Abstract
Heart disease is the leading cause of mortality and morbidity in the Western world. The heart has little regenerative capacity after damage, leading to much interest in understanding the factors required to produce new cardiac myocytes. Despite a robust understanding of the molecular networks regulating cardiac differentiation1,2, no single transcription factor or combination of factors has been shown to activate the cardiac gene program de novo in mammalian cells or tissues. Here we define the minimal requirements for transdifferentiation of mouse mesoderm to cardiac myocytes. We show that two cardiac transcription factors, Gata4 and Tbx5, and a cardiac-specific subunit of BAF chromatin-remodeling complexes, Baf60c, can direct ectopic differentiation of mouse mesoderm into beating cardiomyocytes, including the normally noncardiogenic posterior mesoderm, and the extraembryonic mesoderm of the amnion. Gata4 with Baf60c initiated ectopic cardiac gene expression. Addition of Tbx5 allowed differentiation into contracting cardiomyocytes and repression of noncardiac mesodermal genes. Baf60c was essential for the ectopic cardiogenic activity of Gata4 and Tbx5, partly by permitting binding of Gata4 to cardiac genes, indicating a novel instructive role for BAF complexes in tissue-specific regulation. The combined function of these factors establishes a robust mechanism for controlling cellular differentiation, and may allow reprogramming of new cardiomyocytes for regenerative purposes.
The transcriptional regulation of the developing heart has been well-studied1,2, but the factors sufficient to induce the cardiac program in mammalian cells have remained elusive. Recent work has demonstrated important roles for members of the polymorphic Swi/Snf-like BAF chromatin remodeling complexes in cell-type specification and differentiation3-7. Baf60c, a cardiac-enriched BAF complex subunit, physically links DNA-binding transcription factors to BAF complexes, thereby modulating the transcription of target genes3. Mouse embryos with reduced levels of Baf60c have severe heart defects and defective cardiac differentiation3. Because Baf60c is expressed specifically in precardiac mesoderm, we determined whether it is necessary for the activity of essential cardiac DNA-binding factors in noncardiac cells.
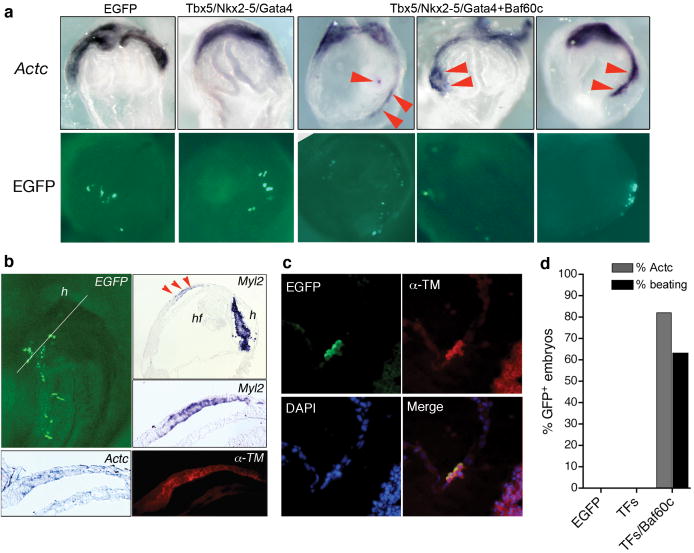
We transiently transfected cultured mouse embryos with expression constructs for Baf60c and combinations of three transcription factors that are important for activation of cardiac genes1,2: the zinc-finger transcription factor Gata4, the homeodomain transcription factor Nkx2-5, and the T-box transcription factor Tbx5 (Fig. 1a). Induction of cardiac differentiation was assessed by expression of the early cardiac marker Actc, encoding alpha cardiac actin. Control transfections with enhanced green fluorescent protein (EGFP)-expressing constructs or Tbx5/Nkx2-5/Gata4 combined did not induce Actc (Fig. 1a, 0/12 and 0/13 embryos, respectively). In contrast, cotransfection of Tbx5/Nkx2-5/Gata4+Baf60c led to markedly expanded and ectopic activation of Actc (Fig. 1a,d, 9/11 embryos). We could reliably induce Actc between E6.5 and E8.75, but later transfections proved ineffective, indicating a limited time window for Actc induction or technical limitations inherent to our approach. Actc induction was not potentiated by myocardin, a transcriptional coactivator that activates some cardiac genes in cell culture8, or the precardiac mesoderm transcription factor Mesp1 (Ref. 9), which can promote cardiac lineages in embryonic stem cells (ESCs)10-12.
Figure 1.
Ectopic induction of cardiac differentiation in mouse embryos. a, Actc in situ hybridization (top) shows endogenous cardiac crescent at E8.0 and ectopic cardiac gene expression (red arrowheads) in embryos transfected with indicated expression constructs. Bottom row shows EGFP signal. b, Ectopic expression of Myl2, α-TM, and Actc in consecutive sections of an embryo transfected with Tbx5/Nkx2-5/Gata4+Baf60c. Top left panel: EGFP signal and plane of section. h: heart; hf: headfold. c, Ectopic α-TM expression (red) is restricted to EGFP+ cells (green). DAPI shows nuclei (blue). d, Percentage of embryos with ectopic Actc-positive foci and beating tissue.
In embryos cotransfected with Tbx5/Nkx2-5/Gata4+Baf60c, we also detected ectopic Myl2 mRNA, and α-tropomyosin (α-TM) and cardiac troponin T (cTnT) proteins, specific markers of the embryonic heart (Figs. 1b, 2a). Induction of cardiac markers was confined to transfected cells, suggesting a cell-autonomous effect (Figs. 1c, 2a,c). Strikingly, ectopic beating cardiac myocytes were observed in normally noncardiogenic mesoderm transfected with Tbx5/Nkx2-5/Gata4+Baf60c (9/16 embryos), suggesting induction of a full cardiac program (Fig. 1d, online movie 1). Ectopic contractile tissue was observed even though the endogenous cardiac field was not yet beating, indicating accelerated cardiac differentiation. Thus, a simple combination of DNA-binding transcription factors and the chromatin-remodeling protein Baf60c induced cardiac differentiation in vivo.
Figure 2.
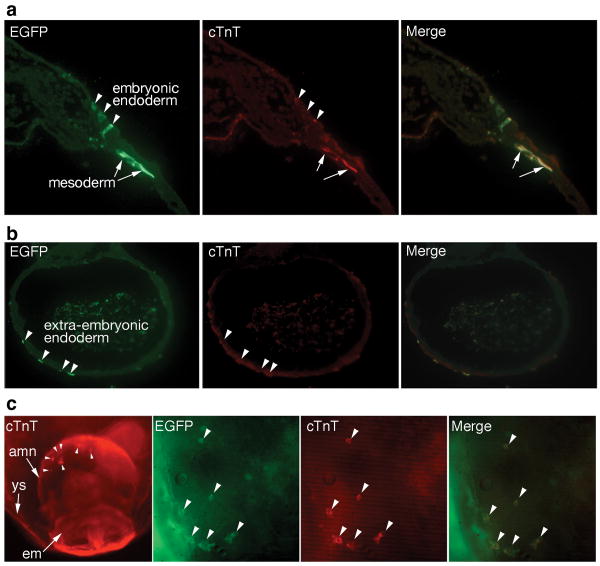
Induction of cardiac differentiation in embryonic and extraembryonic mesoderm. a, Transfected cells were observed in endoderm and mesoderm (EGFP), but induction of cTnT was mainly restricted to mesoderm. b, cTnT was not induced in extraembryonic endoderm. c, Induction of cTnT in mouse amnion by Gata4/Tbx5+Baf60c. Left panel: low-magnification view of a transfected embryo; arrowheads show ectopic cTnT in the amnion. Right panels: close-up of the amnion, showing transfected cells (EGFP), cTnT expression, and merge of both. Arrowheads indicate transfected cTnT+ cells. amn: amnion; em: embryo; ys: yolk sac.
cTnT was induced predominantly in mesoderm, but not in endoderm (Fig. 2a,b). We could not reliably transfect ectoderm. Ectopic cardiac markers were activated in all mesodermal regions, including the posterior and medial mesoderm, which are normally not fated to cardiac lineages (Figs. 1a, 2a, 3a, 4b). Strikingly, ectopic cTnT and beating tissue could be induced in the amnion, an extraembryonic tissue (Fig. 2c). The amnion is composed of a mesoderm layer derived from the primitive streak9,13, and an ectodermal layer, suggesting that endodermal signals were dispensable for cardiogenic induction. Thus, noncardiac mesoderm was directly programmed into cardiac tissue by Tbx5/Nkx2-5/Gata4+Baf60c.
Figure 3.
Minimal requirements for cardiac gene activation in mouse embryos. a, Expression of Actc in embryos transfected with pairwise combinations of transcription factors+Baf60c. Only Gata4+Baf60c induced ectopic Actc (red arrowheads). b, Expression of Nkx2-5; Gata4+Baf60c induced ectopic Nkx2-5 (arrowheads). c, Percentage of Actc-positive and beating embryos for each combination. d, Specificity of cardiogenic factors; percentage of embryos in which Actc was induced is shown.
Figure 4.
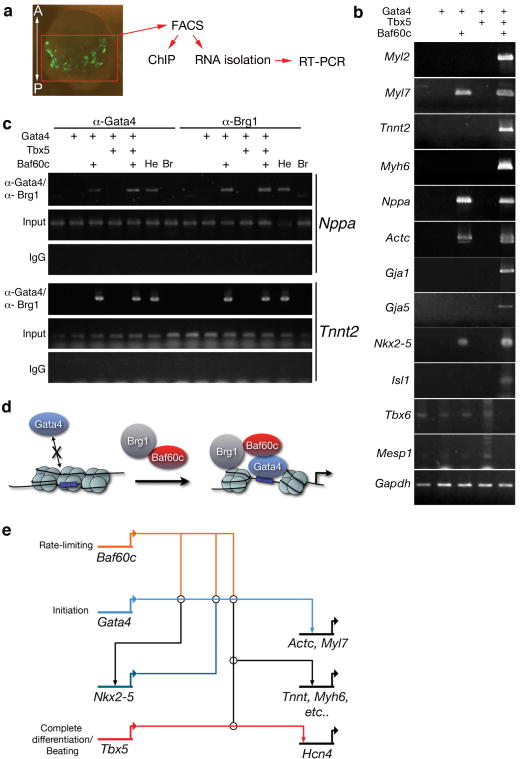
Mechanism for induction of cardiac differentiation. a, Strategy for isolating and analyzing transfected cells. b, RT-PCR of several cardiac markers in RNA isolated from transfected EGFP-positive mouse embryonic cells obtained by FACS. c, Chromatin immunoprecipitation (ChiP) shows that GATA4 and Brg1 bind Tnnt2 and Nppa only in the presence of Baf60c. Br: brain, He: heart, IgG: nonspecific immunoserum. d, Model for action of Baf60c. e, Minimal transcriptional network for the ectopic induction of cardiac differentiation. Made using Biotapestry software.
We sought to define the minimal set of factors required to induce cardiac differentiation. Ectopic Actc was efficiently induced by Gata4+Baf60c (9/11 embryos) but not by Nkx2-5+Baf60c or Tbx5+Baf60c (Figs. 3a,c, 4b). Nkx2-5 was induced by Gata4+Baf60c (Figs. 3b, 4b), but Tbx5 was not (not shown). As Gata4 and Nkx2-5 cooperatively activate numerous cardiac genes1,2, the induction of Nkx2-5 by Gata4+Baf60c provides an important feed-forward mechanism to establish and reinforce the cardiac program.
We substituted the hematopoietic GATA factor Gata1 for Gata4 (Ref. 14) and Baf60a or Baf60b, which are not expressed in the heart3, for Baf60c. Gata1 functionally replaced Gata4 but with reduced potency, and Baf60b (but not Baf60a) replaced Baf60c, again with reduced potency (Fig. 3d). Gata1 and Baf60b together were ineffective at activating Actc. These results indicate a significant degree of tissue-specificity conferred by Gata4 and Baf60c.
Although Gata4+Baf60c initiated cardiac gene expression, they did not induce contractile tissue (Fig. 3c, 0/10). We hypothesized that additional input from Tbx5 was required to complete the cardiac program. To investigate this possibility, we used fluorescence-activated cell sorting (FACS) to isolate EGFP-positive cells from the noncardiogenic posterior mesoderm of transfected embryos and isolated RNA for RT-PCR analysis. Some cardiac genes were induced by Gata4+Baf60c, but Tbx5 was required to induce additional genes required for cardiac function (Figs. 4b,c, S1). Accordingly, transfection of Gata4/Tbx5+Baf60c induced ectopic beating tissue (8/16 embryos, Fig. 3c, online Movie 2). Thus, we defined a minimal set of transcriptional regulators required to induce cardiac differentiation.
Isl1 expression, which marks early cardiac progenitors15, was not observed in transfected cells, indicating that we did not target undifferentiated cardiac precursors (Fig. 4b). However, Isl1 was induced as part of the cardiac program induced by Gata4/Tbx5+Baf60c. Interestingly, expression of the posterior mesoderm markers Tbx6 and Mesp1 was reduced in cells transfected with Gata4/Tbx5+Baf60, a finding that suggests reprogramming of mesoderm (Fig. 4b).
A combination of tissue-restricted BAF complex subunits may be required to enact a specific transcriptional program. The BAF complex subunits Baf45a and Baf45b have been implicated in neural progenitor– and neuron-specific transcription5,7. Baf45c (also known as DPF3) is predominantly expressed in the developing heart6 and recognizes specific histone modifications, suggesting that it may enhance the specificity of target gene recognition6. Expression constructs for Baf45c alone or with Tbx5/Nkx2-5/Gata4 did not induce cardiac gene expression (0/8), and the addition of Baf45c to Tbx5/Nkx2-5/Gata4+Baf60c did not increase the efficiency of induction (5/6 vs 5/6).
We investigated the mechanism by which Baf60c potentiates the function of Gata4. If Baf60c functions primarily to promote transcriptional activity of GATA4 independent of chromatin remodeling, we would expect that a fusion of GATA4 and VP16, which strongly activates transcription by recruiting preinitiation complexes16, would activate cardiac genes. Transfection of GATA4-VP16 alone did not activate Actc (0/7) (Fig. 3c) or Nkx2-5 (0/5), and GATA4-VP16+Baf60c induced Actc efficiently (5/5) but did not induce beating tissue (0/9) (Fig. 3c). Thus Baf60c must have functions separate from the recruitment of the preinitiation complex. We then examined whether Baf60c is important for binding of GATA4 to target genes. We performed chromatin immunoprecipitation on cells isolated by FACS from transfected embryonic posterior mesoderm. GATA4 could not bind Tnnt2 or Nppa regulatory regions in the absence of Baf60c, but inclusion of Baf60c resulted in detectable GATA4 occupancy on both genes (Fig. 4c). The BAF complex ATPase Brg1 was also detected only in the presence of Baf60c (Fig. 4c). Therefore, unlike the “pioneer factor” function of GATA4 in liver development17, Baf60c is required to allow GATA4 to bind these cardiac genes in noncardiac mesoderm. While the mechanism of this action is unknown, it may involve recognition of histone modifications by subunits of Baf60c-associated BAF complexes6,7 or other interacting transcription factors18, followed by remodeling of the chromatin at these loci or cooperative binding to target DNA.
Our results define a combination of three factors that can execute a cardiac transcriptional program and fully induce noncardiac mesoderm to differentiate directly into contractile cardiac myocytes. The direct induction of ectopic beating tissue in vivo in amniotes is unprecedented; application of cardiogenic growth factors to chick embryonic mesoderm only induced early markers of cardiomyocyte lineage, such as Nkx2-519,20. In lower vertebrates such as Xenopus or zebrafish, Mesp1 or gata5 could induce cardiac differentiation, but only at low frequency (<10% transfected embryos)10,21. Mesp1 can promote cardiogenic mesoderm from ESCs10-12, but was ineffective in the mouse, suggesting that it primarily promotes lineage specification rather than inducing differentiation. We may have targeted the in vivo equivalent of epiblast stem cells (EpiSCs)22,23 rather than mesoderm; this is unlikely as these should be rare or absent at stages at which we were able to induce cardiogenesis (E7.0–8.0), and loss of posterior mesoderm markers in induced cardiac cells supports targeting of mesoderm. It has been suggested that permissive cues from adjacent endoderm or ectoderm are important for cardiac differentiation; however in chick and Xenopus, heart tissue could only be induced in explants19,20,24,25, suggesting that strong negative cues prevent ectopic cardiac differentiation in vivo. Our results show that repressive influences are unlikely to be major components of cardiac program regulation or that they can be overcome by our combination of factors.
Overall our data suggest that, in amniotes, an additional layer of tissue-specific gene regulation, in the form of Baf60c, has been superimposed upon cardiac transcriptional regulators. Consistent with loss-of-function experiments26, Gata4 was a key factor in initiating the cardiac program, while Tbx5 was required for full differentiation into contracting cardiomyocytes. Since Gata4+Baf60c induced Nkx2-5, the combined input from Gata4, Nkx2-5, and Tbx5, which synergize on target enhancers1-3, provided sufficient information to activate the cardiac program. Unlike other contexts where DNA-binding factors were sufficient for transdifferentiation27-29, Baf60c was required to potentiate the function of Gata4 and Tbx5, partly by allowing binding of Gata4 to cardiac loci. These findings reveal a novel mechanism for tissue-specific gene regulation by chromatin-remodeling complexes. Harnessing this potential in a permissive cell type could lead to the production of cardiac cells suitable for therapeutic use in diseased myocardium.
Methods Summary
Mouse embryos were transfected with separate expression constructs for each factor and EGFP, and cultured as described30 with modifications mainly in the timing and location of the transfection. Embryos were collected, fixed with 4% paraformaldehyde, and processed for whole-mount in situ hybridization, paraffin sections, or cryosections. Immunohistochemistry was performed on whole embryos, or on sections after antigen retrieval. Chromatin imunoprecipitation was performed on proteins cross-linked to DNA from cells FACS-sorted from 25–30 embryos for each condition.
Supplementary Material
Acknowledgments
We thank K. Koshiba-Takeuchi for assistance with immunohistochemistry, H. Ogawa for advice on ChIP, B.L. Black and D. Srivastava for helpful discussions and expression constructs, G.R. Crabtree, M. Nemer, E.N. Olson, and Y. Saga for expression constructs, and G. Howard and S. Ordway for editorial assistance. This work was supported by Human Frontiers Science Program CDA and short-term fellowships (J.K.T.), MEXT's program for young independent researchers (J.K.T), Mitsubishi Foundation (J.K.T.) and NHLBI (B.G.B.). This work was also supported by an NIH/NCRR grant (J. David Gladstone Institutes), and the William H. Younger Family (B.G.B.).
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
The authors declare competing financial interests.
References
- 1.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Lickert H, Takeuchi JK, von Both I, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 4.Simone C, Forcales SV, Hill DA, et al. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 5.Lessard J, Wu JI, Ranish JA, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange M, Kaynak B, Forster UB, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creemers EE, Sutherland LB, Oh J, et al. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol Cell. 2006;23:83–96. doi: 10.1016/j.molcel.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Saga Y, Miyagawa-Tomita S, Takagi A, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 10.David R, Brenner C, Stieber J, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 11.Bondue A, Lapouge G, Paulissen C, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Lindsley RC, Gill JG, Murphy TL, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinder SJ, Tsang TE, Quinlan GA, et al. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- 14.Pevny L, Simon MC, Robertson E, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 15.Laugwitz KL, Moretti A, Caron L, et al. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 16.Black JC, Choi JE, Lombardo SR, et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Cirillo LA, Lin FR, Cuesta I, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 18.Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 20.Marvin MJ, Di Rocco G, Gardiner A, et al. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiter JF, Alexander J, Rodaway A, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 23.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 24.Small EM, Warkman AS, Wang DZ, et al. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132:987–997. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- 25.Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 26.Zhao R, Watt AJ, Battle MA, et al. Loss of both GATA4 and GATA6 blocks cardiac myocyte diffferentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 28.Xie H, Ye M, Feng R, et al. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, Brown J, Kanarek A, et al. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto M, Saijoh Y, Perea-Gomez A, et al. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature. 2004;428:387–392. doi: 10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.