Abstract
Hydroxyurea is a potent remedy against a variety of ailments and an efficient inhibitor of DNA synthesis, yet its pharmacology is unclear. Hydroxyurea acts in Escherichia coli by the same mechanism as it does in eukaryotes, via inhibition of ribonucleotide reductase. When examining a controversy about concentrations of hydroxyurea that prevent thymineless death in E. coli, we found instability in hydroxyurea solutions which avoided prior detection due to its peculiar nature. In contrast to freshly dissolved hydroxyurea, which did not affect respiration and was bacteriostatic, one-day-old hydroxyurea solutions inhibited respiration and were immediately bactericidal. Respiration was inhibited by two gasses, hydrogen cyanide (HCN) and nitric oxide (NO), whose appearance we detected in “aged” hydroxyurea stocks by GC-MS; however, neither gas was bactericidal. While determining the cause of toxicity, we found that hydroxyurea damages DNA directly. We also demonstrated accumulation of peroxides in hydroxyurea solutions by enzymatic assays, which explains the toxicity, as both NO and HCN are known to kill bacteria when combined with hydrogen peroxide. Remarkably, we found that bactericidal effects of NO + H2O2 and HCN + H2O2 mixtures were further synergistic. Accumulation of decomposition products in solutions of hydroxyurea may explain the broad therapeutic effects of this drug.
Keywords: hydroxyurea, hydrogen cyanide, nitric oxide, hydrogen peroxide, thymineless death
Introduction
Hydroxyurea (HU) is a familiar and potent medicine with an apparently straightforward mechanism of biochemical action towards a well-characterized target, yet with unclear pharmacology 1. The only known cellular target of HU is ribonucleotide reductase, whose inactivation inhibits production of DNA precursors, specifically blocking DNA synthesis in both human and bacterial cells 2–8. The inhibition of chromosomal replication by HU in bacteria is reversible, as no lethality is observed during the first several hours of HU treatment 5; 9; 10. Cytotoxicity of HU in mammalian and human cells is more complex, depending on several parameters and suggesting contribution of additional unknown factors. First, HU kills S-phase cells, but does not kill G1 or G2 cells; second, HU kills continuously-dividing cells, but not the cells that were stimulated to divide; third, HU kills cultured cells better than the cells within an organism 11–15. However, even in the cases of maximal sensitivity, residual survival of HU-treated cell populations is still 20–50%.
Its apparently mild action notwithstanding, HU is therapeutically potent and is a drug of choice against myeloproliferative disorders 16 and painful crises in sickle cell anemia 17. HU also shows promise against recurrent and inoperable meningiomas 18 and as an addition to anti-AIDS cocktails 19. Since HU blocks eukaryotic cells in G1, and G1 cells are more sensitive to DNA damage compared to S- or G2-cells, HU acts as a classic “radiation enhancer” during radiotherapy 20. HU was also used to maintain normal white blood cell counts during chronic myeloid leukemias before modern specific enzyme-targeting treatments appeared 21. HU use in treatment of diverse medical conditions lacking common denominator emphasizes incompleteness of our understanding of the mechanisms of action of this drug. Studies aimed at finding “the active species” among HU decomposition products are quite common 22–26.
Thymine auxotrophic cells (the thyA mutants in bacteria) undergo rapid loss of viability in growth media without thymine (or more frequently used thymidine, the next step in conversion of thymine into the DNA precursor dTTP), a phenomenon known as thymineless death (TLD) 27. TLD has been studied for over five decades, but its mechanism remains an enigma. To examine the role of DNA replication in TLD, HU was used, but results were conflicting 10; 28. In earlier studies, 100 mM HU brought a complete protection to TLD 10; in a more recent report, 75 mM and 100 mM HU offered no protection, while 200 mM HU did 28. The latter authors reported, though, that all HU concentrations blocked DNA synthesis completely, proposing that it is HU’s modest effect on RNA synthesis, which is responsible for the observed protection 28.
The original objective of this study was to test whether TLD is dependent on DNA replication, using HU to inhibit DNA synthesis. Unexpectedly, we discovered a rapid bactericidal effect of aged HU solutions. The picture was complicated by the fact that the surviving cells then became completely resistant to TLD. Since the biological activity (the ability to inhibit DNA synthesis) of HU solutions does not change over a period of several months, HU is considered to be essentially stable in solutions 29–31, and with a few exceptions that specifically used only freshly-made HU 10; 32, experimental reports of HU-treatment of cells do not mention details of HU preparation or storage. At the same time, there are periodic reports of HU instability in aqueous solutions, although they disagree about the resulting species 33–35, which suggests elusive decomposition products. Using gas chromatography followed by mass spectrometry (GC-MS), we identified two gasses, hydrogen cyanide and nitric oxide, in aged HU stocks; using enzymatic assays, we found peroxides. We then confirmed that combining HCN, NO and H2O2 together emulates the killing and TLD-protection effects of aged HU. Since HCN, NO and H2O2 are all physiologically active, these novel decomposition products of hydroxyurea may be relevant to its therapeutic potency and versatility as a medicine. While seeking the source of HU toxicity we also found evidence that both fresh and aged HU damage DNA bases directly.
Results
Aged versus fresh HU in killing and protection against TLD
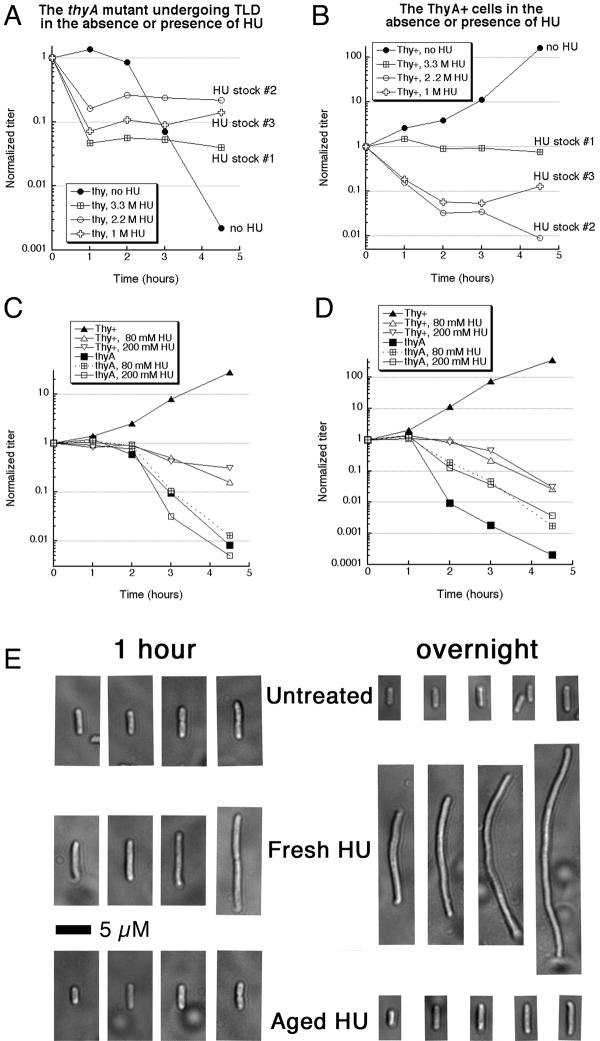
In our conditions, the characteristic rapid loss of titer from TLD begins after 2 hours without thymidine (Fig. 1A), so any loss of titer before 2 hours would be due to other toxic sources, while any reduced killing after 2 hours means alleviation of TLD. Morganroth and Hanawalt reported that 75 mM HU has no effect on TLD, whereas 200 mM HU can relieve TLD 28. In contrast, we found that 80 mM HU already offers protection against TLD, but only after an initial killing period that occurred well before the onset of TLD (Fig. 1A). 200 mM HU had a similar effect (not shown). Surprisingly, HU stocks made on different dates showed significant variation in the extent of the initial bactericidal effect (Fig. 1A). This killing by HU did not depend on thymine limitation in thyA mutants, as it was observed in Thy+ cells as well and was also inconsistent (compare Fig. 1A versus 1B, stock #1). We suspected that this “killing potential” accumulated in HU stocks during storage; consistent with possible changes, our “aged” HU solutions change pH from 5.5 to 6.2.
Fig. 1. Aged versus fresh HU in killing and protection against TLD.
A. Changes in the titer of the thyA mutant (KKW 58) incubated without thymidine in the M9CAA medium. Thymidine was removed at time 0 and, where indicated, HU was added to 80 mM from HU stock solutions. Stock #1 (3.3 M in water) was 5 months old, stock #2 (2.2 M in water) was 3 months old, while stock #3 (1 M in M9CAA medium) was 48 hours old.
B. Changes in the titer of the wild type control (KKW 59). Cells were processed exactly as in “A”, but on a different day.
C. Freshly prepared HU offers minimal protection against TLD, but does not immediately kill either. Method as in “A”. Thy+, KKW 59; thyA, KKW 58.
D. The effect of fresh HU on TLD in Morganroth and Hanawalt (2006) conditions. Cells were grown in Davis minimal medium at 37°C. Fresh hydroxyurea was used in the final concentrations indicated.
E. Microscopic images of cells, treated with either fresh or aged 80 mM HU, for either one hour or overnight. The scale bar is 5 μM.
As mentioned in the introduction, with a few exceptions that specifically used only freshly-made HU 10; 32, experimental reports of HU-treatment of cells do not give details of HU preparation or storage. We repeated our experiments by dissolving crystal HU in the growth medium (M9CAA) just before addition to bacterial cultures. We found that such a freshly-prepared HU does not relieve TLD at any concentration tested (up to 400 mM), while showing a mildly bactericidal effect on Thy+ cells after longer incubations (Fig. 1C), the latter being consistent with the literature reports 5; 9; 10. To test if the discrepancy between our results and those of Morganroth and Hanawalt was due to the difference in the growth media (M9 or MOPS-minimal-P versus Davis) or incubation temperatures (30°C versus 37°C), we repeated the experiment using their exact conditions. Fresh HU in Davis Medium does alleviate TLD in our hands, but only moderately (Fig. 1D), which disagrees with the original report of complete protection 28. Hence, under both growth conditions, freshly prepared HU at low or high concentration does not relieve TLD, but it also does not kill immediately. We also found that the cells treated with “fresh” HU would continue to elongate (as was reported before 10), whereas cells treated with “aged” HU stop elongating (Fig. 1E), suggesting that their metabolism was inhibited. We conclude that, in our case, only the HU preparations that stop the cell growth, and sometimes kill the cells, can protect against TLD. These killing/protecting characteristics of aged HU preparations suggested accumulation of toxic decomposition products in aqueous solutions of HU. Irreproducibility of our results suggested elusive (volatile) substances, forcing us to standardize HU aging conditions (see “Materials and Methods”).
Inhibition of DNA synthesis by HU versus TLD
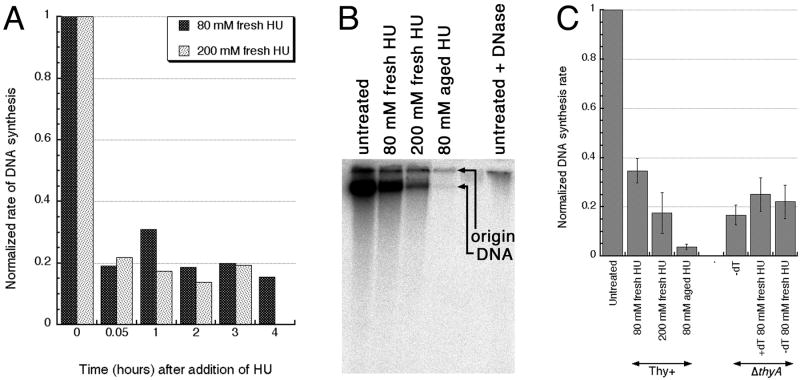
Morganroth and Hanawalt measured DNA synthesis by following cumulative label incorporation and found that both 200 mM HU (that protected against TLD) and 75 mM HU (that failed to protect) inhibit DNA synthesis completely 28. Therefore, these authors proposed that HU inhibits TLD because in the higher concentrations it can also inhibit RNA synthesis. Suspecting that aged HU would inhibit DNA synthesis stronger than fresh HU, we decided to measure DNA synthesis during HU inhibition more precisely, by following the rate of incorporation. The standard protocol of measuring 3H-thymidine incorporation over 1–2 minute intervals 36; 37 showed that fresh HU-treated cells still retain around 20% of the DNA synthesis capacity of untreated cells (Fig. 2A). However, for comparison of the inhibition of DNA synthesis by HU with the one during TLD, the standard protocol is unsuitable, for the following reason.
Fig. 2. Inhibition of DNA synthesis by fresh versus aged HU in ThyA+ cells and during TLD in thyA mutants.
A. Inhibition of DNA synthesis by fresh HU, measured by thymidine incorporation. The data points are averages of two independent measurements. The tdk mutant (KKW23), unable to utilize thymidine for DNA incorporation (the negative control) showed no incorporation over the background (not shown).
B. Alkaline agarose gel to measure DNA synthesis in cultures growing in the presence of various preparations of HU by 30 minute 32P-orthophosphate incorporation. The combined signal from the DNA band plus origin is quantified by PhosphorImager.
C. 30 minute 32P-orthophosphate incorporation into the chromosomal DNA, detected as in “B”, in thymidine-starved thyA mutant (KKW 58) or in its Thy+ counterpart (KKW59), normalized to untreated or unstarved cultures. The data points are averages of two-to-seven independent measurements ± SE.
Measurements of DNA synthesis inhibition during TLD is non-trivial, because the only DNA-specific label, (deoxy)thymidine, is also the nutrient for which thyA mutant cells are starved, and its addition, even in small amounts, relieves starvation, making the resulting DNA synthesis rates misleadingly high. Thus, to detect true replication rates during thymine starvation, a different label has to be used, requiring additional separation (because it will not be a DNA-specific label). We chose 32P-orthophosphate, because we used it successfully for chromosomal DNA labeling 38; 39 and we can separate DNA from other phosphate-incorporating species, like RNA, LPS and poly-phosphates (Luciana Amado and A.K., unpublished) (Fig. 2B). There is a second complication of the standard protocol to measure DNA synthesis rates, this time for HU-treated cells. The DNA synthesis rate is normalized to O.D. of the culture, to enable comparison with untreated, growing cultures, but cells treated with fresh HU elongate, while cells treated with aged HU do not (Fig. 1E), invalidating the comparison. Thus, the measurements have to be taken from specific volume of HU-treated or thymidine-starving cultures independently of their actual OD and then normalized to the level of incorporation in equal volume of the untreated or unstarved aliquot of the same culture.
Using such 32P-orthophosphate labeling, we found that 1) during the first 30 minutes of thymine starvation, the incorporation is only 10% of the unstarved culture; 2) 200 mM fresh HU inhibits replication to the same extent as TLD, while 80 mM fresh HU still allows 40% of replication compared to untreated cultures (Fig. 2C). Therefore, the rate of DNA synthesis in fresh HU-treated cells was not low enough to block the residual DNA synthesis during TLD, explaining the failure of fresh HU to prevent TLD (Fig. 1C). In contrast, aged HU blocked DNA synthesis almost completely (Fig. 2C), which was consistent with its protection against TLD (Fig. 1A).
The observed incomplete inhibition of DNA synthesis by fresh HU is consistent with the incomplete inhibition by HU of the ribonucleotide reductase activity in cell extracts, which does not exceed 70–80% 2; 3; 5. Moreover, the significant residual replication in HU-treated cells, if measured as the rate of DNA synthesis, was reported before 5; 40; 41. Our result also shows that the DNA synthesis-inhibiting ability of HU does not disappear in aged stocks and is, in fact, enhanced. Finally, the observation that HU reduces DNA synthesis rates stably for up to several hours (Fig. 2A) argues against a significant decomposition of HU in the presence of bacterial cells in the course of our experiments.
DNA damage by HU
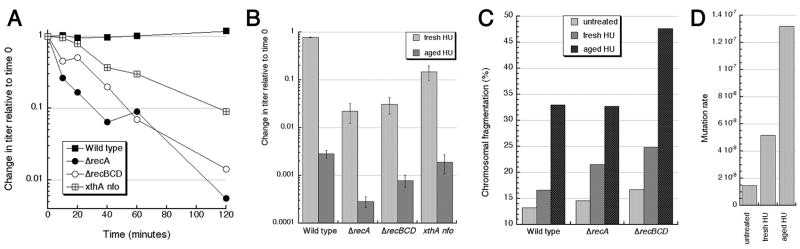
We next checked if the cell killing by aged HU is due to DNA damage. Although long incubation with HU are known to break both chromosomal DNA in E. coli cells 41, as well as pure DNA in solutions 34, no loss of viability with fresh HU in our conditions argued for, at most, a minimal DNA damage that is efficiently repaired in wild type cells. To detect DNA damage upon treatments with fresh versus aged HU, we employed sensitive physical and genetic assays. First, since a small fraction of any DNA damage is converted by DNA replication into double-strand DNA breaks, the recA and recBC mutants that are deficient in repair of these replication-dependent one-ended double-strand breaks are rapidly killed by any DNA damage (reviewed in 42), offering a sensitive way to detect consequences of a broad spectrum of DNA lesions. We found that the recA and recBC mutants are moderately sensitive to acute treatments with fresh HU (Fig. 3A), and quite sensitive to aged HU (Fig. 3B). Although acute HU treatments of the rec mutants were never reported before, the sensitivity of the recA and recBC mutants to chronic HU treatments is known and can be explained by reversal/breakage of inhibited replication forks in combination with the inability of these mutants to repair resulting double-strand ends 43; 44. However, the increased killing of the recA and recBCD mutants by aged HU must have a different nature. Remarkably, the difference between the fresh versus aged HU killing (~100-fold) was the same for wild type cells and for both rec mutants (Fig. 3B), suggesting that the additional killing is due to either a non-repairable DNA damage (for example, replication-independent two-ended double-strand breaks in the unreplicated portion of the chromosome) or due to a non-DNA damage to the cell.
Fig. 3. DNA damage by 80 mM hydroxyurea.
Wild type, KKW59; ΔrecA, KKW60, ΔrecBCD, KKW62, xthA nfo, RPC 501.
A. Time course of the sensitivity of DNA repair mutants to fresh HU. The values are averages of three independent measurements conducted on different days.
B. Sensitivity of wild type cells and DNA repair mutants to fresh versus aged HU. The method was essentially as in “A”, but the time of treatment was fixed at 1 hour. The values are averages of five independent measurements conducted on different days ± SE.
C. Chromosomal fragmentation in untreated and HU-treated cells, as measured by pulsed-field gel electrophoresis. The data are averages of two independent measurements.
D. Mutation rate to Rifampicin-resistance in untreated and HU-treated cells (KKW59). The rate was determined from the median mutant numbers from 11-culture sets.
Two-ended double-strand breaks fragment chromosomal DNA, and HU treatment is a known clastogen (induces chromosomal fragmentation) 44. To test whether aged HU induces additional chromosomal fragmentation relative to fresh HU, we employed pulsed-field gels to detect broken chromosomal DNA. This assay showed that both fresh HU and aged HU cause increased chromosomal fragmentation, but to a different extent (Fig. 3C). In wild type cells, as well as in the recA and recBCD mutants, fresh HU caused a 3–8% increase in chromosomal fragmentation compared to the corresponding untreated controls, whereas aged HU increased fragmentation by 18–30% over untreated cells (Fig. 3C). Increased clastogenesis in fresh HU-treated cells was explained by the replication fork reversal model 44. The additional clastogenesis in aged HU-treated cells is consistent with much deeper inhibition of DNA replication by aged HU.
As a control against the possibility that HU treatment damages DNA directly via DNA base modification 26 we employed perhaps the most sensitive indicator of DNA base modification, which is increased mutagenesis. To this end, we measured the level of mutagenesis towards rifampicin resistance in fresh HU-treated versus aged HU-treated cells. Mutation rate was increased over the untreated control 3-fold in fresh HU-treated cells and 9-fold in aged HU-treated cells (Fig. 3D), suggesting increasing level of base modifications. Modified DNA bases are subject to excision by several DNA glycosylases, with the formation of the same toxic intermediate — the abasic site, — which is nicked by an abasic site endonuclease to complete the repair 45. E. coli has two abasic site endonucleases, exonuclease III and endonuclease IV, and the xthA nfo double mutant that lacks both is exquisitely sensitive to DNA base modifications, being poisoned by the accumulating abasic sites 46. We found that the xthA nfo double mutant is unexpectedly sensitive to fresh HU (Fig. 3A) and is ~100-fold more sensitive to aged HU relative to fresh one (Fig. 3B). Together with the mutagenesis data (Fig. 3D), these xthA nfo results indicate that 1) treatment with fresh HU damages chromosomal DNA at least partially through direct modification of DNA bases, and this damage is removable by base-excision repair; 2) products of decomposition in HU solutions kill via a different route, perhaps via non-repairable DNA damage — for example, by replication-independent two-ended double-strand breaks in the unreplicated portion of the chromosome. We conclude that fresh HU damages DNA directly, but non-lethally for DNA-repair-proficient cells; meanwhile, at least part of the killing potential of aged HU is realized via increased lethal DNA damage, specifically chromosomal fragmentation.
The concentration- and time-dependence of killing by HU
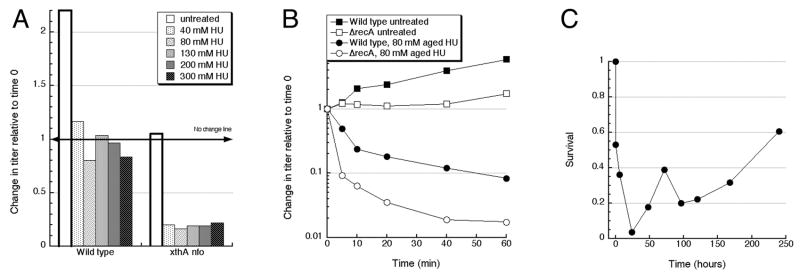
If HU kills by itself, this killing should be dependent on HU concentration. However, we found no correlation of the extent of xthA nfo killing and the concentration of fresh HU (Fig. 4A), indicating that the killing capacity of fresh HU is already saturated in 40 mM solutions. We did not explore lower concentrations of HU.
Fig. 4. The concentration- and time-dependence of killing by HU and kinetics of HU aging.
A. Concentration-dependence of HU-killing. The titer of growing cultures of the indicated genotype was determined by serial dilution of aliquots with 1 % NaCl and plating on LB, and then the cultures were continued being incubated with aeration at 30°C, with various concentrations of fresh HU or without the drug, for 1 hour, after which the titer was again measured, as above, and normalized to the original titer.
B. Time course of killing by aged HU. After taking an aliquot for titer at time 0 hour, HU from “aged” 1 M stock was added into the culture so that the final HU concentration was 80 mM. Aged HU was prepared in water as 1 M stock (protocol #1) and was 3 months aged at the time of experiment. Cultures were then kept at 30°C with aeration, and at the indicated times 30 μl of cultures were aliquotted and kept on ice until plating. Titer counts were normalized to the initial count at time 0 hour (right before HU were added).
C. Kinetics of HU aging under oil. HU was aged for the indicated amount of time at 37°C in loosely-capped tubes under oil, and aliquots of the same bacterial culture of wild type cells (KKW59) were mixed with these HU stocks of various age to give the final concentration of 80 mM and shaken at 30°C for 20 minutes. The titer of the treated cultures was then normalized to the untreated control. The values are averages of two or three independent measurements.
To characterize the dynamics of killing, as well as to detect a possible generation of the DNA-damaging species in the course of our treatment, we took a time course of treatment with aged HU, comparing a wild type strain to the recA mutant. We found that both the wild type and recA mutant cells significantly lose viability within the first 10 minutes of exposure to aged HU (Fig. 4B), with a slower decrease in titer afterwards. This may be due to several reasons: 1) the killing species are volatile (dissolved gasses) and rapidly escape from the cell suspension; 2) the killing species are titrated (inactivated) by the cells’ inert component; 3) cells may become resistant to the killing species. Apparently, further accumulation of these species during the treatment is minimal, otherwise we would have observed a continuous killing. The absence of additional killing during bacterial culture incubation is also consistent with the idea that the species are volatile. The already mentioned evidence that also pointed towards unstable or volatile species was the frustrating inconsistency of the killing extent with various aged HU preparations (Fig. 1AB and see Methods).
We used the rapid killing by aged HU to explore the time course of HU “aging” under mineral oil, which we started adding to inhibit the escape of possible gasses. It should be noted that mineral oil would only inhibit, without completely preventing, gas escape, and we specifically allowed the possibility (using non-air-tight tubes) and plenty of time for the gas to escape in this experiment — in part to indirectly confirm the presence of gasses. HU solutions starts killing after six hours at 37°C, with accumulation of the killing species maximal by one-two days of incubation, but then the killing potential of aged HU eventually decreases and completely disappears after ten days (Fig. 4C), which is consistent with it being dependent on gasses.
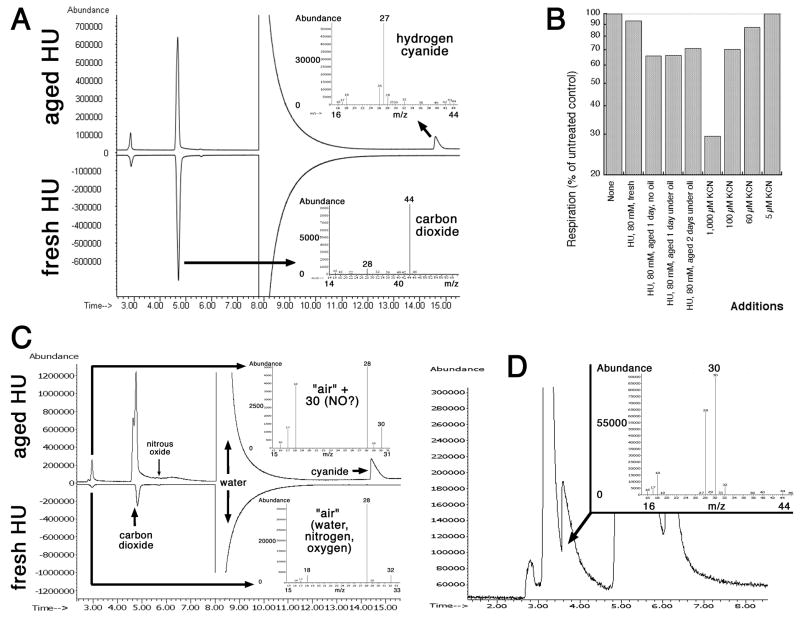
GC-MS reveals HCN and NO in aged HU
So far, our experiments on treating E. coli cells with stocks of HU of different age indicated generation of toxicants in HU solutions, among them possible gasses, that both kill cells via DNA damage and, at the same time, protect survivors against TLD, probably by inhibiting general metabolism. We tried to use liquid chromatography with mass spectrometry (LC-MS) to identify these toxicants in HU stock solutions, but the poor ionization of HU made identification of even HU itself difficult by this technique (besides, the lowest detection limit of our LC-MS machine was only m/z=50, while some decomposition products were expected to be smaller). We next tried several settings and conditions for gas chromatography (liquid-phase column) with mass spectrometry (GC-MS), with better results (Fig. S1). However, when initially we used non-derivatized HU solutions and typical GC injection temperatures (300°C), we could not even detect HU in our samples. Instead, there were urea and cyanuric acid (Fig. S1A) that were definitely forming during the heating of underivatized HU for GC-MS, because a sensitive indicator for cyanuric acid in solutions, melamine (detection limit 400 μM of cyanuric acid in our hands), failed to precipitate any in our 1M HU stocks (not shown). When we lowered the injection temperature to 200°C, both urea and cyanuric acid disappeared, and now we could see a broad peak of HU, accompanied by a strong peak of hydroxylamine (Fig. S1B) that, again, was likely to be an artifact of heating, as our subsequent experiments with boiling of HU stocks suggested (Fig. S2). To reduce the possibility of high-temperature artifacts, we derivatized HU with MSTFA (trimethylsilyl groups), but found formamide that was somehow non-derivatized (Fig. S1C). Testing these various artifacts of thermal HU decomposition in our TLD assay showed that 80 mM cyanuric acid had no effect (not shown), but the other two blocked TLD, either completely (formamide) or almost completely (hydroxyalamine) (Fig. S1D). A scheme of how these artifacts of thermal HU instability could have formed is shown in Fig. S1E. Since all these species were found in both fresh and aged HU, and none of them was immediately toxic to E. coli, even if some of them were not artifacts, we deemed them irrelevant for the effects of aged HU on TLD.
Since we suspected that aged HU toxicants were volatile, we then used GC-MS with a gas-detecting column. Indeed, we found two gasses in the aged HU that were absent from fresh HU: hydrogen cyanide (HCN) (Fig. 5A) and nitric oxide (NO) (Fig. 5C and D, Fig. S3-S6). Yet another gas, carbon dioxide (CO2), was present in both fresh and aged HU (Fig. 5A and C). Nitric oxide is the major product of enzymatic HU decomposition in vivo 31, and its detection in minute amounts in HU stocks in vitro was not surprising. To confirm the presence of HCN and/or NO in aged HU stocks, we tested how treatment with fresh or aged HU inhibit respiration, which is sensitive to 10 μM NO or to 200 μM cyanide 47; 48. We found that fresh HU does not inhibit respiration in inverted vesicles, whereas aged HU does, at the level of inhibition of 100 μM cyanide (Fig. 5B), indicating the presence of cyanide and/or NO in aged HU. Since inhibition of E. coli respiration by NO is strong only in low oxygen pressure and is minimal under fully aerobic conditions 47; 48, the observed level of respiration inhibition could be mostly ascribed to cyanide. However, cyanide is also known to greatly potentiate inhibition of respiration by NO 47, so we interpret this result to mean the presence of both cyanide and nitric oxide in aged HU preparations. By modifying gas chromatography conditions, we were eventually able to reliably separate the small NO peak from the dominant air peak (Fig. 5D, Fig. S4–6). At the same time, our attempts to directly detect cyanide in HU solutions with sensitive iron-based analytical color assays (formation of prussian blue) 49 were unsuccessful due to the formation of bright-magenta HU-iron complexes 50; structurally, hydroxyurea is a close relative of hydroxamic acids, which are known siderophores 51.
Fig. 5. Detection of hydrogen cyanide (HCN) and nitric oxide (NO) in aged HU by GC-MS with the gas-specific column.
A. Total ion profile from the gas-specific column reveals HCN. The first three peaks, both in fresh and in aged HU, from left to right are: 1) air (the smallest one), 2) carbon dioxide, for which the m/z spectrum is shown on the lower right, 3) water (the dominant peak). The HCN peak is present only in aged HU (the m/z spectrum on the upper right).
B. Aged HU inhibits respiration. Respiration was monitored as oxidation of NADH by membrane vesicles. As a positive control for respiration inhibition, various concentrations of KCN were also used in the same assay.
C. Traces of NO (m/z=30) in the air peak. Parameters were exactly like in “A”.
D. Separation of NO peak from the air peak by a gas-specific column. HU was prepared as a 1M stock and aged under oil for 1 day at 37°C.
Influence of decomposition products on TLD and bacterial survival
Thus, we have three sets of data, obtained by very different methodologies. First, using survival in bacterial cultures as a read-out, we characterized two distinct effects of aged HU (Fig. 1AB): 1) rapid killing; 2) conferring resistance to TLD on surviving cells, probably by inhibiting general metabolism. Fresh HU does not have either effect (Fig. 1CD). Second, using DNA-associated processes as a read-out (Fig. 2 and 3), we found that: 1) fresh HU moderately inhibits DNA synthesis and also damages DNA bases directly; 2) aged HU blocks DNA synthesis completely, perhaps through inhibition of respiration and, in addition to base damage, is also more clastogenic than fresh HU. Third, employing GC-MS, we found two gasses in aged HU solutions, — hydrogen cyanide and nitric oxide, — that were absent in freshly-made HU solutions (Fig. 5).
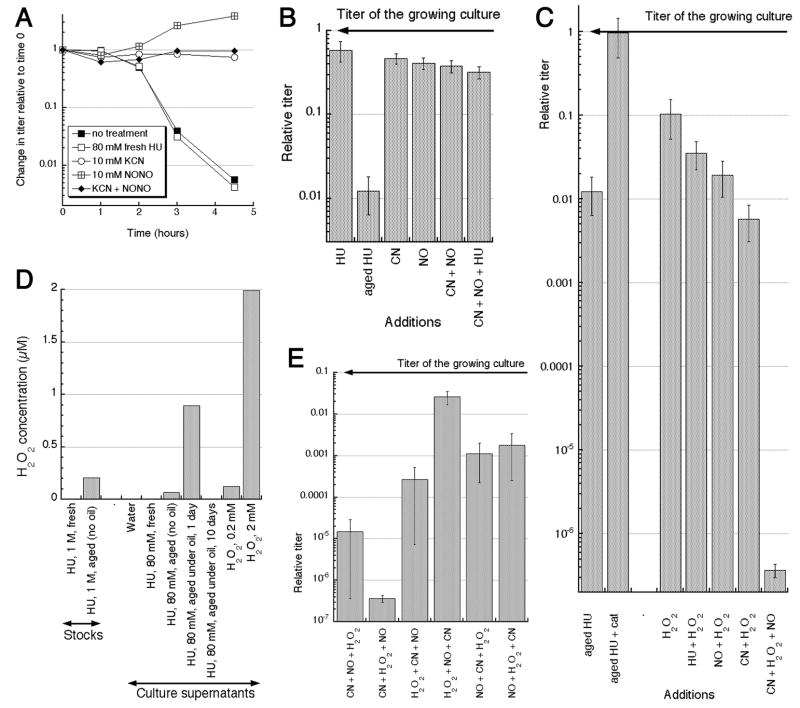
Next, we tested the effect of the two gasses on cells undergoing TLD. We confirmed that hydrogen cyanide does not kill E. coli and completely saves thyA mutants from TLD (Fig. 6A), as was reported before 52. NO has a similar static/protecting effect (Fig. 6A). The mixture of NO + CN, or addition of both NO and CN to fresh HU, did not kill either (Fig. 6A and B). We concluded that, by itself, accumulation of CN and NO in aqueous HU stocks upon storage could not explain the bactericidal effect of aged HU; at least one more HU breakdown product was, apparently, escaping detection by GC-MS.
Fig. 6. The effect of HCN and/or NO on bacteria, and detection of H2O2 in aged HU.
A. Influence of NO or KCN on TLD. The indicated concentrations of fresh HU, KCN or DEA-NONOate were added to the thyA mutant cells incubated in the growth medium without thymidine.
B. CN, NO or their mixture do not kill. Incubation was for 1 hour. Concentrations used: HU, 80 mM; CN (KCN), 10 mM; NO (DEA-NONOate), 10 mM.
C. CN or NO kill in the presence of H2O2. As in “B”, but H2O2 concentration was 2 mM, and ~290 units of catalase (cat) was added where indicated. Shown in the same scale as “B” to facilitate comparison.
D. Measurement of the peroxide concentration by the HRP/AR assay, either in HU stocks, or in supernatants of bacterial cultures, treated for 40 minutes.
E. The effect of the order of addition of DEA-NONOate, KCN and H2O2 to the bacterial culture on the cell survival. The method was as in “B”, except that the three substances were aliquotted into individual glass tubes, and then bacterial cultures were added to them consecutively and transferred from tube to tube in the indicated order.
Remarkably, addition of catalase (the enzyme that degrades hydrogen peroxide) to aged HU completely prevented the cell killing (Fig. 6C). Although both cyanide and NO are bacteriostatic, they become strongly bactericidal if combined with otherwise mildly bactericidal concentrations of hydrogen peroxide (53–57 and Fig. 6C). By employing the HRP/AR assay, we detected peroxides in both aged HU stocks and especially in bacterial cell cultures treated with these stocks (Fig. 6D). Interestingly, we would not have been able to detect peroxides with GC-MS due to instability of these species at high temperatures.
Since it was proposed that NO + H2O2 kills by the same mechanism as CN + H2O2 57, we anticipated the same level of killing from the CN + H2O2 + NO three-component mixture. Unexpectedly, this three-component mixture killed bacterial cultures completely, to the level of detection (Fig. 6C), when added in certain order and above certain concentrations. Since cyanide is quite reactive, while NO and H2O2 are extremely reactive, it was not surprising to find that the killing power of the three-component mixture critically depended on the order of addition, with at least two combinations out of six total showing enhanced killing (Fig. 6E). We conclude that aged HU solutions are bactericidal due to slow decomposition to yield low amounts of peroxides, nitric oxide and hydrogen cyanide, the substances that, at these concentration, are bacteriostatic on their own, but kill bacteria in pairwise combinations and especially as a combination of all three.
Discussion
It is currently thought that hydroxyurea (HU) is essentially stable in aqueous solutions, does not modify DNA directly and have a single target in the cell, — the enzyme ribonucleotide reductase. However, by seeking to resolve a disagreement in the literature about whether HU, a specific and efficient inhibitor of DNA synthesis, saves E. coli cells from thymineless death in concentrations that do not significantly affect transcription 10; 28, we unexpectedly found that:
Even at the highest concentrations, HU allows significant residual DNA synthesis, which is at least the same as the level of synthesis in thymine-starved cells — this is probably why freshly-prepared HU has no influence on TLD;
In addition to its indirect effect on DNA via inhibition of replication, treatments with both fresh and aged HU modify or remove DNA bases directly, and in some cases also break duplex DNA.
There is decomposition in aqueous HU solutions, — as a result, aged HU solutions immediately kill most of the cells, with the surviving cells becoming resistant to TLD. GC-MS reveals two gasses, nitric oxide and hydrogen cyanide, in aged HU solutions. Enzymatic assays reveal peroxides that, due to their thermal instability, would be missed by GC-MS.
Although CN, NO or H2O2 are bacteriostatic or modestly bactericidal by themselves, NO + H2O2, CN + H2O2 and especially CN + H2O2 + NO mixtures are strongly bactericidal, explaining both effects of aged HU on TLD.
Below we will discuss all four findings under the corresponding numbers.
1. Does HU completely block DNA synthesis, or does it not?
Our finding of significant levels of residual DNA synthesis in HU-treated cells may sound surprising, but in fact is in good agreement with the previous results. Indeed, if the overall DNA accumulation is followed, no new synthesis in the presence of HU is detected 5; 9; 10; 28. In contrast, determination of the rate of DNA synthesis yields values that are only 3–6 times lower than the uninhibited ones (5; 40; 41, our results). We propose that the latter values better reflect the actual situation, since HU inhibition of the ribonucleotide reductase activity in crude extracts is never complete 2; 3; 5. One could suspect that the reason for the residual DNA synthesis is the presence of an alternative, HU-insensitive ribonucleotide reductases in the E. coli cells. E. coli has two HU-sensitive ribonucleotide reductases, NrdAB and NrdHIEF, and one HU-insensitive enzyme, NrdDG 58, but the latter enzyme works only anaerobically. Although the reason for the incomplete inhibition of the ribonucleotide reductase activity in the extracts of aerobically-growing cells is still unclear, the inefficiency of quenching of the tyrosyl radical in the enzyme active center due to a limited accessibility to HU 59; 60 may play a role.
What could be the reason for the discrepancy in determination of HU-inhibited DNA synthesis by cumulative incorporation versus the rate of incorporation? In (fresh) HU-treated WT cells, degradation of chromosomal DNA is not observed 10 but HU-treated recA mutant cells degrade their DNA more than untreated cells 32. It could be that, during HU treatment, while some DNA synthesis is going on, the stability of this newly-synthesized DNA is low, — hence, there is little cumulative DNA increase in cells treated with HU, in contrast to the still significant rate of DNA synthesis. We conclude that the high residual rates of DNA synthesis invalidate the idea that HU treatment blocks DNA replication completely.
2. Does HU treatment directly damage DNA bases?
We interpret sensitivity of the xthA nfo double mutant to HU as an indirect evidence that HU treatment modifies or removes DNA bases. The products of these two genes are, correspondingly, exonuclease III and endonuclease IV — the only known dedicated abasic site endonucleases in E. coli 45. Nicking of abasic sites by these enzymes is a critical step in the base-excision repair that removes modified DNA bases. Sensitivity of the double xthA nfo mutant of E. coli to an agent in question is taken to mean that the agent modifies or removes DNA bases directly 61. Sensitivity of the recA and recBC mutants to hydroxyurea is consistent with replication inhibition by the drug. However, the rapid loss of viability of the recA mutant to fresh HU (Fig. 2A) suggests that the DNA damaging species are present in solution right from the start, whereas HU is not continuously decomposing to re-generate these species (at least not in the “right” order and proportion, critical for the killing (Fig. 4E)). This is also consistent with the constant inhibitory level of DNA synthesis offered by HU throughout the time course of the experiments.
The only currently known metabolic effect of hydroxyurea (HU) is specific and efficient block of DNA synthesis due to poisoning of the nucleotide reductase and the resulting depletion of the DNA precursor pools 2–8. The complete reversibility of this block 4 dovetails with the reported bacteriostatic nature of HU treatment during the first several hours 5; 10. However, longer treatments with HU were found to be bactericidal 5; 9, prompting Rosenkranz and colleagues to question ribonucleotide reductase as the only target of HU and to propose an alternative target, chromosomal DNA 41. Rosenkranz indeed found that HU breaks pure DNA in solutions upon co-incubation at elevated temperatures, most likely by producing intermediate reactive species, like carbamoyloxyurea 25; 34. Although pure carbamoyloxyurea kills E. coli faster than HU, this killing is still too gradual 25 to explain the immediate bactericidal effects of aged HU that we observed in this study.
In contrast to the prolong treatments, killing by fresh HU in acute treatments, like the one we observed in recA, recBC and xthA nfo mutants, has never been reported before. The recA, recB, recG and ruv mutants, defective in double-strand break repair, were shown to be more sensitive to low concentrations of HU during chronic treatments on plates (although see 62 for an exception), which was taken as the consequence of the inability to maintain inhibited replication forks 43; 44. Inhibited replication forks can reverse, turning into Holliday junctions, with subsequent (hypothesized) replication fork breakage via Holliday junction resolution by RuvABC resolvasome 63. Indeed, both chromosomal fragmentation in, and killing of, HU-treated recB mutant cells are suppressed by inactivation of the RuvABC resolvasome, supporting the replication fork reversal mechanism 44. For the cells to survive fork reversal and breakage, disintegrated replication forks need to be reassembled by recombinational repair 63; 64, and the sensitivity of recA and recBC mutants to acute treatments with HU (32; 44, this work) supports this scenario. However, our finding that the base-excision repair-deficient mutant xthA nfo is also sensitive to fresh HU challenges the idea that the sensitivity of the rec mutants to HU is exclusively due to replication fork reversal with subsequent breakage and suggests that HU damages or removes DNA bases directly. Interestingly, there is a recent in vitro evidence of direct DNA damage by HU 26. The resistance of WT cells to fresh HU indicates that these DNA lesions are well-repaired. Separately and contrary to the claim in a single previous report 65, we found that the recA mutants are exquisitely sensitive to hydroxylamine (not shown), a major HU decomposition product in vivo; this result could have been expected, since hydroxylamine is widely used as an in vitro mutagen and therefore modifies DNA bases directly.
3. Instability of HU in aqueous solutions
It is currently unclear whether HCN, NO and peroxides are the products of limited decomposition of HU itself or of impurities in HU preparations. Our finding of these additional species in aqueous HU solutions should not be that surprising considering the ease of enzymatic breakdown of HU in vivo. In the human body, several enzymes are known to act on HU to produce hydroxylamine, nitroxyl (HNO) and nitric oxide (NO) 31. In fact, at one point it was suggested that the mode of HU’s therapeutic action is via its hydrolysis to hydroxylamine 22, but now NO has taken the lead as the most potent HU-breakdown product in the human body 31. In contrast to the in vivo situation, HU is considered to be essentially stable in neutral aqueous solutions 29–31, although there is anecdotal evidence to the contrary 33, especially at elevated temperatures 34, sometimes with outcomes as dramatic as industrial explosions 35. The belief about HU stability in vitro is reflected in the fact that most publications dealing with HU do not even mention how HU solutions were prepared and stored.
With the help of GC-MS, we revealed hydrogen cyanide and nitric oxide in aging HU solutions. We also found urea, cyanuric acid, formamide, carbon dioxide and hydroxylamine, but they all appear to be products of thermal decomposition of HU during subsequent gas chromatography, rather than of HU aging per se. It is interesting to note that NO, CO2, formamide and hydroxylamine are all generated during enzymatic HU decomposition in vivo 31. The finding of CN, NO and peroxides in aqueous solutions of HU should be an important consideration when HU is infused intravenously using pre-made aqueous solutions, which are stored before use for up to one week 66–70 or are prepared beforehand as liquid formulation for infants 30.
4. The bactericidal power of the CN + H2O2 + NO mixture
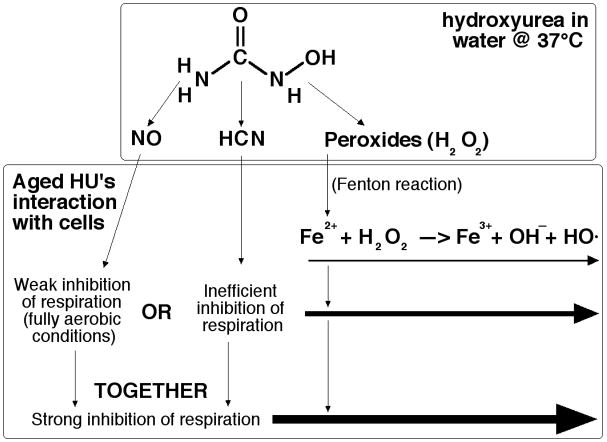
Our original observation of the bactericidal effect of aged HU can be understood in terms of the known enhanced bacterial killing by either NO + H2O2 53; 55; 57 or by KCN + H2O2 54; 56, although we do not currently know the source(s) and the nature of the peroxide(s) in aged HU — whether this is just hydrogen peroxide or some HU-based peroxides. It was assumed that the two pairs of substances (NO + H2O2 and KCN + H2O2) kill bacteria by the same mechanism, via increasing availability of electron donors and thus promoting the Fenton reaction via reduction of the free intracellular iron 56; 57, so our finding that the two mechanisms act synergistically may seem surprising. However, by themselves, both nitric oxide or hydrogen cyanide in our conditions will inhibit respiration only partially (reflecting intermediate bactericidal potency — Fig. 7) — nitric oxide due to the fully aerobic conditions, hydrogen cyanide due to the relatively low concentrations 47. On the other hand, these concentrations of hydrogen cyanide are known to greatly potentiate respiration inhibition by NO in fully aerobic conditions 47, providing a possible explanation to the observed synergistic killing (Fig. 7).
Fig. 7. A possible explanation for the synergistic killing of bacteria by CN + H2O2 + NO mixture.
Hydrogen peroxide kills (thin arrow) by generating hydroxyl radicals (HO·) via interaction with reduced iron (Fe2+). Inhibition of respiration is proposed to stimulate formation of Fe2+ 56; 57. Either nitric oxide (NO) or hydrogen cyanide (HCN) inhibit respiration, but only partially (NO — due to our fully aerobic conditions, while inhibition by HCN is generally inefficient 47; 48). Thus, enhanced killing by hydrogen peroxide (thicker arrow in the middle). However, together NO + HCN inhibit respiration synergistically 47, which, we propose, causes the synergistic killing of bacteria in the tri-component mixture with hydrogen peroxide (the thick arrow).
Our finding that NO and HCN complement each other in potentiating the bactericidal power of peroxides may have the real life counterpart in operation of the immune system. Currently, phagocytic immune cells are known to employ only NO + H2O2 bactericidal mechanism 71; 72. In this respect it is interesting to note that neutrophils, which are the major antimicrobial phagocytes, are known to induce production of hydrogen cyanide from the relatively abundant serum thiocyanate in response to antigenic activation in general 73 and to bacteria in particular 74–77. The neutrophil enzyme involved in this production, myeloperoxidase, is better known for its conversion of thiocyanate into hypothiocyanite (OSCN-) 78; 79, another bactericidal agent. Thus, we speculate that the human immune system could use both NO and HCN to synergistically potentiate bactericidal effects of peroxides.
Could decomposition products of HU contribute to its therapeutic potency?
Our finding of hydrogen cyanide and nitric oxide in HU solutions may be also relevant for the current medical use of HU to relieve symptoms associated with sickle cell anemia 17. HU reduces the frequency of the painful episodes and the acute chest syndrome in sickle cell anemia patients, yet the therapeutic basis of its action is still unclear 80. Painful episodes during sickle-cell anemia are caused by polymerization of the mutant hemoglobin inside erythrocytes, which dramatically reduces their plasticity while increasing adhesion to the walls of blood vessel, — the combination that results in blood vessel occlusion. Anti-sickling agents, decreasing this blood vessel clogging, belong to four distinct classes depending on their target. The first class acts at the gene level, stimulating the production of the fetal hemoglobin, which decreases polymerization of the sickle hemoglobin. The second class modifies the red cell membrane to increase the volume of erythrocytes, keeping sickle hemoglobin more dilute. The third class are vasodilators that transiently open up blood vessels. Finally, the fourth class covalently modifies the sickle hemoglobin protein itself, reducing its polymerization. HU was always considered to be an example of class I agent, because it does increase the production of fetal hemoglobin 81; surprisingly however, its anti-sickling effect was found to be poorly correlated with the level of fetal hemoglobin expression 80. In addition, HU was found to also increase the volume of erythrocytes 80, apparently acting as a class II agent. Our finding of nitric oxide in HU solutions, and, even more importantly, ready enzymatic conversion of HU into nitric oxide in the organism 31 positions HU as a class III agent, via generation of a known vasodilator (NO) 23. Finally, our finding of hydrogen cyanide in HU solutions suggest that HU can also act as a class IV agent.
Human body readily detoxifies small amounts of cyanide by converting it into an essentially inert thiocyanate with the help of rhodanese enzyme of liver (reviewed in 82; 83). Although the effect of thiocyanate on sickle-cell anemia was never addressed systematically, anecdotal evidence indicates that thiocyanate has a dramatic anti-sickling effect in patients 84 and a pronounced ameliorating effect on the sickle erythrocytes in vitro 85; 86. There is also a compelling evidence of much lower incidents of development of the sickle cell anemia and a decreased severity of the disease in parts of Africa where people maintain a traditional African diet rich in thyiocyanate-generating nitrilosides 87–89 (although see 90). Finally, a chemically related compound, cyanate, was at one time a promising treatment for sickle cell anemia 91; 92, because the resulting carbamylation of sickle hemoglobin prevents its polymerization 93. Importantly, chronic sublethal dietary ingestion of cyanide increases both the levels of serum thiocyanate and carbamylated hemoglobin in the animal model 94. All this allows us to speculate that HU, via cyanide and thiocyanate, modifies the sickle hemoglobin directly. If HU indeed covalently modifies sickle hemoglobin, better agents for this reaction (for example, thiocyanate) should be tested.
Concluding remarks
Returning to the original question that initiated this project, our results made the previous studies of TLD using HU inconclusive, since one cannot be sure now that the only effect of HU is direct DNA synthesis inhibition via block to DNA precursor synthesis. First, it is likely that the 30% rate of DNA synthesis during HU inhibition is still enough for DNA damage via replication to occur during TLD. Moreover, in contrast to common belief, our study indicates that even fresh HU directly damages DNA, while aged HU induces additional double-strand DNA breaks. Both DNA lesions cause cellular responses that may interfere with thymine starvation. In summary, it is too early to conclude that DNA replication does not play a role in thymineless death. Separately, it is important to consider the elusive yet potent toxicants in HU solutions during therapeutic use.
Materials and Methods
Strains
All E. coli strains are K-12 and are derivatives of AB1157 (Table 1). Alleles were moved between strains by P1 transduction 95; 96. Precise deletion-replacement alleles of selected genes were created by the Datsenko and Wanner method 97 and confirmed by PCR and phenotypic tests. The deo mutants were confirmed by their sensitivity to 5-fluorodeoxyuridine 98. The recA and recBCD mutants were confirmed by their characteristic sensitivity to UV irradiation and to tert-butyl-hydroperoxide. The xth nfo double mutants were confirmed by their sensitivity to H2O2 and tert-butyl-hydroperoxide 46. The tdk mutants were confirmed by their inability to incorporate exogenous thymidine 99.
Table 1.
Strains.
| Strain source and name | Relevant genotype or description | Reference or derivation |
|---|---|---|
| AB1157a | Rec+ | 107 |
| AAM1 | ΔthyA71::cat | Deletion-replacement |
| AK4 | Δ(srlR-recA)306::Tn10 | 108; 109 |
| JB1 | ΔrecBCD3::kan | 96 |
| AK141 | Δtdk6::kan | 39 |
| RPC501 | nfo-1::kan (xthA-pncA)90 | 46 |
| (s406) | ΔrecA635::kan | 97 |
| KKW23 | Δtdk7 (this work) | AK141; kan removed by pCP20 |
| KKW37 | ΔthyA72 (this work) | AAM1; cat removed by pCP20 |
| KKW47 | ΔdeoCABD1::cat (this work) | Deletion-replacement |
| KKW58 | ΔthyA72 deoCABD2 (this work) | KKW 37 x P1 KKW 58, cat removed by pCP20 |
| KKW59 | ΔdeoCABD2 (this work) | KKW 57; cat removed by pCP20 |
| KKW60 | ΔdeoCABD2 (srlR- recA)306::Tn10 (this work) | KKW 59 x P1 AK 4 (supplemented with p-recA+) |
| KKW62 | ΔdeoCABD2 recBCD3::kan (this work) | KKW 59 x P1 JB 1 (supplemented with p-recBCD+) |
AB1157 complete genotype: F- lambda- rac- thi-1 hisG4 Δ(gpt-proA)62 argE3 thr-1 leuB6 kdgK51 rfbD1 araC14 lacY1 galK2 xylA5 mtl-1 tsx-33 supE44(glnV44) rpsL31(strR)
Growth conditions
All strains were kept on LB plates with appropriate antibiotics, supplemented with 10 μg/ml thymidine if strains were thyA mutants. LB broth per 1 L contains: 10 g tryptone, 5 g Yeast extract, 5 g NaCl, pH 7.4 with 250 μl 4M NaOH; LB agar contained 15 g agar per 1 liter of LB broth; M9CAA medium contain 1xM9 salts (Difco), 2 mM MgSO4, 0.1 mM CaCl2, 10 mg/l thiamine (B1), 0.2 % glucose and 0.2 % Casamino acids (Difco). Davis minimal medium per 1 L contained: 1 g glucose, 7 g Dipotassium Phosphate, 2 g Monopotassium Phosphate, 0.5 g Sodium Citrate, 0.1 g Magnesium Sulfate, 1 g Ammonium Sulfate, supplemented with 0.2 % Casamino acids (Difco). MOPS-minimal phosphate medium was according to 100. Antibiotics were used in the following concentrations: kanamycin – 50 μg/ml, ampicillin – 100 μg/ml, tetracycline – 10 μg/ml, chloramphenicol – 12.5 μg/ml.
Cells were grown at 30°C in M9CAA medium until mid-logarithmic phase (OD600 ~ 0.2) followed by HU treatment and/or thymineless death conditions, unless noted otherwise. Removal of supplemented thymidine from thyA mutant cultures to induce thymineless death was performed by collecting cells on nitrocellulose membrane filters (type 0.22 μm GS, Millipore), washing with two volumes of 1% NaCl, followed by one volume of a pre-warmed medium without thymidine and resuspending in one volume of the same medium. Viability of cultures was measured by spotting 10 μl of serial dilutions (in 1% NaCl) of cultures on LB plates (supplemented with 10 μg/ml thymidine for thy mutants). Colonies formed at each spot were counted under the stereomicroscope after an overnight incubation of the plates at 22°C.
Chemicals
KCN (Mallinckrodt) was used as a source of CN−; 1 M KCN stocks were prepared immediately before each experiment. DEA-NONOate (Sigma) is an NO donor with a half-life in water of 2 minutes; because of its short half-life, it was added as crystals directly to the bacterial culture. Bovine liver catalase (16 mg/ml) and hydrogen peroxide (30%) were also from Sigma; both were diluted to working stocks freshly for every experiment.
Hydroxyurea preparation and treatment
We used hydroxyurea interchangeably from either Sigma, or Calbiochem, or MP Biomedicals, because we found no difference between them in our assays. The 1M stocks were prepared by dissolving HU crystals in sterile deionized water (we also tried HPLC-grade water, with no difference) and were added to bacterial cultures to achieve the desired HU concentrations. “Fresh” HU was prepared immediately before use. Preparation of “aged” HU went though several revisions to minimize inconsistencies (sometimes an “aged” stock would show an effect and sometimes it would not (compare Fig. 1A versus 1B, stock #1) before the protocol was established that made it work all the time. Protocol #1: at first, ‘aging” HU stocks were prepared in regular glass bottles with tightened screw caps, dated and stored at 4°C for weeks and months. The results were highly inconsistent, suggesting that something was escaping from the bottles upon their opening. Protocol #2: to trap the suspected gas decomposition products, we began aging HU stocks in closed microfuge tubes for 1–2 days at 37°C, — this made results more reproducible. Protocol #3: to further reduce the inconsistency, we switched to aging HU in standard addition aliquots (80 μl) in glass tubes under a 3 mm layer of mineral oil (Sigma), adding bacterial cultures directly to such a tube. Mineral oil emulsifies upon shaking and does not interfere with subsequent culture growth. Protocol #4: alternatively, in cases when no direct addition to bacterial cultures was necessary (enzymatic reactions, GC-MS) HU aging was for 1 day at 37°C in a closed 1.5 ml microfuge tube under mineral oil.
Method: measuring the rates of DNA synthesis by 3H-thyminidine incorporation
Cells (KKW59) were grown in M9CAA at 30°C until the exponential phase and treated with indicated concentrations of fresh HU. At different times, 200 μl aliquots of the cultures being treated were mixed with an equal volume of M9CAA supplemented with 1 μCi/ml of 3H-thymidine and were incubated at 37°C for 1 minute. 5 ml of ice-cold 5% TCA was then added to stop the reaction, and the TCA-killed cells were filtered through a 25 mm Fisher G6 glass fiber filter, using manifold, to remove the unincorporated label. The filters were washed with 5 ml of 5% TCA, followed by 5 ml of ethanol and drying. 100 μl of 0.1 M KOH were deposited on each filter to quench the intrinsic fluorescence during subsequent counting. After drying, the filters were soaked in a scintillation cocktail overnight, and the counts were determined in the LS 6500 Beckman scintillation counter. Counts were first normalized to OD600 taken at each time point, then to the CPM/OD600 of the initial value of each culture.
Measuring the rate of DNA synthesis by 32P-orthophosphate incorporation
Cells were grown in MOPS-minimal phosphate medium 100 to ensure robust 32P-orthophosphate incorporation. Kinetics and extent of TLD is not changed in the MOPS-minimal phosphate medium compared to M9 (not shown). Overnight cultures were diluted 100x and grown at 30°C to about OD600 = 0.1, then either thymidine was removed (from thyA mutants) by filtering and resuspending in thymidine-free medium (for TLD) or HU was added to thyA+ cells. All of the cultures from the same experiment were made to start with approximately the same OD600 so that the amount of cells at the beginning of each treatment was the same. After addition of 40 μCi of 32P-orthophosphate to each culture, they were continued shaking at 30°C. At indicated times, 150 μl aliquots were taken into microcentrifuge tubes, the cells were washed by pelleting and resuspending in 150 μl of TE, and were finally resuspended in 60 μl of TE buffer and kept at 37°C. Cells were converted into agarose plugs by adding 2.5 μl of 20 mg/ml proteinase K (NEB), 65 μl of molten 1.5% agarose in 0.2x lysis buffer (1% sarcosine, 50 mM Tris HCl pH 8.0, 25 mM EDTA), and transferring the mixture into a plug mold. Plugs were incubated with 1x lysis buffer overnight at 60°C, and then with alkaline electrophoresis loading buffer (50 mM NaOH, 1 mM EDTA) at room temperature with shaking for 2 hours, then inserted into the well of the 1.1% alkaline agarose gel (made on 30 mM NaOH, 1 mM EDTA) which was run for 900 minutes (15 hours) at 1.5 V/cm. The gel was then washed with 1M Tris-HCl pH 7.8 for 30 mins, vacuum-dried onto a piece of chromatography paper (Fisher) for 2 hours at 80°C and exposed to a PhosphorImager screen overnight for quantification.
Since the top band at the gel origin does not disappear upon DNase treatment, only the chromosomal DNA band was used for quantification. The “current interval” incorporation was calculated by subtracting cumulative incorporation until the previous time point from the cumulative incorporation at this time point. Then it was divided by the interval length (in hours) and, finally, normalized to the incorporation in the untreated control during the first 30 minutes, to obtain the relative rate of DNA synthesis.
Detection of chromosomal fragmentation by Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis was run as before 39; 101 with some minor modifications: all strains were grown in M9CAA; overnight cultures were diluted 100x and were grown with 10 μCi of 32P-orthophosphoric acid for 3 hours at 30°C; OD600 was taken to determine the volume of aliquots to be used so that, within the experiment, the cell mass per lane was the same for all samples; 2.5 μl of proteinase K (20 mg/ml) and 65 μl of 1.2% agarose in lysis buffer were added and mixed by pipetting before casting the suspension in the plug molds.
Measuring mutation rates in HU treated cells
Independent cultures were shaken in M9CAA at 30°C till OD600 = 0.2. HU was added to 80 mM, and shaking at 30°C continued for 2 hours, after which the cultures were spun down and resuspended in the same medium without HU. Cultures were then allowed to outgrow overnight to saturation. Cultures were serially diluted in 1% NaCl for spotting on LB to obtain titer and were spread on LB + 100 μg/ml Rifampicin plates to select for mutants. Colonies were scored after 4 days of incubation at 22°C. Untreated controls received the same handling except that water was added instead of HU. To calculate mutation rates, the Lea and Coulson method of the medians, as explained by Rosche and Foster 102, was used. The mutation rate = m/(1.44Nt) where m is the number of mutations per culture and Nt is the final number of cells in a culture.
GC-MS Analysis
Gas chromatography was performed on Agilent 6890 gas chromatographer equipped with an Agilent 5973 mass selective detector and Agilent 7683B autosampler. Specific columns used are identified in figure legends. Perfluorotributylamine (PFTBA) was used to autotune the system daily. Operation of the MSD was in the electronimpact (EI) mode at 70 eV. While the injection temperature varied between 200°C and 300°C depending on the specifics of the run, the interface temperature was always set to 250°C, and the ion source temperature was always adjusted to 230°C. The helium carrier gas (99.995% purity) was set at a constant flow rate between 1.3 and 3.5 ml min−1, depending on the specifics of the run. Mass spectra were evaluated using the HP Chemstation (Agilent, Palo Alto, CA) and AMDIS (NIST, Gaithersburg, MD) programs.
Derivatization of HU for GC-MS
50 μl of 1 M stocks of HU and HA were prepared in water, then dried in a vacuum centrifuge. The 1:1 ratio of MSTFA (Fluka) and pyridine were then added to the reaction and incubated at 40°C for 1 hour. Samples were stored at − 80°C until injection.
H2O2 detection
The procedure followed the protocol of Seaver and Imlay 103, with modifications. In the presence of H2O2, horseradish peroxidase (HRP) (type II, from Sigma) oxidizes amplex red (AR) (Molecular Probes) to the fluorescent product resorufin. HU- or H2O2-treated bacterial cultures (KKW 59) were collected by centrifugation after 40 mins of treatment at 30°C, and the supernatant was recovered for the assay. One milligram of AR was dissolved in 0.78 ml of DMSO, and 0.75 ml of this solution was then diluted into 18 ml of 50 mM potassium phosphate (pH 7.8) to generate a 200 μM stock solution. The bottle with this solution was wrapped in foil to shield it from light. HRP was dissolved in 50 mM potassium phosphate (pH 7.8) to 0.02 mg/ml. To measure H2O2, 0.45 ml of a sample (HU stocks or supernatant from cultures) was mixed with 0.25 ml of AR and 0.25 ml of HRP stocks. Fluorescence was then measured in a Shimadzu RF Mini-150 fluorometer and converted to H2O2 concentration using a calibration curve obtained from standard H2O2 serial dilutions after accounting for the background (value obtained from the reaction with water only was set to zero).
Inhibition of respiration in cells extracts
The procedure followed the protocol of Imlay and Fridovich 104, with modifications 105. Respiration rate was monitored spectrophotometrically by the oxidation of NADH by membrane vesicles that were inverted and could oxidize respiratory substrates and reduce O2 on the external surface. The vesicles were obtained from 100 ml of wild type cells, grown in M9CAA and collected at mid-exponential phase, and were centrifuged and resuspend in 3 ml of cold 50 mM phosphate buffer (pH 7.8). Cells were then lysed with a French press and centrifuged for 20 min at 17,000 x g at 4°C to remove cell debris. Vesicles were resuspended in 1.5 ml of cold phosphate buffer and kept on ice. Assay was performed at 37°C in 50 mM phosphate buffer, 5 mM NADH (Sigma), containing the indicated concentrations of KCN and HU, with 100 μl of isolated vesicles. NADH oxidation was monitored for 15 mins by the decrease in absorbance of NADH at 340 nm in the presence of KCN or HU using a Beckman DU 640 Spectrophotometer. The kinetics of NADH oxidation reflects the activity of the terminal oxidases in the respiratory chain; the rate of respiration was obtained by the slope of the decreasing curve. The rate of respiration calculated from untreated vesicles was set to be 100% active, and the levels of respiration in all other samples were normalized accordingly.
Differential Interference Contrast (DIC) microscopy
The procedure followed the protocol of Gut et al. 106, with modifications. The 20 μl aliquots were removed from indicated cultures and fixed with 4% formaldehyde (Sigma) for 30 min at 37 °C followed by mounting on glass slides in 20% glycerol (Sigma). Differential Interference Contrast (DIC) microscopy images were collected using an Applied Precision assembled DeltaVision EpiFluorescence microscope containing an Olympus Plan Apo 100x oil objective with NA 1.42 and a working distance of 0.15 mm. The images were processed using SoftWoRX Explorer Suite (Issaquah, WA).
Supplementary Material
Acknowledgments
We want to thank Dr. Alex Ulanov from Roy J Carver Metabolomics center at UIUC for his help with Mass Spectrometry, Jim Imlay, Sergei Korshunov, Soo Jin Jang and Mianzhi Gu for their help with various enzymatic assays and Steven Blanke and Ian Gut for their help with microscopy. This work was supported by grant # RSG-05-135-01-GMC from the American Cancer Society and by grant # GM 073115 from the National Institutes of Health.
Abbreviations
- HU
hydroxyurea
- TLD
thymineless death
- DIS#N
discussed in the Supplement, section #N
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Navarra P, Preziosi P. Hydroxyurea: new insights on an old drug. Crit Rev Oncol Hematol. 1999;29:249–255. doi: 10.1016/s1040-8428(98)00032-8. [DOI] [PubMed] [Google Scholar]
- 2.Elford HL. Effect of hydroxyurea on ribonucleotide reductase. Biochem Biophys Res Commun. 1968;33:129–135. doi: 10.1016/0006-291x(68)90266-0. [DOI] [PubMed] [Google Scholar]
- 3.Krakoff IH, Brown NC, Reichard P. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 1968;28:1559–1565. [PubMed] [Google Scholar]
- 4.Reichard P, Ehrenberg A. Ribonucleotide reductase — a radical enzyme. Science. 1983;221:514–519. doi: 10.1126/science.6306767. [DOI] [PubMed] [Google Scholar]
- 5.Sinha NK, Snustad DP. Mechanism of inhibition of deoxyribonucleic acid synthesis in Escherichia coli by hydroxyurea. J Bacteriol. 1972;112:1321–1334. doi: 10.1128/jb.112.3.1321-1334.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarbro JW. Mechanism of action of hydroxyurea. Semin Oncol. 1992;19, suppl 9:1–10. [PubMed] [Google Scholar]
- 7.Young CW, Hodas S. Hydroxyurea: inhibitory effect on DNA metabolism. Science. 1964;146:1172–1174. doi: 10.1126/science.146.3648.1172. [DOI] [PubMed] [Google Scholar]
- 8.Young CW, Schochetman G, Hodas S, Balis ME. Inhibition of DNA synthesis by hydroxyurea: structure-activity relationships. Cancer Res. 1967;27:535–540. [PubMed] [Google Scholar]
- 9.Rosenkranz HS, Carr HS. Studies with hydroxyurea. II Prolonged exposure of Escherichia coli to hydroxyurea. J Bacteriol. 1966;92:178–185. doi: 10.1128/jb.92.1.178-185.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenkranz HS, Garro AJ, Levy JA, Carr HS. Studies with hydroxyurea. I The reversible inhibition of bacterial DNA synthesis and the effect of hydroxyurea on the bactericidal action of streptomycin. Biochim Biophys Acta. 1966;114:501–515. [PubMed] [Google Scholar]
- 11.Bradley MO, Kohn KW, Sharkey NA, Ewig RA. Differential cytotoxicity between transformed and normal human cells with combinations of aminonucleoside and hydroxyurea. Cancer Res. 1977;37:2126–2131. [PubMed] [Google Scholar]
- 12.Farber E, Baserga R. Differential effects of hydroxyurea on survival of proliferating cells in vivo. Cancer Res. 1969;29:136–139. [PubMed] [Google Scholar]
- 13.Grdina DJ, Sigdestad CP, Peters LJ. Phase-specific cytotoxicity in vivo of hydroxyurea on murine fibrosarcoma cells synchronized by centrifugal elutriation. Br J Cancer. 1979;39:152–158. doi: 10.1038/bjc.1979.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawicki SG, Godman GC. On the differential cytotoxicity of actinomycin D. J Cell Biol. 1971;50:746–761. doi: 10.1083/jcb.50.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair WK. Hydroxyurea: differential lethal effects on cultured mammalian cells during the cell cycle. Science. 1965;150:1729–1731. doi: 10.1126/science.150.3704.1729. [DOI] [PubMed] [Google Scholar]
- 16.Rice L, Baker KR. Current management of the myeloproliferative disorders: a case-based review. Arch Pathol Lab Med. 2006;130:1151–1156. doi: 10.5858/2006-130-1151-CMOTMD. [DOI] [PubMed] [Google Scholar]
- 17.Lanzkron S, Strouse JJ, Wilson R, Beach MC, Haywood C, Park H, Witkop C, Bass EB, Segal JB. Systematic review: hydroxyurea for the treatment of adults with sickle cell disease. Ann Intern Med. 2008;148:939–955. doi: 10.7326/0003-4819-148-12-200806170-00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton HB, Scott SR, Volpi C. Hydroxyurea chemotherapy for meningiomas: enlarged cohort with extended follow-up. Br J Neurosurg. 2004;18:495–499. doi: 10.1080/02688690400012392. [DOI] [PubMed] [Google Scholar]
- 19.Lori F, Foli A, Groff A, Lova L, Whitman L, Bakare N, Pollard RB, Lisziewicz J. Optimal suppression of HIV replication by low-dose hydroxyurea through the combination of antiviral and cytostatic (‘virostatic’) mechanisms. AIDS. 2005;19:1173–1181. doi: 10.1097/01.aids.0000176217.02743.d1. [DOI] [PubMed] [Google Scholar]
- 20.Schilsky RL. Biochemical pharmacology of chemotherapeutic drugs used as radiation enhancers. Semin Oncol. 1992;19 (4 Supple 11):2–7. [PubMed] [Google Scholar]
- 21.Frame D. Chronic myeloid leukemia: standard treatment options. Am J Health Syst Pharm. 2006;63 (23 Suppl 8):S10–S14. doi: 10.2146/ajhp060525. [DOI] [PubMed] [Google Scholar]
- 22.Fishbein WN, Carbone PP. Hydroxyurea: mechanism of action. Science. 1963;142:1069–1070. doi: 10.1126/science.142.3595.1069. [DOI] [PubMed] [Google Scholar]
- 23.Jiang J, Jordan SJ, Barr DP, Gunther MR, Maeda H, Mason RP. In vivo production of nitric oxide in rats after administration of hydroxyurea. Mol Pharmacol. 1997;52:1081–1086. doi: 10.1124/mol.52.6.1081. [DOI] [PubMed] [Google Scholar]
- 24.King SB. N-hydroxyurea and acyl nitroso compounds as nitroxyl (HNO) and nitric oxide (NO) donors. Curr Top Med Chem. 2005;5:665–673. doi: 10.2174/1568026054679362. [DOI] [PubMed] [Google Scholar]
- 25.Rosenkranz HS. Some biological effects of carbamoyloxyurea, an oxidation product of hydroxyurea. J Bacteriol. 1970;102:20–23. doi: 10.1128/jb.102.1.20-23.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakano K, Oikawa S, Hasegawa K, Kawanishi S. Hydroxyurea induces site-specific DNA damage via formation of hydrogen peroxide and nitric oxide. Jpn J Cancer Res. 2001;92:1166–1174. doi: 10.1111/j.1349-7006.2001.tb02136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 28.Morganroth PA, Hanawalt PC. Role of DNA replication and repair in thymineless death in Escherichia coli. J Bacteriol. 2006;188:5286–5288. doi: 10.1128/JB.00543-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fishbein WN, Winter TS, Davidson JD. Urease catalysis. I Stoichiometry, specificity and kinetics of a second substrate: hydroxyurea. J Biol Chem. 1965;240:2402–2406. [PubMed] [Google Scholar]
- 30.Heeney MM, Whorton MR, Howard TA, Johnson CA, Ware RE. Chemical and functional analysis of hydroxyurea oral solutions. J Pediatric Hematol Oncol. 2004;26:179–184. doi: 10.1097/00043426-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 31.King SB. Nitric oxide production from hydroxyurea. Free Radic Biol Med. 2004;37:737–744. doi: 10.1016/j.freeradbiomed.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 32.Shimada K, Shibata Y, Takagi Y. Bacteriocidal action of hydroxyurea on Escherichia coli K12 recA. Japan. J Microbiol. 1975;19:72–74. doi: 10.1111/j.1348-0421.1975.tb00850.x. [DOI] [PubMed] [Google Scholar]
- 33.El-Yazigi A, Al-Rawithi S. Analysis of hydroxyurea in capsules and aqueous solution and stability study with capillary gas chromatography and thermionic (N–P) specific detection. Pharm Res. 1992;9:115–118. doi: 10.1023/a:1018996130753. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs SJ, Rosenkranz HS. Detection of a reactive intermediate in the reaction between DNA and hydroxyurea. Cancer Res. 1970;30:1084–1094. [PubMed] [Google Scholar]
- 35.Lunghi A, Aloni C, Gigante L, Mazzei N, Cardillo P. Hydroxyurea explosion: a thermoanalytical and calorimetric study. Journal of Loss Prevention in the Process Industries. 2002;15:489–495. [Google Scholar]
- 36.Drlica K, Engle EC, Manes SH. DNA gyrase on the bacterial chromosome: possibility of two levels of action. Proc Natl Acad Sci USA. 1980;77:6879–6883. doi: 10.1073/pnas.77.11.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuzminov A, Schabtach E, Stahl FW. χ-sites in combination with RecA protein increase the survival of linear DNA in E. coli by inactivating ExoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kouzminova EA, Kuzminov A. Fragmentation of replicating chromosomes triggered by uracil in DNA. J Mol Biol. 2006;355:20–33. doi: 10.1016/j.jmb.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 39.Kouzminova EA, Rotman E, Macomber L, Zhang J, Kuzminov A. RecA-dependent mutants in E. coli reveal strategies to avoid replication fork failure. Proc Natl Acad Sci USA. 2004;101:16262–16267. doi: 10.1073/pnas.0405943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill WE, Fangman WL. Single-strand breaks in deoxyribonucleic acid and viability loss during deoxyribonucleic acid synthesis inhibition in Escherichia coli. J Bacteriol. 1973;116:1329–1335. doi: 10.1128/jb.116.3.1329-1335.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenkranz HS, Jacobs SJ, Carr HS. Studies with hydroxyurea, VIII. The deoxyribonucleic acid of hydroxyurea-treated cells. Biochim Biophys Acta. 1968;161:428–441. [PubMed] [Google Scholar]
- 42.Kuzminov A, Stahl FW. Overview of homologous recombination and repair machines. In: Higgins NP, editor. The Bacterial Chromosome. ASM Press; Washington, D.C.: 2005. pp. 349–367. [Google Scholar]
- 43.Beam CE, Saveson CJ, Lovett ST. Role for radA/sms in recombination intermediate processing in Escherichia coli. J Bacteriol. 2002;184:6836–6844. doi: 10.1128/JB.184.24.6836-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guarino E, Salguero I, Jiménez-Sánchez A, Guzmán EC. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J Bacteriol. 2007;189:5782–5786. doi: 10.1128/JB.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 46.Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevanin TM, Ioannidis N, Mills CE, Kim SO, Hughes MN, Poole RK. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo’ or bd, from nitric oxide. J Biol Chem. 2000;275:35868–35875. doi: 10.1074/jbc.M002471200. [DOI] [PubMed] [Google Scholar]
- 48.Yu H, Sato EF, Nagata K, Nishikawa M, Kashiba M, Arakawa T, Kobayashi K, Tamura T, Inoue M. Oxygen-dependent regulation of the respiration and growth of Escherichia coli by nitric oxide. FEBS Lett. 1997;409:161–165. doi: 10.1016/s0014-5793(97)00494-8. [DOI] [PubMed] [Google Scholar]
- 49.Dunbar KR, Heintz RA. Chemistry of Transition Metal Cyanide Compounds: Modern Perspectives. Progress in Inorganic Chemistry. 1997;45:283–391. [Google Scholar]
- 50.Kujundzic N, Nigovic B, Sankovic K. Reaction of Hydroxyurea with Iron(III): products and the stoichiometry of the redox reaction. Z Anorg Allg Chem. 2004;630:2749–2753. [Google Scholar]
- 51.Miller MJ. Syntheses and therapeutic potential of hydroxamic acid based siderophores and analogues. Chem Rev. 1989;89:1563–1579. [Google Scholar]
- 52.Nakayama H, Hanawalt PC. Sedimentation analysis of deoxyribonucleic acid from thymine-starved Escherichia coli. J Bacteriol. 1975;121:537–547. doi: 10.1128/jb.121.2.537-547.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brunelli L, Crow JP, Beckman JS. The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch Biochem Biophys. 1995;316:327–334. doi: 10.1006/abbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- 54.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 55.Pacelli R, Wink DA, Cook JA, Krishna MC, DeGraff W, Friedman N, Tsokos M, Samuni A, Mitchell JB. Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J Exp Med. 1995;182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodmansee AN, Imlay JA. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J Biol Chem. 2002;277:34055–34066. doi: 10.1074/jbc.M203977200. [DOI] [PubMed] [Google Scholar]
- 57.Woodmansee AN, Imlay JA. A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol Microbiol. 2003;49:11–22. doi: 10.1046/j.1365-2958.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- 58.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 59.Karlsson M, Sahlin M, Sjöberg BM. Escherichia coli ribonucleotide reductase. Radical susceptibility to hydroxyurea is dependent on the regulatory state of the enzyme. J Biol Chem. 1992;267:12622–12626. [PubMed] [Google Scholar]
- 60.Sneeden JL, Loeb LA. Mutations in the R2 subunit of ribonucleotide reductase that confer resistance to hydroxyurea. J Biol Chem. 2004;279:40723–40728. doi: 10.1074/jbc.M402699200. [DOI] [PubMed] [Google Scholar]
- 61.Spek EJ, Wright TL, Stitt MS, Taghizadeh NR, Tannenbaum SR, Marinus MG, Engelward BP. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J Bacteriol. 2001;183:131–138. doi: 10.1128/JB.183.1.131-138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sutera VAJ, Lovett ST. The role of replication initiation control in promoting survival of replication fork damage. Mol Microbiol. 2006;60:229–239. doi: 10.1111/j.1365-2958.2006.05093.x. [DOI] [PubMed] [Google Scholar]
- 63.Michel B, Grompone G, Florès MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuzminov A. Instability of inhibited replication forks in E. coli. BioEssays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 65.Ishii Y, Kondo S. Comparative analysis of deletion and base-change mutabilities of Escherichia coli B strains differing in DNA repair capacity (wild-type, uvrA-, polA-, recA-) by various mutagens. Mutat Res. 1975;27:27–44. doi: 10.1016/0027-5107(75)90271-7. [DOI] [PubMed] [Google Scholar]
- 66.Blumenreich MS, Kellihan MJ, Joseph UG, Lalley KA, Sherrill EJ, Sullivan DM, Hamm JT, Gentile PS, Sheth SP, Seeger J, Woodcock TM. Long-term intravenous hydroxyurea infusions in patients with advanced cancer. A phase I trial . Cancer. 1993;71:2828–2832. doi: 10.1002/1097-0142(19930501)71:9<2828::aid-cncr2820710924>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 67.Kaubisch A, Kaleya R, Haynes H, Rozenblit A, Wadler S. Phase II clinical trial of parenteral hydroxyurea in combination with fluorouracil, interferon and filgrastim in the treatment of advanced pancreatic, gastric and neuroendocrine tumors. Cancer Chemother Pharmacol. 2004;53:337–340. doi: 10.1007/s00280-003-0727-4. [DOI] [PubMed] [Google Scholar]
- 68.Newman EM, Carroll M, Akman SA, Chow W, Coluzzi P, Hamasaki V, Leong LA, Margolin KA, Morgan RJ, Raschko JW, Shibata S, Somlo G, Tetef M, Yen Y, Ahn CW, Doroshow JH. Pharmacokinetics and toxicity of 120-hour continuous-infusion hydroxyurea in patients with advanced solid tumors. Cancer Chemother Pharmacol. 1997;39:254–258. doi: 10.1007/s002800050569. [DOI] [PubMed] [Google Scholar]
- 69.Smith DC, Vaughan WP, Gwilt PR, Trump DL. A phase I trial of high-dose continuous-infusion hydroxyurea. Cancer Chemother Pharmacol. 1993;33:139–143. doi: 10.1007/BF00685331. [DOI] [PubMed] [Google Scholar]
- 70.Swinnen LJ, Rankin C, Carraway H, Albain KS, Townsend JJ, Budd GT, Kish JA, Rivkin SE, Blumenthal DT. A phase II study of cisplatin preceded by a 12-h continuous infusion of concurrent hydroxyurea and cytosine arabinoside (Ara-C) for adult patients with malignant gliomas (Southwest Oncology Group S9149) J Neurooncol. 2008;86:353–358. doi: 10.1007/s11060-007-9483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hampton MB, Kettle AJ, Winterbourn CC. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood. 1998;92:3007–3017. [PubMed] [Google Scholar]
- 72.Vazquez-Torres A, Fang FC. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 2001;9:29–33. doi: 10.1016/s0966-842x(00)01897-7. [DOI] [PubMed] [Google Scholar]
- 73.Graham GG, Whitehouse MW, Bushell GR. Aurocyanide, dicyano- aurate (I), a pharmacologically active metabolite of medicinal gold complexes. Inflammopharmacology. 2008;16:126–132. doi: 10.1007/s10787-007-0020-y. [DOI] [PubMed] [Google Scholar]
- 74.But PG, Muraviev RA, Fomina VA, Rogovin VV. Antimicrobial activity of mieloperoxidase from neutrophilic peroxisome. Biology Bulletin. 2002;29:212–215. [Google Scholar]
- 75.Stelmaszynska T. Formation of HCN by human phagocytosing neutrophils-- 1. Chlorination of Staphylococcus epidermidis as a source of HCN. Int J Biochem. 1985;17:373–379. doi: 10.1016/0020-711x(85)90213-7. [DOI] [PubMed] [Google Scholar]
- 76.Stelmaszynska T. Formation of HCN and its chlorination to ClCN by stimulated human neutrophils--2. Oxidation of thiocyanate as a source of HCN. Int J Biochem. 1986;18:1107–1114. doi: 10.1016/0020-711x(86)90084-4. [DOI] [PubMed] [Google Scholar]
- 77.Zgiczynski JM, Stelmaszynska T. Hydrogen cyanide and cyanogen chloride formation by the myeloperoxidase-H2O2-Cl- system. Biochim Biophys Acta. 1979;567:309–314. doi: 10.1016/0005-2744(79)90116-5. [DOI] [PubMed] [Google Scholar]
- 78.Furtmüller PG, Burner U, Obinger C. Reaction of myeloperoxidase compound I with chloride, bromide, iodide, and thiocyanate. Biochemistry. 1998;37:17923–17930. doi: 10.1021/bi9818772. [DOI] [PubMed] [Google Scholar]
- 79.Thomas EL, Fishman M. Oxidation of chloride and thiocyanate by isolated leukocytes. J Biol Chem. 1986;261:9694–9702. [PubMed] [Google Scholar]
- 80.Charache S. Mechanism of action of hydroxyurea in the management of sickle cell anemia in adults. Semin Hematol. 1997;34:15–21. [PubMed] [Google Scholar]
- 81.Charache S. Fetal hemoglobin, sickling, and sickle cell disease. Adv Pediatr. 1990;37:1–31. [PubMed] [Google Scholar]
- 82.Faust RA OAK RIDGE RESERVATION ENVIRONMENTAL RESTORATION PROGRAM. Toxicity Summary for Cyanide 1994 [Google Scholar]
- 83.World Health Organization. Hydrogen Cyanide and Cyanides: Human Health Aspects. In: Petrova F, Fishbein L, editors. Concise international chemical assessment document. 2004. p. 61. [Google Scholar]
- 84.Torrance EG, Schnabel TG. Potassium sulphocyanate: a note on its use for the painful crises in sickle cell anemia. Ann Intern Med. 1932;6:782–788. [Google Scholar]
- 85.Malomo SO, Oyewole OI. Cations Content And Membrane Properties Of Human Sickle Blood Incubated With Hydroxyurea, Tellurite And Thiocyanate. Egyptian J Biochem Mol Biol. 2008;26:117–125. [Google Scholar]
- 86.Oyewole OI, Malomo SO, Adebayo JO. Comparative studies on antisickling properties of thiocyanate, tellurite and hydroxyurea. Pakistan J Med Sci. 2008;24:18–22. [Google Scholar]
- 87.Agbai O. Anti-sickling effect of dietary thiocyanate in prophylactic control of sickling cell anemia. J Natl Med Assoc. 1986;78:1053–1056. [PMC free article] [PubMed] [Google Scholar]
- 88.Houston RG. Sickle cell anemia and dietary precursors of cyanate. Am J Clin Nutr. 1973;26:1261–1264. doi: 10.1093/ajcn/26.11.1261. [DOI] [PubMed] [Google Scholar]
- 89.Houston RG. Letter: Dietary nitriloside and sickle cell anemia in Africa. Am J Clin Nutr. 1974;27:766–769. doi: 10.1093/ajcn/27.8.766. [DOI] [PubMed] [Google Scholar]
- 90.Lambotte C. Letter: Sickle cell anemia and dietary precursors of cyanate. Am J Clin Nutr. 1974;27:765–766. doi: 10.1093/ajcn/27.8.765. [DOI] [PubMed] [Google Scholar]
- 91.Cerami A, Allen TA, Graziano JH, DeFuria FG, Manning JM, Gillette PN. Pharmacology of cyanate. I General effects on experimental animals. J Pharmacol Exp Ther. 1973;185:653–666. [PubMed] [Google Scholar]
- 92.Gillette PN, Manning JM, Cerami A. Increased survival of sickle-cell erythrocytes after treatment in vitro with sodium cyanate. Proc Natl Acad Sci USA. 1971;68:2791–2793. doi: 10.1073/pnas.68.11.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cerami A, Manning JM. Potassium cyanate as an inhibitor of the sickling of erythrocytes in vitro. Proc Natl Acad Sci USA. 1971;68:1180–1183. doi: 10.1073/pnas.68.6.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jackson LC, Oseguera M, Medrano S, Kim YL. Carbamylation of hemoglobin in vivo with chronic sublethal dietary cyanide: implications for hemoglobin S. Biochem Med Metab Biol. 1988;39:64–68. doi: 10.1016/0885-4505(88)90059-x. [DOI] [PubMed] [Google Scholar]
- 95.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 96.Miranda A, Kuzminov A. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics. 2003;163:1255–1271. doi: 10.1093/genetics/163.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zalkin H, Nygaard P. Biosynthesis of Purine Nucleotides. In Escherichia coli and Salmonella. In: Neidhardt FC, editor. Cellular and Molecular Biology. ASM Press; Washington, D.C.: 1996. pp. 561–579. [Google Scholar]
- 99.Hiraga S, Igarashi K, Yura T. A deoxythymidine kinase-deficient mutant of Escherichia coli, I. Isolation and some properties. Biochim Biophys Acta. 1967;145:41–51. doi: 10.1016/0005-2787(67)90652-1. [DOI] [PubMed] [Google Scholar]
- 100.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rotman E, Kuzminov A. The mutT defect does not elevate chromosomal fragmentation in Escherichia coli because of the surprisingly low levels of MutM/MutY-recognized DNA modifications. J Bacteriol. 2007;189:6976–6988. doi: 10.1128/JB.00776-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 105.Korshunov S, Imlay JA. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol. 2006;188:6326–6334. doi: 10.1128/JB.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gut IM, Prouty AM, Ballard JD, van der Donk WA, Blanke SR. Inhibition of Bacillus anthracis spore outgrowth by nisin. Antimicrob Agents Chemother. 2008;52:4281–4288. doi: 10.1128/AAC.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bachmann BJ. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In Escherichia coli and Salmonella typhimurium. In: Neidhardt FC, editor. Cellular and Molecular Biology. American Society for Microbiology; Washington, D.C.: 1987. pp. 1190–1219. [Google Scholar]
- 108.Czonka LN, Clark AJ. Deletions generated by the transposon Tn10 in the srl-recA region of the Escherichia coli K-12 chromosome. Genetics. 1979;93:321–343. doi: 10.1093/genetics/93.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shi IY, Stansbury J, Kuzminov A. Defect in Acetyl-CoA<—>Acetate Pathway Poisons Recombinational Repair-deficient mutants of Escherichia coli. J Bacteriol. 2005;187:1266–1275. doi: 10.1128/JB.187.4.1266-1275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.