-resistant MCF cells compared with that in tamoxifen-sensitive MCF cells by microarray analysis. However, by qRT-PCR analysis, they have demonstrated that there is an over 10-fold increase in miR-221 and miR-222 expression in tamoxifen-resistant cells. qRT-PCR might be better than microarray when we quantify a specific miRNA. Another important reason is that the samples for microarray analysis and for qRT-PCR are from different group of rats. In addition, the rats used for macroarray data are older (≈320g) than these used for qRT-PCR (≈250g). The age might also be a potential reason for the difference. The effect of age on miRNA expression is also supported by recent reports. For example, miR-21 expression is significantly increased in neonatal rat cardiac myocytes compared with that in adult rat cardiac myocytes (27).
Another finding regarding miR-221 and miR-222 expression is that the extent of miR-221 and miR-222 upregulation in vivo in balloon-injured rat arteries is larger than that in vitro in cultured VSMCs stimulated with either PDGF or serum. The result indicates that there are multiple stimuli in vivo that could synergistically determine the up-regulation of miRNAs.
It is well established that tissue and cell-specific expression is one important characteristic of miRNA expression. One miRNA may be highly expressed in one type of cell, but has no or low expression in another type of cell. To investigate the role of miR-221 and miR-222 in VSMC biology, we initially determined their cellular distribution in the vascular walls to see if they were located in VSMCs. The results from in situ hybridization clearly showed that both miR-221 and miR-222 were localized in VSMCs of the injured vascular wall.
Cellular functions of a miRNA are cell-type specific. For example, miR-21 has an anti-apoptotic function in glioblastoma cell and VSMCs (14, 15, 23), but has no such a function in HeLa cells (15). In cultured cells, we found for the first time that miR-221 and miR-222 have a pro-proliferative effect on VSMCs. The cellular effect of miR-221 and miR-222 on VSMCs is consistent with that on cancer cells.
One surprising result from the study is that the inhibitor against miR-222 also suppresses miR-221 expression. To further confirm that the 2'OMe-miR-222-mediated inhibition is miR-221 and miR-222 specific, we have determined the effect of the miRNA inhibitor on other unrelated miRNAs such as miR-24 and miR-146. No inhibitory effect has been found on these unrelated miRNAs (online Figure V). Although the reason that 2'OMe-miR-222 can inhibit both miR-221 and miR-222 is not clear, we think the similarity of the sequence might explain the finding.
miRNAs have multiple mRNA targets that are responsible for miRNA-mediated biological functions. First, based on the pro-proliferative effect of miR-221 and miR-222 on VSMCs, we hypothesize that their target gene(s) might have negative effect on cell growth. p27(Kip1) and p57(Kip2) are tumor suppressors and cell cycle inhibitors (21) and might be the target genes of miR-221 and miR-222 based on recent cancer cell studies (16–21). Computational analysis for miRNA target prediction demonstrates that p27(Kip1) and p57(Kip2) have binding sites for both miR-221 and miR-222. The direct binding and inhibitory functions of miR-221 and miR-222 on p27(Kip1) and p57(Kip2) are validated in HEK 293 cells using luciferase constructs, in which the 3’-UTR fragments of these target gene mRNAs are included. To further confirm that p27(Kip1) and p57(Kip2) are target genes of miR-221 and miR-222 in VSMCs, both gain-of-function and loss-of-function approaches are applied. We have confirmed that both genes are targets of miR-221 and miR-222 in our experimental cells. Furthermore, both p27(Kip1) and p57(Kip2) are involved in miR-221 and miR-222-mediated effect on VSMC proliferation as the decreased inhibitory effect on VSMC proliferation is found in these gene deficient VSMCs. However, miR-221 and miR-222-mediated effect on cell proliferation is only partially inhibited in both p27(Kip1) and p57(Kip2) depleted cells, suggesting that other unidentified gene targets should be determined to completely understand their molecular mechanisms.
miRNA binds to complementary sites in the mRNA target to negatively regulate gene expression via degradation or translational inhibition of their target mRNAs. To further understand the detailed mechanisms of miR-221 and miR-222-mediated regulatory effect on expression of p27(Kip1) and p57(Kip2), the expression of the two target genes at mRNA level is determined. We have found that overexpression of miR-221 or miR-222 decreases p27(Kip1) mRNA level, but has no effect on p57(Kip2) mRNA level. Recent studies have revealed that degradation of the target gene mRNA is binding site dependent (28). Target gene mRNA with multiple binding sites even with partial complementarity is easy to be depredated (28). Our results match the results from computational analysis in which p27(Kip1) has 4 binding sites with partial complementarity, whereas p57(Kip2) has only one binding site with partial complementarity, for both miR-221 and miR-222 at their 3’ UTR.
In addition to the effect of miR-221 and miR-222 on p27(Kip1) and p57(Kip2) expression, other miRNAs may be also involved in the regulation of p27(Kip1) and p57(Kip2) expression in injured arteries via indirect mechanisms. For example, in our previous study we have found that balloon-injury of injury is able to cause an increase in miR-21 expression and that PTEN is one of the targets of miR-21 (23). PTEN is reported to be able to reduce the ubiquitinization of p27 (29), so it could be possible that the reduced PTEN levels could cause a reduction of p27 levels.
VSMC proliferation is the critical cellular event in vascular neointimal lesion formation and p27(Kip1) and p57(Kip2) are negative regulators for VSMC proliferation in vivo (30, 31). In the current study, we have demonstrated that p27(Kip1) and p57(Kip2) are two gene targets of miR-221 and miR-222 in rat carotid artery in vivo. Finally, downregulation of the overexpressed miR-221 and miR-222 significantly decreases VSMC proliferation and neointimal lesion formation in rat carotid arteries after angioplasty.
Neointimal formation does not only rely on VSMCs but also is related to re-endothelialization and inflammation. Our result demonstrated miR-221 and miR-222 are expressed in both VSMCs and endothelial cells, which is consistent with other recent studies showing that miR-221 and miR-222 are also expressed in endothelial cells and hematopoietic cells (32, 33). miR-221 and miR-222 are found to be able to inhibit endothelial cell migration (33). Thus, the biological effects of miR-221 and miR-222 on endothelial cells may be also involved in miR-221 and miR-222-mediated effect on vascular neointimal growth. Currently, the role of hematopoietic cells in miR-221 and miR-222–mediated vascular effect is unclear and needs to be investigated in future studies.
In summary, the current study reveals that the non-coding small miRNAs, miR-221 and miR-222 are novel regulators of VSMC proliferation and vascular neointimal lesion formation via their target genes p27(Kip1) and p57(Kip2). These novel findings may have extensive implications for the therapy of a variety of proliferative vascular diseases.
Acknowledgments
The authors thank Dr. Kimberly Louis in our department for editing assistance.
Sources of Funding
This work was supported by a National Institutes of Health Grant HL080133.
Footnotes
Disclosures: None
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 3.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 7.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 8.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Lee CT, Risom T, Strauss WM. MicroRNAs in mammalian development. Birth Defects Res C Embryo Today. 2006;78:129–139. doi: 10.1002/bdrc.20072. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci (Lond) 2008;114:699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 12.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 14.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 15.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Carè A. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 18.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 19.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 20.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 21.Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:812–819. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 24.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc. Natl Acad Sci USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 26.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 28.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 29.Lee MH, Yang HY. Negative regulators of cyclin-dependent kinases and their roles in cancers. Cell Mol Life Sci. 2001;58:1907–1922. doi: 10.1007/PL00000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D, Krasinski K, Sylvester A, Chen J, Nisen PD, Andrés V. Downregulation of cyclin-dependent kinase 2 activity and cyclin A promoter activity in vascular smooth muscle cells by p27(KIP1), an inhibitor of neointima formation in the rat carotid artery. J Clin Invest. 1997;99:2334–2341. doi: 10.1172/JCI119414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano N, Urasawa K, Takagi Y, Saito T, Kaneta S, Ishikawa S, Higashi H, Tsutsui H, Hatakeyama M, Kitabatake A. Downregulation of cyclin-dependent kinase inhibitor; p57(kip2), is involved in the cell cycle progression of vascular smooth muscle cells. Biochem Biophys Res Commun. 2005;338:1661–1667. doi: 10.1016/j.bbrc.2005.10.093. [DOI] [PubMed] [Google Scholar]
- 32.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]