Summary
MITF (Microphthalmia-associated transcription factor) is involved in melanocyte cell development, pigmentation and neoplasia. To determine whether MITF is somatically mutated in melanoma, we compared the sequence of MITF from primary and metastatic lesions to patient matched normal DNA. In the 50 metastatic melanoma tumor lines analyzed, we discovered four samples that had genomic amplifications of MITF and four had MITF mutations in the regions encoding the transactivation, DNA binding or basic, helix-loop-helix domains. Sequence analysis for SOX10, a transcription factor which both acts upstream of MITF and synergizes with MITF, identified an additional three samples with frameshift or nonsense mutations. MITF and SOX10 were found to be mutated in a mutually exclusive fashion, possibly suggesting disruption in a common genetic pathway. Taken together we found that over 20% of the metastatic melanoma cases had alterations in the MITF pathway. We show that the MITF pathway is also altered in primary melanomas: 2/26 demonstrated mutations in MITF and 6/55 demonstrated mutations in SOX10. Our findings suggest that altered MITF function during melanomagenesis can be achieved by MITF amplification, MITF single base substitutions or by mutation of its regulator SOX10.
Keywords: 5-7 MITF, SOX10, melanoma, mutations, p21, sequencing
Introduction
Malignancy of pigment-producing cells known as melanocytes results in melanoma, an aggressive and often fatal cancer whose incidence is increasing worldwide. Melanocytes arise from the neural crest during embryonic development, and throughout their maturation migrate widely and proliferate extensively prior to their terminal differentiation and entry into the epidermis and hair follicle. In addition, melanocytes undergo multiple cycles of regeneration, in which melanocyte stem cells give rise to new populations of melanocytes.
Microphthalmia-associated Transcription Factor (MITF), a basic, helix-loop-helix, leucine-zipper transcription factor, plays a central role in regulation of the differentiation, growth, and survival of cells of the melanocytic lineage. MITF regulates the transcription of a large number of downstream targets that are involved in melanocyte differentiation, proliferation, and survival (Levy et al., 2006, Hou and Pavan, 2008, Steingrimsson et al., 2004). Therefore, changes in the expression levels of MITF and subsets of its various downstream targets can result in widely different outcomes, such as cellular growth or death.
Although melanomas are heterogeneous in nature, recent studies of gene expression indicate that melanoma progression appears to involve notable changes in the expression of genes shown to be important for melanocyte development and regeneration (Ryu et al., 2007, White and Zon, 2008). The multifaceted role of MITF in supporting differentiation, proliferation or cell death in normal melanocytes suggests that, depending on the downstream targets affected, changes in MITF signaling could result in downstream changes that would support melanoma formation. For example, MITF has been shown to be amplified in 10–20% of melanoma metastases and to potentially act as a dominant oncogene (Garraway et al., 2005). Conversely, MITF regulates expression of the cell cycle gene p21, and mutations of melanoma-associated genes that would reduce MITF stability or transcriptional activity could potentially remove MITF's tight control of the cell cycle (Carreira et al., 2005, Loercher et al., 2005). Recent melanoma gene expression profiling supported a model in which highly proliferative but weakly metastatic melanomas displayed a “neural crest-like” gene expression pattern, in which MITF expression was activated by the WNT pathway; however, melanomas that have progressed to a highly metastatic and weakly proliferative state had expression patterns that resembled TGFbeta-like signaling, with a positive feedback loop and an inhibition of WNT pathway, resulting in downregulation of neural crest genes including MITF (Hoek et al., 2006).
Given the evidence supporting a central role for MITF in melanoma progression reflective of its vital role in melanocyte biology, and because somatic mutations are relevant to the progression of cancer, we examined melanoma tumor DNA for the presence of somatic mutations in MITF. Here we demonstrate that somatic mutation of MITF occurs in melanoma metastases. Strikingly, genetic evaluation of the transcription factor SOX10, which acts upstream, activates MITF expression and also synergizes with MITF to activate downstream targets (Potterf et al., 2000, Potterf et al., 2001, Bondurand et al., 2000, Jiao et al., 2004, Ludwig et al., 2004), shows that SOX10 is also mutated in melanoma. Our genetic analyses reveal that previously unknown mutations in MITF and SOX10 occur in over 9% of primary melanoma tumors and 22% of metastatic melanoma tumors.
Results
MITF is somatically mutated in melanoma metastases
MITF has recently been shown to be amplified in 10-20% of melanoma cases (Garraway et al., 2005). However, no study has evaluated whether melanoma-specific intragenic mutations occur in MITF. We therefore examined 13 MITF exons that encompass the full coding sequences of all six MITF isoforms. The exons were polymerase chain reaction (PCR) amplified and directly sequenced from genomic DNA isolated from 50 melanoma metastases. Any observed changes were evaluated against genomic DNA from patient-matched normal DNA to identify somatic, tumor-specific mutations. A total of six changes, including four non-synonymous point mutations and one splice-site alteration, were identified (Table 1 and Supplementary Figure 1). These mutations occurred at evolutionarily conserved residues within distinct functional domains.
| Mutations identified in MITF | |||||||
|---|---|---|---|---|---|---|---|
| Exon | Nucleotide | Amino acid | Functional domain | Zygosity | Sample | Primary/metastases | Histologic type |
| 2 | G259A | E87R | Activation domain 1 | Heterozygous | 4T | Metastases | N/A |
| 2 | A260G | E87R | Activation domain 1 | Heterozygous | 4T | Metastases | N/A |
| 2 | A260G | Splice site | Exon 2B donor splice site | Heterozygous | 4T | Metastases | N/A |
| 4 | T403G | L135V | Activation domain 1 | Heterozygous | 13T | Metastases | N/A |
| 4 | G426C | L142F | Activation domain 1 | Heterozygous | 127 | Primary | SSM |
| 7 | A/T (-3) | splice site | Basic helix loop helix (Basic domain) | Heterozygous | D196 | Primary | ALM |
| 8 | G712A | G244R | Helix loop helix (helix 2) | Heterozygous | 21T | Metastases | N/A |
| 9 | G1120A | D380N | Activation domain 3 | Heterozygous | 85T | Metastases | N/A |
| Mutations identified in SOX10 | |||||||
| Exon | Nucleotide | Amino acid | Functional domain | Zygosity | Sample | Primary/metastatic | Histologic type |
| 1 | C373T | Q125X | HMG box | Heterozygous | 47T | Metastases | N/A |
| 1 | 44-62del | Pro14fsX10 | Truncation before HMG domain | Heterozygous | 68T | Metastases | N/A |
| 1 | G128A | R43Q | HMG domain | LOH | AM141 | Primary | Mucosal |
| 3 | C1082T | A361V | TAD | LOH | MX41 | Primary | LMM |
| 3 | G1237A | G413S | TAD | LOH | MX4 | Primary | LMM |
| 3 | G1238A | G413D | TAD | Heterozygous | MX67 | Primary | LMM |
| 3 | C1240T | H414Y | TAD | Heterozygous | MX67 | Primary | LMM |
| 3 | C1271T | A424V | TAD | LOH | 114 | Primary | SSM |
| 3 | 1346-1353del | Ser449SerfsX66 | TAD | LOH | 59T | Metastases | N/A |
Exon number with nucleotide and amino acid change resulting from mutation. Nucleotide position refers to position within coding sequence, where position 1 corresponds to the first position of the start codon. Tumors 4T and MX67 contained two somatic alterations. In addition to the 16 nonsynonymous mutations recorded in the table, we detected 1 synonymous alteration. The splice site alteration in MITF was in position 3 of the donor site of exon 2B. A total number of 50 cutaneous melanoma tumors were used for the MITF and SOX10 studies, A total number of 26 primary tumors were used to screen for MITF mutations and a total number of 55 primary tumors were used to screen for SOX10 mutations. ALM, acral lentiginous melanoma, LMM, lentigo malignant melanoma, SSM, superficial spreading melanoma.
To test whether MITF is altered by amplification in the same set of 50 melanoma samples, quantitative PCR was performed. Four out of the 50 samples showed amplification of MITF (data not shown). No MITF mutations were present in these 4 samples, demonstrating that MITF mutations and amplifications occurred in a mutually exclusive pattern (Supplementary Table 1). Taken together, these results show that MITF is somatically altered in 16% (8/50) of melanoma metastases.
SOX10 is somatically mutated in melanoma metastases
Since MITF mutations were found in the melanoma samples analyzed, we proposed that other genes in the MITF pathway might also be altered. SOX10 is a transcription factor known to act upstream of MITF (Bondurand et al., 2000, Lee et al., 2000, Potterf et al., 2000, Verastegui et al., 2000) and to synergize with MITF in the transactivation of genes such as Tyrosinase (TYR) (Murisier et al., 2007), and Dopachrome tautamerase (DCT) (Jiao et al., 2004, Potterf et al., 2001). We therefore evaluated the presence of intragenic mutations in SOX10 in the same 50 metastatic melanoma samples. A total of three mutations were identified in SOX10. Two of the mutations would be predicted to result in truncation prior to the HMG box, DNA binding domain. The third mutation, (Ser449Serfsx66), would be predicted to result in an additional 66AA of 3′UTR encoded peptide to be produced instead of the terminal 16AA of WT SOX10 due to an 8bp nucleotide deletion (Table 1 and Supplementary Figure 2). Again, we observed that the tumor samples, which harbored these SOX10 mutations, did not contain either MITF amplifications or intragenic mutations. It is also of note that the 59T cell line that contained the SOX10 mutation (Ser449Serfsx66) was derived from a tumor in a patient exhibiting de novo cellular immune reactivity against SOX10 (Khong and Rosenberg, 2002). Presumably this newly identified SOX10 mutation was the cause for the tumor infiltrating lymphocyte (TIL) reactivity found in this patient.
BRAF and NRAS mutation status
Mutations affecting the MAPK pathway via either the serine/threonine kinase, BRAF, or the small guanine-nucleotide binding protein, NRAS, are frequently observed in metastatic melanoma (Davies et al., 2002, Demunter et al., 2001, Omholt et al., 2002, Omholt et al., 2003). Thus we next characterized the mutation status of both BRAF and NRAS in our panel of 50 metastatic tumor samples. In this panel of 50 metastatic tumors we found BRAF activating mutations in 38/50 (76%) of the tumors, while we observed oncogenic NRAS mutations in 6/50 (12%). Correlation of these results with the 11 tumors harboring mutations in the MITF pathway found 10 of the 11 tumors presenting with activating mutations in BRAF while none of the 11 tumors were found to harbor mutations in the NRAS (Supplementary Table 1). This correlation between mutations altering the MITF pathway and BRAF mutations is consistent with what has been observed previously, as all NCI60 cell lines harboring MITF amplifications were also found to harbor BRAF (V600E) mutations (Garraway et al., 2005).
MITF and SOX10 mutations occur at early stage melanoma
To evaluate whether somatic mutations in MITF and SOX10 occur at earlier stages of melanoma progression, we also evaluated primary melanoma samples. This revealed a total of 2 MITF mutations in 26 primary melanoma samples (7.7%), both of which occurred in similar genomic locations to the mutations found in the melanoma metastases samples (Figure 1). No MITF amplifications were found in the primary melanomas. The SOX10 screen in the primary melanoma samples identified six (R34Q, A361V, G413D, G413S, H414Y and A424V) SOX10 mutations in five of 55 primary melanomas (9%) (Table 1 and Supplementary Table 2). Analysis of sequence traces across the SOX10 amplified regions indicated that for four of the five tumors in which mutations were identified, also exhibited a loss of heterozygosity at this locus (Table 1). None of the MITF or SOX10 mutations were observed in a panel of lymphocyte DNA from 150 Caucasian controls (data not shown), suggesting that these mutations arose somatically.
Figure 1. MITF and SOX10 mutations in primary and metastaic melanoma.
Schematic representing the protein domains of MITF and SOX10 show mutations found in primary melanoma (blue arrows) and metastatic melanoma (red arrows). Functional domains of MITF include AD1 (activation domain 1), AD2 (activation domain 2), AD3 (activation domain 3) and bHLHZip (basic helix-loop-helix leucine-zipper domain). Functional domains of SOX10 include HMG (High Mobility Group) and TAD (transcriptional activation domain).
MITF 4TΔ2B mutation exhibits increased transcriptional activity
The positions of mutations within MITF implied that they were likely to alter its transcriptional activity, because no truncating mutations were observed, all the mutations were heterozygous and the mutations occurred in clearly defined domains. To test the effect these mutations have on MITF activity, each was cloned individually for transactivation studies. RNA derived from mutant MITF cell lines was amplified, and corresponding cDNA cloned into a mammalian expression plasmid. When amplifying the cDNA of mutant 4T, several transcript bands were seen (Figure 2A), and these were subsequently cloned and sequenced. Interestingly, the mutations in sample 4T (Figure 2B) had two effects on mRNA sequence. First, heterozygous guanidine to adenosine and adenosine to guanidine mutations within exon 2 resulted in a substitution of glutamine to arginine at codon 87. Second, analysis of the genomic sequence revealed that the adenosine to guanidine mutation was located at the splice donor site for exon 2. This mutation resulted in the abrogation of the splice donor site (AGGT), which caused deletion of exon 2B in the mutant transcript (Figure 2C). Thus full length 4T had the E87R amino acid alteration, and one of the truncated cDNA versions had a 168bp deletion corresponding to deletion of exon 2B (Figure 2D). We named this mutation 4TΔ2B.
Figure 2. Somatic mutation in tumor sample 4T results in MITF splice variant.
A. RT-PCR analysis of MITF expression in tumor samples. The product of the wild type MITF band is 1260bp, the 4T sample products are 1260bp, 1193bp and 1092bp. The 1193bp transcript represents a variant with a 67bp deletion from exon 2B that did not express protein by western (data not shown). The 1092bp transcript encodes the 4TΔ2B MITF variant, reduced in size by 168bp. B. Somatic mutation of the MITF gene in sample 4T. The top sequence chromatogram was obtained from normal tissue and the lower sequence chromatogram from the 4T melanoma sample. Arrow indicates the location of the mutation. C. The MITF mutation A to G transition in nucleotide 260 is located in the splice donor and leads to a loss of exon 2B (shaded area) and an mRNA molecule that is 168 nucleotides shorter than wild-type message. D. Sequence analysis of the 1260bp cDNA.
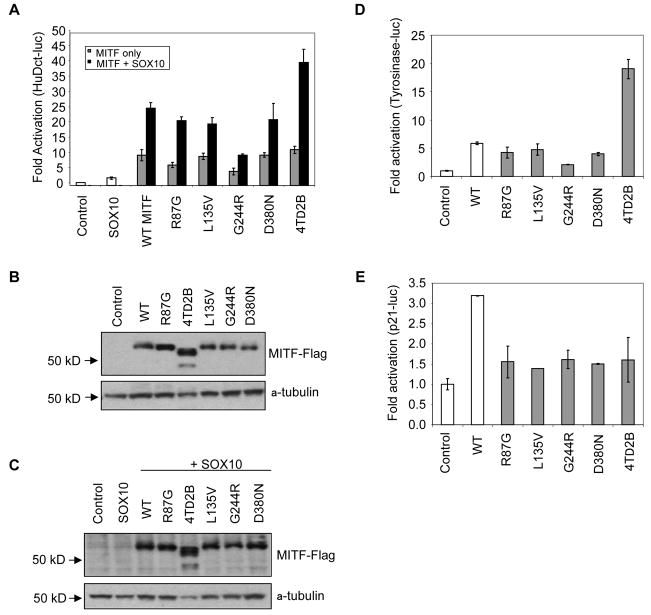
To test the effects of these MITF mutations on transcriptional activity, we expressed FLAG epitope fusion constructs of wild type (WT) MITF or five of the identified MITF mutants in HEK 293T cells and assessed their transcriptional activity. We chose this non-melanocytic cell line because it does not express endogenous MITF, allowing us to eliminate potential confounding effects. We transfected the MITF constructs, together with the MITF-responsive promoter HuDct-luc, in the presence or absence of WT SOX10. As seen previously (Jiao et al., 2004, Ludwig et al., 2004, Potterf et al., 2001) WT MITF synergized with SOX10 to activate expression of the HuDct-luc reporter gene, and similar levels of stimulation were observed with four out of five of the MITF mutants. However, expression of the 4TΔ2B MITF mutant conferred approximately two-fold increased activity compared to WT protein when co-expressed with WT SOX10 (Figure 3A). Similar results were obtained when the MITF mutants were transfected into the retinal pigment epithelial cell line ARPE-19, where only the 4TΔ2B MITF mutation showed greater synergy with SOX10 relative to WT MITF (Supplementary Figure 3).
Figure 3. Mutant forms of MITF activate HuDct and Tyrosinase luciferase but not p21 luciferase.
A. HEK 293T cells were transiently transfected with the indicated constructs together with the HuDct-luciferase reporter construct. Firefly luciferase activities in samples were normalized to Renilla luciferase activities in the same samples. Ratios between firefly and Renilla luciferase activities are shown. B. and C. Immunoblots of immunoprecipitated MITF derived from transfected HEK 293T cells probed with the indicated antibodies. D. HEK 293T cells were transiently transfected with the WT or mutant MITF, or empty vector together with the Tyrosinase-luciferase reporter construct. Fold activation was calculated as in A. E. HEK 293T cells were transiently transfected with the WT or mutant MITF, or empty vector together with the p21-luciferase reporter construct. Fold activation was calculated as in A.
To examine if the increased transcriptional activity of the 4TΔ2B MITF mutant was due to increased protein expression, we evaluated MITF levels in lysates from HEK 293T cells. The protein levels of 4 of the 5 mutant proteins were comparable to WT, while the level of MITF 4TΔ2B was higher than both WT MITF and the other MITF mutants (Figure 3B). The increased expression of the 4TΔ2B mutant was also observed when WT SOX10 was co-expressed (Figure 3C). This suggests that this variant of MITF confers increased protein stability, and may explain its increased ability to activate HuDct-luc. Of note, the lack of serine 73 (transcribed from exon 2B), which is phosphorylated by ERK2 resulting in enhanced proteosome-mediated degradation of MITF (Steingrimsson et al., 2004, Wu et al., 2000, Xu et al., 2000), may be the underlying reason for the increased protein stability.
To examine if mutations in MITF had any affect on other known MITF target gene promoters, we transiently transfected HEK 293T cells with WT MITF or mutant forms of MITF, and measured their transcriptional activity with either p21-luc or Tyr-luc reporter (Bentley et al., 1994) constructs. MITF has been reported to transactivate both of these reporters in cell based assays (Carreira et al., 2005). Expression of WT MITF resulted in a 6-fold increase in activation of the Tyr-luc reporter compared to empty vector control (Figure 3D). Most MITF mutants (R87G, L135V, G244R and D380N) showed activation of Tyr-luc similar to the WT MITF protein. However, expression of the 4TΔ2B MITF mutant conferred an increase in activation 4-fold greater than WT protein (Figure 3D). In contrast, when transactiviation assays were evaluated within the context of the p21-luc reporter, all 5 MITF mutants showed little to no p21-luc activation (Figure 3E). These results suggest that while somatic mutations of MITF encode proteins that can act upon the tyrosinase promoter even in the absence of SOX10, these mutant MITF proteins appear unable to activate the p21 promoter. These results might allow us to postulate how the lack of p21 activation by mutated MITF in melanoma cells could result in escape from p21-dependent cell cycle arrest.
To test the effects of the SOX10 mutations, a HuDct-luc transactivation assay was performed using NIH3T3 cells that over-express WT MITF together with either WT SOX10 or each of five SOX10 mutants identified in primary melanomas (R34Q, G413S, G413D, H414Y and A424V) and the three SOX10 mutants identified in metastatic melanoma (Pro14fsX10, Q125X, or Ser449SerfsX66). A 7- to 10-fold activation of HuDct-luc was observed when WT MITF was co-transfected with either WT SOX10 or the five primary melanoma SOX10 mutants (Figure 4) in comparison to WT SOX10 alone. With respect to the three SOX10 mutations observed in the primary melanoma samples, two mutations Pro14fsX10 and Q125X would be predicted to result in early termination of SOX10 protein prior to regions associated with nuclear localization properties (Sudbeck and Scherer, 1997). Co-expression of WT MITF with these two SOX10 mutations revealed these mutations were unable to transactivate the HuDct-luc reporter (Figure 4). Furthermore, immunofluorescence analysis of these SOX10 mutants demonstrated their aberrant localization to the cytoplasm (data not shown), consistent with previous data and underlying their reduced transactivation abilities. Ser449SerfsX66 mutation, although capable of transactivating the HuDct-luc promoter in combination with MITF does so at reduced efficiency relative to wild type (p-value = 0.0012).
Figure 4. Truncated versions of SOX10 lack the ability to transactivate HuDct-Luciferase.
NIH3T3 cells were transiently transfected with the indicated constructs together with the HuDct-luciferase reporter construct. Firefly luciferase activities in samples were normalized to Renilla luciferase activities in the same samples. Ratios between Firefly luciferase and Renilla luciferase activities are shown.
Discussion
In this study we have used DNA sequence analysis to determine whether MITF and SOX10 are targets of somatic mutation during melanomagenesis. Sequencing of all MITF and SOX10 coding exons in a panel of primary and metastatic melanoma samples identified sixteen de novo mutations. In total, the primary and metastatic melanoma samples analyzed harbored non-synonymous (NS) mutations and amplifications with frequencies of 13.2% (10/76) in MITF and 8.6% (9/105) in SOX10 (Supplementary Table 2). Although the sample size was insufficient to document statistical significance, the mutually exclusive nature of the MITF and SOX10 mutations could indicate that their effects are functionally redundant. As many of the patients from which the metastatic tissues were derived have been treated with various chemotherapeutic regimens, the cytotoxic drugs might have caused some of the observed mutations. However, the finding of similar mutations in some of the primary melanomas mitigates this concern. Taken together, this data provides the first evidence for the presence of NS mutations in both MITF and SOX10 in both primary and metastatic melanoma samples underscoring the involvement of the MITF transcriptional network in melanoma tumorigenesis (Carreira et al., 2005; Carreira et al., 2006; Levy et al., 2006).
It is interesting to note that unlike what has been observed for metastatic melanoma, no amplifications of MITF were observed in the primary tumor samples suggesting that amplification may be a late stage alteration taking place in a context dependent genome that can tolerate increased activity of MITF. This is consistent with prior analysis using arrays CGH primary melanomas (Curtin et al., 2005) The non-synonymous MITF mutations identified in both primary and metastatic samples were always found as heterozygous for a wild type copy of MITF. The mutations were throughout the protein, causing amino acid substitutions in conserved residues in defined functional domains (Figure 1). Two of the MITF mutations (L135V and L142F) occurred in the conserved acidic activation domain (AD1) of MITF which has been shown to be necessary for MITF transcriptional activity (Supplementary Figure 2). Two of the five MITF mutations G244R and 4TΔ2B have corresponding alleles in mouse, Mib and Mibws (Hallsson et al., 2000, Steingrimsson et al., 1996), respectively. In the Mib “brownish” mouse model the same amino acid, G244, is found mutated from a small, non-polar AA to a large, charged AA side chain (G244E). The amino acid location of G244 is at the junction of the loop and helix 2 where the occurrence of a small, non-charged or hydrophobic amino acid and is conserved in all basic helix-loop-helix, leucine-zipper proteins (Steingrimsson et al., 1996). Functional analysis of the Mib allele demonstrated an MITF protein with reduced DNA binding activity, either as a homodimer or as a heterodimer in combination with TFE3. The second mouse model, Mibws, is similar to the 4TΔ2B mutation in that it ultimately results in exon skipping and deletion of exon 2B. This animal exhibits a reduction in melanocyte cell numbers, as reflected by the black and white spotted (bws) coat phenotype (Hallsson et al., 2000). Within this exon is Ser73, whose phosphorylation status can mediate MITF protein turnover (Wu et al., 2000, Xu et al., 2000). Consistent with this finding, we observed increased protein expression for the 4TΔ2B allele mutated in melanoma. Subsequent analysis will be required to determine whether the MITF mutations present in these animal models, Mib and Mibws, confer increased frequency of melanoma metastasis as measured by intercrossing with murine melanoma-prone models.
Given the mutation spectrum and the results from corresponding mouse alleles it might be surprising that most of the mutations had minimal effects on MITF's ability to transactivate the melanogenic target genes, DCT and TYR. However, MITF provides transcriptional regulation for a wide array of genes, mediating transcriptional control over both differentiation (i.e., DCT and TYR) and cell cycle progression pathways (Bismuth et al., 2005). Our analysis suggests that these MITF mutations may lead to alterations in the capacity of MITF to regulate gene expression in a promoter specific manner, as all five of the mutations analyzed exhibited reduced transactivation activity of the p21 promoter, an MITF target gene previously shown to regulate cell cycle progression (Carreira et al., 2005). The uncoupling of MITF's ability to regulate differentiation and proliferation is consistent with studies by Bismuth et al., (2005) where they showed specific isoforms and mutations of MITF can have different effects on cell proliferation, DNA binding and transcriptional activity. It will be interesting to explore how these melanoma associated mutations affect different aspects of cell cycle and target gene regulation.
The mutation spectrum we observed for SOX10 suggests that SOX10 might be acting as a tumor suppressor gene. In metastatic melanoma samples two of the three mutations generated frameshifts predicted to result in protein truncations before the DNA binding domain and thus a product with inactivation of SOX10 function. Consistent with this, these mutations did not transactivate the promoter of a known SOX10 target gene, DCT. The third mutation is predicted to produce a truncation and frameshift at the c-terminus of SOX10. This product demonstrated reduced activity on the DCT promoter, and in the tumor, this mutation was associated with LOH. Data from primary melanoma also support a SOX10 tumor suppressor gene model where we found that of the five samples that harbored somatic mutations, four also exhibited LOH and only retained the NS mutated allele. The fifth sample demonstrated two NS mutations, though it is not known if these are in trans or if a wild type copy remains and is expressed. All but one (R43Q) of the NS mutations in SOX10 that were identified in primary melanoma occurred within its transactivation domain, and strikingly two different melanoma samples had mutations affecting the same residue. However, these mutations did not alter SOX10's ability to transactivate DCT activity. It is possible that the melanomagenic activity for these SOX10 alleles is due to a reduced SOX10 function that is not observable on the DCT promoter under conditions used. While these data support a hypothesis of SOX10 as a tumor suppressor, further functional studies and assessment of additional mutations (for example gene amplifications) needs to be explored before a mechanistic model can be confirmed.
Alternatively, these alleles may promote differential effects on specific target genes similar to what we have shown for the MITF melanoma alleles ability to transactivate TYR and p21 differently. As one of the known SOX10 target genes is MITF it is possible that the various genetic alterations identified in this study have a direct or indirect effect on altering MITF activity, selected for downregulation of MITF activity to adjust to a favorable tumorigenic outcome. As the mutations of MITF and SOX10 are mutually exclusive in the melanoma samples we analyzed, our results support the “biological rheostat” model (Carreira et al., 2006) where tight regulation of MITF levels is essential for balancing melanocyte growth and arrest.
In summary, this is the first study demonstrating that melanoma patients harbor somatic mutations in MITF and SOX10. Our findings indicate that a substantial fraction of melanomas acquire genetic alterations in the MITF pathway, occurring at both the primary and metastatic stages and suggest that altered MITF function during melanomagenesis can be achieved by MITF amplification, MITF single base substitutions or by mutation of its regulator SOX10.
Methods
Tumor tissues and generation of melanoma cell lines
A panel of pathology-confirmed metastatic melanoma tumor resections, paired with apheresis-collected peripheral blood mononuclear cells, was collected from 80 patients enrolled in IRB-approved clinical trials at the Surgery Branch of the National Cancer Institute, with informed consent from all human subjects. Pathology-confirmed melanoma cell lines were derived from mechanically or enzymatically dispersed tumor cells, which were then cultured in RPMI 1640 + 10% FBS at 37°C in 5% CO2 for 5-15 passages. For all samples, matching between germline and tumor DNA was verified by direct sequencing of 26 single nucleotide polymorphisms (SNP) at 24 loci. Cytopathology was used to determine the percentage of melanoma antigen expressing cells.
PCR, sequencing and mutational analysis
Melanoma metastases samples and their matched normal samples were provided by Dr. Steven Rosenberg (NCI). Genomic DNA was isolated using DNeasy Blood & Tissue kits (Qiagen, Valencia, CA). For all samples, matching between germline and tumor DNA was verified by direct sequencing of 26 single nucleotide polymorphisms (SNP) at 24 loci (data not shown). PCR and sequencing primers were designed using Primer 3 (http://www-genome.wi.mit.edu/cgibin/primer/primer3_www.cgi) and synthesized by Invitrogen (Carlsbad, CA). PCR amplification, sequencing and analysis was performed as previously described (Samuels et al., 2004) using the primers listed in Supplementary Table 3.
To increase our confidence that the five mutations for which no matched normal DNA sample was available (L142F, A361V, G413D, G413S, A424V) did not represent germline polymorphisms, we sequenced the corresponding exons of MITF and SOX10 in 149 normal DNA samples and detected no abnormalities.
DNAs from paraffin embedded primary human melanoma samples were obtained from the collaborators at the University of California, San Francisco and were part of a prior study (Curtin et al., 2005).
Construction of the MITF expression vectors and other expression plasmids used
Mammalian expression vectors containing either the WT or mutant MITF cDNAs were constructed by PCR amplification of the corresponding region from cDNA of melanoma metastases cell lines and subcloning into the Flag-Tag Vector 4 (Stratagene, La Jolla, CA) with EcoRI and HindIII restriction sites. Forward and reverse PCR primer sequences were 5′ TGACCAGAATTCATGCTGGAAATGCTAGAATA 3′ and 5′ TGGTCAAAGCTTACAAGTGTGCTCCGTCTCTT 3′ respectively. Clones were sequenced and prepped by Qiagen Maxiprep Kit (Qiagen, Valencia, CA). Tyr-luc and p21-luc constructs provided by Colin Goding.
SOX10 mutation constructs
SOX10 mutations were generated using an overlapping two-fragment PCR mediated strategy using Phusion High-Fidelity polymerase (New England Biolabs, Ipswich, MA). Minor modifications of this protocol for specific clones are indicated below. In general, forward and reverse CDS mutation-containing oligos of 21-26bp in length were designed with the corresponding mutation centrally located within the oligonucleotide. These primers were used in the two independent PCR reactions with hSOX10-pSport2 cDNA template as follows (1) SOX10 5′ATGGCGGAGGAGCAGGACCTA3′ and Mutation-R primer pairs and (2) Mutation-F SOX10-3R 5′TTAGGGCCGGGACAGTGTCGT3′. Primers were removed using Microcon YM-30 column (Millipore, Bedford, MA) and corresponding generated 5′ and 3′ SOX10 PCR containing fragments were together used as template for PCR using ATTB1-Sox10–5′GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGCGGAGGAGCAGGACCTA3′ and AttB2-SOX10 -5′GGGGACCACTTTGTACAAGAAAGCTGGGTTTTAGGGCCGGGACAGTGTCGT3′ PCR primers to generate full length, Gateway- BP PCR products compatible for cloning into pDonr221 entry vector (Invitrogen, Carlsbad, CA). Sequence verified entry clones were subsequently transferred into pcDNA3.1 NHA-GW using standard Gateway protocols (Invitrogen, Carlsbad, CA).
Minor modifications to the PCR cloning strategies as outlined above include: (1) For the del1346-1353 mutation the following primers modifications were used. The 3′ SOX10 primer was replaced with 1832R primer (5′TCATCAGGGCAGTGAGCCAGAC3′) and the corresponding mutation primers were longer: Del 1346-1353 F- 5′CAGCCCCTCAGGGCCCCAGTCCCACACACTGGGAGCAGCC3′; Del 1346-1353 R- 5′GCTGCTCCCAGTGTGTGGGACTGGGGCCCTGAGGGGCTG3′. (2) For the C373T containing mutations a single PCR was performed using B1F-HSOX10 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGCGGAGGAGCAGGACCTA3′, with either B2R-HSOX10-C373T 5′GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAGTCCGCGAGCTTCCTGCGCG3′, or B2R-HSOX10- C372T-;C373T 5′GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAATCCGCGAGCTTCCTGCGCG3′ as appropriate. (3) Del 44 mutation was generated using mutation primers HSOX10-del44-62 F 5′GGAGGTGGAGCTGAGCCCCGCTGCCTGTCCCCGGGGAGC3′ and HSOX10-del44-62 R 5′GCTCCCCGGGGACAGGCAGCGGGGCTCAGCTCCACCTCC3′.
Destination vector pcDNA3.1 NHA-GW was generated by PCR amplification using primers RCAS YF- 5′GAGCTGACTCTGCTGGTGGC3′ and RCAS-YR 5′CAGATACGCGTATATCTGGC3′ to amplify a fragment containing the N-terminal HA epitope, in frame with Gateway ccdB selection cassette fragment from RCAS-Y NHA (Loftus et al., 2001) and subsequent cloning into pcDNA3.1/CTGFP-TOPO (Invitrogen, Carlsbad, CA).
Quantitative real-time PCR for genomic DNA templates
Genome content differences were determined by quantitative real-time PCR performed on an iCycler apparatus (Bio-Rad, Hercules, CA). Each tumor DNA was quantified by comparing the MITF locus to the reference locus LINE1, a repetitive element whose copy numbers per haploid genome are similar among all human normal and neoplastic cells. PCR was done in 25 μL with the following components: 15.2 μL sterile tissue culture grade double-distilled water (Invitrogen, Carlsbad, CA), 2.5 μL of 10× PCR buffer, 2.5 μL of 10 mmol/L deoxynucleotide triphosphates (Promega, Madison, WI), 15 μL of DMSO (Sigma, St. Louis, MO), 0.5 μL of SYBR Green I solution (Invitrogen, Carlsbad, CA) diluted 1:1,000 in double-distilled water, 0.5 μL of 50 μmol/L forward and reverse primers (Invitrogen; desalted, 25 nmol scale), 0.25 μL of Platinum taq DNA polymerase (Invitrogen, Carlsbad, CA), and 2 μL of purified DNA. The 10× PCR buffer contained 670 mmol/L of Tris-HCl (pH 8.8), 67 mmol/L of MgCl2, 166 mmol/L of (NH4)2SO4, and 100 mmol/L of 2-mercaptoethanol. The reaction was monitored on an iCycler (Bio-Rad) with the following cycling conditions: (94°C, 2 min) × 1, (94°C, 10 s; 57°C, 15 s; 70°C, 15 s) × 35. The threshold cycle number was determined using Bio-Rad analysis software (version 3.0) with the PCR baseline subtracted. PCRs were done in triplicate with the following primers: 5′-AAACCCCACCAAGTACCACA-3′ for MITF forward, 5′-ACATGGCAAGCTCAGGAC-3′ for MITF reverse, 5′-AAAGCCGCTCAACTACATGG-3′, for LINE1 forward, and 5′-TGCTTTGAATGCGTCCCAGAG-3′ for LINE1 reverse. The MITF gene amplification rate in tumor DNA (ARTu-DNA) was determined by using the formula ARTu-DNA = 2−ΔΔCT with ΔΔCT being [CT (MITF;tumor) − CT (average LINE1;tumor)]. An increase of 0.5 in the amplification rate indicates a gain of one MITF gene copy.
Cell Culture and transfections
Melanoma metastases lines were maintained as previously described (Chappell et al., 1999). HEK293T and ARPE-19 lines purchased from ATCC (Manassas, VA), and maintained in complete DMEM medium under standard conditions. Cells were transfected with Lipfectamine2000 reagent (Invitrogen, Carlsbad, CA). Melanoma cell lines were grown in T2 medium (RPMI-1640 supplemented with 10% fetal bovine serum, 2.5 mM HEPES, pH 7.5, 2 mM L-glutamine, 1X Pen/Strep, 10 mg/ml Ciprofloxan, 10 mg/ml Genetamicin, and 1.25 mg/ml Fungizone).
Immunoblotting
Protein extracts prepared as described previously (Samuels et al., 2005). Blots were blocked in 10% non-fat dry milk, 1×TBST (25 mM Tris, 150mM NaCl, 0.1% Tween20) for 1 hour at room temperature before incubation with primary antibody in block (1 hour to overnight). Blots were washed 2 times before the addition of HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) diluted in block. Blots were washed again before developing with Amersham ECL Blotting Kit (GE Healthcare, Piscataway, NJ). Primary antibodies used were monoclonal MITF (C5 Thermo Scientific, Fremont, CA), monoclonal a-Tubulin (Calbiochem, San Diego, CA), Anti-FLAG M2 monoclonal peroxidase-conjugate (Sigma, Saint Louis, MO).
Luciferase Assay
HEK 293T or ARPE-19 cells were cultured in 12-well plates for 24 hours prior to transfection. Cells were co-transfected with 300ng HuDct-luc or 100ng Tyrosine-Luc or p21-Luc reporter plasmids; 60ng-100 ng of WT or mutant HuMITF; 60ng CMV-SOX10 or pSPORT empty vector; and 10-25 ng phRG-TK Renilla luciferase plasmid (Promega, Madison, WI). Cells were cultured for 24 hours before lysis; the resulting extracts were assayed for luciferase activity using the Dual-Luciferase reporter Assay System (Promega, Madison, WI). All experiments were done in duplicate or triplicate. NIH3T3 cells grown in a 24-well plate format, with 8 ng pCMV-RL renilla expression vector (Promega, Madison, WI), 400ng HuDct-luc, 400 ng SOX10 construct or empty vector, with 2ul Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) per well. Luciferase assay was performed using Dual Luciferase Reporter 1000 Assay System (Promega,Madison, WI) using a Fluoroskan Ascent FL fluorimeter (Thermo Fisher Scientific, Waltham, MA). All experiments were done in triplicate.
Supplementary Material
In each case, the top sequence chromatogram was obtained from normal tissue and the lower sequence chromatogram from the indicated tumors. Arrows indicate the location of missense mutations. The nucleotide and amino acid alterations are indicated below the tumor chromatograms.
In each case, the top sequence chromatogram was obtained from normal tissue and the lower sequence chromatogram from the indicated tumors. The nucleotide and amino acid alterations are indicated below the tumor chromatograms.
ARPE-19 cells were transiently transfected with the indicated constructs together with the HuDct-luciferase reporter construct. Firefly luciferase activities in samples were normalized to Renilla luciferase activities in the same samples. Ratios between firefly and Renilla are shown as fold activation.
Acknowledgments
We thank Drs. Heinz Arnheiter, Tom Hornyak and Paul Meltzer for their helpful comments on the manuscript. Funded by the National Human Genome Research Institute and National Cancer Institute, National Institutes of Health.
Footnotes
Significance: Melanoma is an aggressive malignancy of the pigment-producing cells, melanocytes, and accounts for the majority of deaths resulting from skin cancer. Contributing factors to the initiation and progression of melanoma include genetic susceptibility, environmental insults such as UV exposure and somatic mutations. Identifying the cellular pathways that when altered by somatic mutations promote the progression of melanoma can provide insights into developing diagnostics and treatments. In this study we investigated whether two genes of an important melanocyte transcriptional pathway, MITF and SOX10, are somatically mutated in melanoma. We found that over 20% of metastatic melanoma tumors and over 7% of primary melanomas have somatic alterations in one of these two genes. Taken together, our data suggest a substantial fraction of melanomas acquire genetic alterations in the MITF pathway, occurring at both the primary and metastatic stages.
References
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bismuth K, Maric D, Arnheiter H. MITF and cell proliferation: the role of alternative splice forms. Pigment Cell Res. 2005;18:349–359. doi: 10.1111/j.1600-0749.2005.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes & Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell DB, Zaks TZ, Rosenberg SA, Restifo NP. Human melanoma cells do not express Fas (Apo-1/CD95) ligand. Cancer Res. 1999;59:59–62. [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, Leboit PE, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Demunter A, Stas M, Degreef H, De Wolf-Peeters C, Van Den Oord JJ. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J Invest Dermatol. 2001;117:1483–1489. doi: 10.1046/j.0022-202x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Favor J, Hodgkinson C, Glaser T, Lamoreux ML, Magnusdottir R, Gunnarsson GJ, Sweet HO, Copeland NG, Jenkins NA, et al. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics. 2000;155:291–300. doi: 10.1093/genetics/155.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–1176. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- Jiao Z, Mollaaghababa R, Pavan WJ, Antonellis A, Green ED, Hornyak TJ. Direct interaction of Sox10 with the promoter of murine Dopachrome Tautomerase (Dct) and synergistic activation of Dct expression with Mitf. Pigment Cell Res. 2004;17:352–362. doi: 10.1111/j.1600-0749.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- Khong HT, Rosenberg SA. The Waardenburg syndrome type 4 gene, SOX10, is a novel tumor-associated antigen identified in a patient with a dramatic response to immunotherapy. Cancer Res. 2002;62:3020–3023. [PMC free article] [PubMed] [Google Scholar]
- Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem. 2000;275:37978–37983. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Watkins-Chow D, Church DM, Pavan WJ. Generation of RCAS vectors useful for functional genomic analyses. DNA Res. 2001;8:221–226. doi: 10.1093/dnares/8.5.221. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Rehberg S, Wegner M. Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett. 2004;556:236–244. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- Murisier F, Guichard S, Beermann F. The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res. 2007;20:173–184. doi: 10.1111/j.1600-0749.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- Omholt K, Karsberg S, Platz A, Kanter L, Ringborg U, Hansson J. Screening of N-ras codon 61 mutations in paired primary and metastatic cutaneous melanomas: mutations occur early and persist throughout tumor progression. Clin Cancer Res. 2002;8:3468–3474. [PubMed] [Google Scholar]
- Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9:6483–6488. [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Mollaaghababa R, Hou L, Southard-Smith EM, Hornyak TJ, Arnheiter H, Pavan WJ. Analysis of SOX10 function in neural crest-derived melanocyte development: SOX10-dependent transcriptional control of dopachrome tautomerase. Dev Biol. 2001;237:245–257. doi: 10.1006/dbio.2001.0372. [DOI] [PubMed] [Google Scholar]
- Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Nii A, Fisher DE, Ferre-D'amare AR, Mccormick RJ, Russell LB, Burley SK, Ward JM, Jenkins NA, Copeland NG. The semidominant Mi(b) mutation identifies a role for the HLH domain in DNA binding in addition to its role in protein dimerization. Embo J. 1996;15:6280–6289. [PMC free article] [PubMed] [Google Scholar]
- Sudbeck P, Scherer G. Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J Biol Chem. 1997;272:27848–27852. doi: 10.1074/jbc.272.44.27848. [DOI] [PubMed] [Google Scholar]
- Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem. 2000;275:30757–30760. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3:242–252. doi: 10.1016/j.stem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp Cell Res. 2000;255:135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In each case, the top sequence chromatogram was obtained from normal tissue and the lower sequence chromatogram from the indicated tumors. Arrows indicate the location of missense mutations. The nucleotide and amino acid alterations are indicated below the tumor chromatograms.
In each case, the top sequence chromatogram was obtained from normal tissue and the lower sequence chromatogram from the indicated tumors. The nucleotide and amino acid alterations are indicated below the tumor chromatograms.
ARPE-19 cells were transiently transfected with the indicated constructs together with the HuDct-luciferase reporter construct. Firefly luciferase activities in samples were normalized to Renilla luciferase activities in the same samples. Ratios between firefly and Renilla are shown as fold activation.