Abstract
Aim
To evaluate the importance of epidermal growth factor receptor (EGFR) protein overexpression and gene amplification in carcinogenesis of glottic cancer.
Method
In order to evaluate EGFR expression at protein and gene level, immunohistochemical (IHC) analysis and fluorescent in situ hybridization (FISH) were performed on tissue microarrays of laryngeal tissue (145 samples) – 38 samples of normal mucosa, 46 samples of hyperplastic lesions, and 61 samples of cancerous lesions.
Results
Membranous (mEGFR) and cytoplasmic (cEGFR) EGFR expression was significantly different between the analyzed groups. The differences were most striking in the suprabasal-transforming zone. IHC evaluation showed that high and low mEGFR staining contributed to the differentiation of dysplastic lesions, simple hyperplasia, and cancerous tissue, as well as between different degrees of atypia in hyperplastic lesions (P < 0.050). EGFR gene amplification was not found in simple and abnormal hyperplastic lesions, but it was confirmed in 2/21 atypical hyperplasias, indicating that gene amplification can facilitate identification of malignant potential in hyperplastic lesions. In cancerous tissue, EGFR gene amplification was found in 8/50 samples. EGFR gene amplification was found in preinvasive cancer in one patient. In invasive carcinomas, gene amplification was not associated with stage or grade. Carcinomas with gene amplification showed significantly higher cEGFR expression (basal layer P = 0.003; suprabasal layer P = 0.002).
Conclusions
This study confirmed an increase in EGFR protein expression and gene amplification with the increase in biological aggressiveness of glottic lesions. A correlation between EGFR gene amplification and protein expression was established. Gene amplification proved to be an early event in glottic carcinogenesis, indicating its importance for glottic cancer prevention, early detection, and protocol selection.
Epidermal growth factor receptor (EGFR), a 170 kDa transmembrane tyrosine kinase receptor, is a member of the EGFR family of cell surface receptors (1,2). The EGFR gene is located on the chromosome 7p12 (1). The most researched family member is human epidermal growth factor receptor 2 (HER-2), which is the target of routine diagnostic and therapeutic protocols for breast cancer (3). Even though less investigated than HER-2, EGFR protein is overexpressed in many solid head and neck tumors. It is connected with advanced and aggressive tumors and poor prognoses (1,2,4-6).
The relation between protein overexpression and gene amplification still remains unclear. Although immunohistochemical (IHC) protein analysis of EGFR has been extensively researched, there is a lack of studies on gene amplification which seems to be important for new anti-EGFR therapies in some tumor types (2,7-14). Two kinds of drugs have become the part of cancer therapy protocols: monoclonal antibodies that act against the ligand-binding domain and small molecules that inhibit the tyrosine-kinase activity of the receptor. They have been approved for use in some types of EGFR-dependent cancers (eg, colon, lung, and pancreas) (2,7-10).
Understanding the EGFR signaling pathway and its implication in tumorigenesis is crucial in the selection of patients who could benefit from EGFR-targeted therapies. The selection of patients suitable for treatment is based on biomarkers that can predict the effectiveness of new therapeutic agents. Intensive studies of some type of lung cancers showed that gene amplification and mutations were more precise markers of treatment response than EGFR protein expression (8,11,12). Research on colonic cancer demonstrated that EGFR gene amplification and protein overexpression were insufficient in predicting therapy response even though they were linked to prognosis. Patients demonstrating gene amplification showed better response to monoclonal antibody treatment in some studies, but downstream molecules such as v-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue seem to be even more important in predicting response to therapy (9,13,14).
Although EGFR protein overexpression has been studied in glottic lesions, there is a lack of information on gene amplification for this specific area. There is the need for further research of EGFR involvement in glottic carcinogenesis in order to optimize treatment protocols for this tumor type. Laryngeal cancer is the most prevalent malignant head and neck tumor in the male population in Croatia (15), with one of the highest mortality rates among cancers of the head and neck region (15,16). The glottic region is the most common site of origin of laryngeal cancers. This cancer was chosen for research because the follow-up of atypical hyperplastic lesions of vocal cords and other precancerous formations is especially difficult and this cancer considerably affects patients' quality of life.
The aim of this study was to investigate the influence of EGFR protein overexpression and gene amplification on cancerogenesis of glottic cancer and their possible role in improvement of follow-up and treatment protocols of precancerous and cancerous glottic lesions.
Patients and methods
Clinicopathological data
The research was performed on 145 biopsy/resection samples of glottic tissue from patients treated at the Department of Pathology and Otorhinolaryngology of the Clinical Hospital Center Rijeka between 2003 and 2008. The study was approved by the Ethics Committee of Clinical Hospital Center Rijeka and patients signed consent forms before enrolment. Based on classic pathohistological findings of hematoxylin-eosin slides, 3 main groups were formed: control patients (n = 38), patients with hyperplastic lesions (n = 46), and patients with cancerous lesions (n = 61). Hyperplastic lesions were graded according to the Gale-Kambič-Lenart Ljubljana classification (17). Carcinomas were graded and staged according to the classification criteria of the World Health Organization and the tumor-node-metastasis system. Subsequently, they were divided into preinvasive (tumor in situ [Tis]) and invasive, and early (Tis-T1T2N0M0) and advanced (all above T2N0M0) carcinomas (18).
The control group comprised patients treated for benign glottic lesions (nodule, polyp) who had simple hyperplasia as the greatest mucosal change. Hyperplastic lesions included 22 abnormal and 24 atypical hyperplastic lesions. All malignant cases were squamous cell carcinomas. Forty-seven invasive, 14 preinvasive, 32 early, and 27 advanced laryngeal carcinomas were analyzed.
There were 123 biopsy samples taken from male patients and 22 taken from female patients. The patients were aged from 38 to 81 years. Mean age of the control group was 51.5 ± 12.1, mean age of patients with precancerous lesions was 58.1 ± 11.8, and mean age of patients with glottic cancer was 56.9 ± 11.3 years. There were 78% smokers in the control group, 76% smokers among patients with precancerous lesions, and 70% of smokers among patients with glottic cancer.
Tissue microarray (TMA) construction
Hematoxylin-eosin stained sections of glottic mucosa were used to mark the areas with surface epithelial layers, avoiding areas of necrosis. Three tissue cores, each 1 mm in diameter, were placed in a recipient paraffin block using a manual tissue arrayer (Alphelys, Plaisir, France). The final TMA blocks contained 38 cores with tissue specimens that served as controls, 46 cores with hyperplastic lesions, and 61 cores of tumor samples. Normal liver tissue was used for slide orientation. Cores were spaced at intervals of 0.5 mm in the x- and y-axes. One section from each TMA block was stained with hematoxylin-eosin for morphological assessment. Serial sections were cut from TMA blocks for IHC staining. Five micrometers thick sections were placed on adhesive glass slides (Capillary Gap Microscope Slides, 75 μm, Code S2024, DakoCytomation, Glostrup, Denmark), left to dry overnight at 37°C, and stored in the dark at +4°C.
Immunohistochemical staining and evaluation of EGFR protein expression
Following deparaffinization in xylene and rehydration in alcohol, heat-induced epitope retrieval was achieved by immersing the slides in Tris-EDTA buffer (pH 9.0) and boiling them for 10 minutes in a water bath. Slides were allowed to cool over 45 minutes and then pre-incubated for 30 minutes with blocking solution containing normal donkey (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or goat serum (DakoCytomation). Sections were incubated with pre-diluted primary monoclonal antibodies for EGFR (EGFR pharmDx, Monoclonal Mouse antihuman EGFR [clone2-18C9]IgG1). Antibody immunohistochemistry was performed in Dako Autostainer Plus (DakoCytomation Colorado Inc, Fort Collins, CO, USA) according to the manufacturer’s protocol. All reagents for automated IHC were purchased from DakoCytomation (ChemMate TM Detection Kit K5001, ChemMate TM Envision HRP detection Kit K5007). Slides were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. Negative control slides were prepared by substituting Dako ChemMate antibody diluent for a secondary antibody. Moreover, for positive and negative controls, colonic cancer tissue sections overexpressing EGFR and apparent normal colonic mucosa were used.
An analysis of EGFR protein expression was performed. Membranous and cytoplasmic staining was evaluated. Membranous staining (mEGFR) was divided into membrane high – continuous linear and membrane low – discontinuous staining. For positive cytoplasmic EGFR (cEGFR) staining, an intense homogenous staining of the cytoplasm was necessary. There was no cEGFR without membrane staining. The cells were analyzed by a method adjusted to multilayer squamous epithelium, where changes occur in the layers. The basal zone was defined as cells with stromal contact, the superficial zone represented surface cells with applanated nuclei, while the suprabasal zone was defined as the transforming layer between them. Cell-counting was performed by 2 pathologists. The results were analyzed on automated image analyzer using Issa software (VamsTec, Zagreb, Croatia) by an experienced pathologist. Statistical analysis was performed counting all cells on 0.01 mm2 in defined zones of control, hyperplastic, or cancerous tissue, and calculating the percentages of positive cells with mEGFR and cEGFR expressions.
Fluorescent in situ hybridization
After deparaffinization, the slides were pretreated with Paraffin Pretreatment Kit (Vysis, Downers Grove, IL, USA) using protease to digest proteins for 10 minutes. FISH was performed by applying EGFR probes (Vysis), LSI EGFR SpectrumOrange and CEP 7 SpectrumGreen for the respective centromeric region at chromosome 7. The LSI EGFR Dual Color probe hybridizes to the band region 7p12 in Spectrum Orange and the CEP 7 to the 7p11.1-q11, 1, D7Z1 locus in Spectrum Green. Denaturation of the probe mixture was performed at 95°C for 5 minutes in Thermobrite (Abbott Molecular Inc, Des Plaines, IL, USA) followed by overnight hybridization at 37°C. The following day, cover glass was removed and slides were washed in post-hybridization washing-buffer (Vysis) at 72°C for 2 minutes. After drying, DAPI (Vysis) was applied and slides were protected from light at -20°C until reading. Automated scanning, capture, and scoring of interphase FISH was analyzed by Cytovision System (Applied Imaging Corp., San Jose, CA, USA). EGFR gene amplification was defined as a ratio between EGFR gene (orange) and centromere (green) signals in tumor cell nuclei over 2.2 or the presence of EGFR gene clusters.
Statistical analysis
Statistical analysis among 3 groups was carried out using Kruskal-Wallis or χ2 tests as appropriate. For differences between single group pairs, the Mann-Whitney test was applied. The level of statistical significance was set at P < 0.05. Data in tables are given as mean values ± standard deviations or percentages. Data were analyzed using SPSS, version 15.0, for Windows (SPSS Inc., Chicago, IL, USA).
Results
Immunohistochemical evaluation of EGFR protein expression
The percentage of positive cells found by IHC analysis in distinct zones of multilayer squamous epithelium was compared within and between the groups (Figure 1-4).
Figure 1.
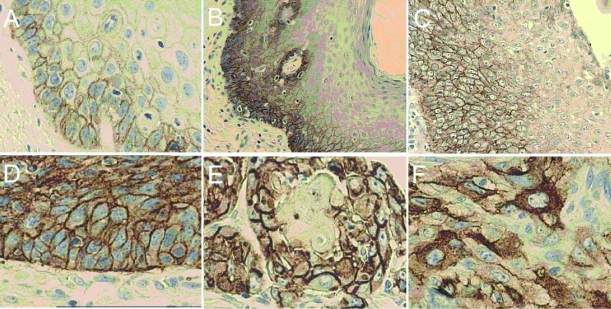
Immunohistochemical (IHC) analysis of epidermal growth factor receptor (EGFR) protein expression in glottic lesions. (A) Simple hyperplasia (control group) displaying predominantly basal (high and/or low) and suprabasal (mainly low) membranous epidermal growth factor receptor (mEGFR) staining. Gradual increase of mEGFR with degrees of atypia from abnormal (B) to atypical hyperplastic lesions (C): in intensity (from low to high) and in extent (through epithelial thickness toward superficial layers). mEGFR expression in the entire epithelial thickness of preinvasive (D) and invasive (E) glottic cancer. (F) Striking example of cytoplasmic epidermal growth factor receptor (cEGFR) expression in glottic cancer. All pictures are photographed at magnification ×40, except atypical lesions ( × 20).
Figure 4.
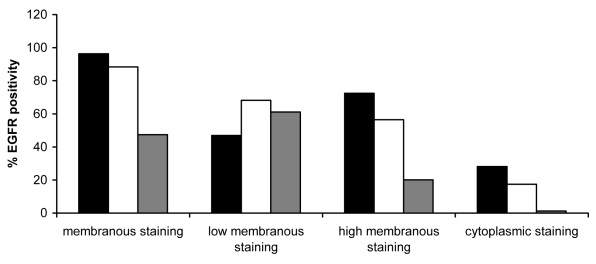
Percentages of membranous (low and/or high) and cytoplasmic positive cells for epidermal growth factor receptor (EGFR) in glottic cancer. Membranous epidermal growth factor receptor (mEGFR) staining in cancerous tissue appeared throughout all layers of squamous epithelium. An increase in high mEGFR staining was noted, especially in the suprabasal and superficial layers, in comparison with controls (suprabasal layer P < 0.001; superficial layer P < 0.001; Mann-Whitney U-test) and hyperplastic lesions (suprabasal layer P = 0.052; superficial layer P = 0.009; Mann-Whitney U-test). Cytoplasmic staining was also much stronger and appeared even in the superficial layer. Closed bars – basal layer; open bars – suprabasal layer; and gray bars – superficial layer.
Although all 3 groups of patients had very similar mEGFR staining in the basal layer, hyperplastic and cancerous lesions group had an increase in high membranous staining vs simple hyperplasia of the control group.
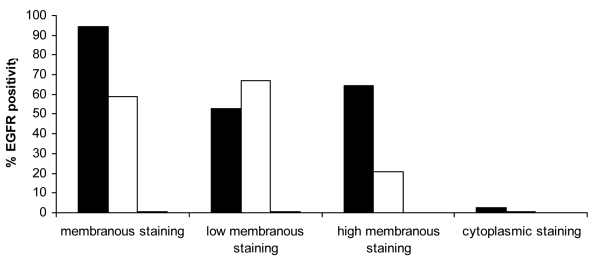
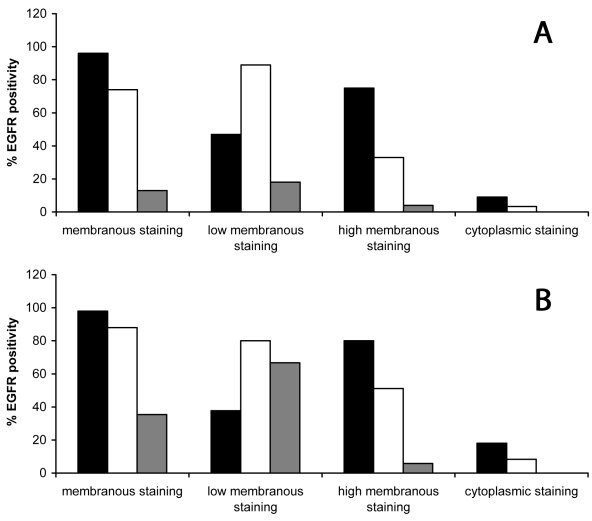
The control group (Figure 1A and 2) had mainly weak mEGFR staining in the suprabasal layer (67%). In dysplastic lesions compared with simple hyperplasia group, an increase in high mEGFR staining was found (P = 0.001; Mann-Whitney U-test), as well as between abnormal and atypical lesions (P = 0.035; Mann-Whitney U-test). Moreover, suprabasal mEGFR expression increased from weak to strong with the degree of atypia in hyperplastic lesions group. In abnormal hyperplastic lesions group (Figure 1B and 3A), 33.2% of cells showed high mEGFR, as opposed to 51.1% in atypical lesions group (Figure 1C and 3B). Values were even higher in cancerous tissue, where 56.5% of cells had high mEGFR expression (Figure 1D, 1E, and 4).
Figure 2.
Percentages of membranous (low and/or high) and cytoplasmic positive cells for epidermal growth factor receptor (EGFR) in the control group. The control group showed mainly membranous staining of basal and suprabasal layers. The staining of the suprabasal layer was predominantly low. Cytoplasmic staining appeared in very low intensity of basal layers. Closed bars – basal layer; open bars – suprabasal layer; and gray bars – superficial layer.
Figure 3.
(A) Percentages of membranous (low and/or high) and cytoplasmic positive cells for epidermal growth factor receptor (EGFR) in abnormal and (B) atypical hyperplastic lesions. Hyperplastic lesions had more positive suprabasal cells than the control group (P = 0.001; Mann-Whitney U-test). Within this group, a gradual change in membranous staining was noted, from weak positive in abnormal hyperplasia to strong positive in atypical lesions. The differences were most striking in the suprabasal-transforming zone. Superficial layers showed mainly low membranous staining and only a small number of cells manifested high staining intensity. Cytoplasmic staining was mostly basal and suprabasal, stronger than in the control group (basal layer P < 0.001; suprabasal layer P < 0.001; Mann-Whitney U-test) but weaker than in cancerous lesions group (basal layer P < 0.001; suprabasal layer P < 0.001; Mann-Whitney U-test). Closed bars – basal layer; open bars – suprabasal layer; and gray bars – superficial layer.
In the superficial layer, no high membranous staining was noticed in the control group, and the percentage of low mEGFR positive cells was lower than 1% (Figure 2). Low mEGFR was noticed in 18% of abnormal (Figure 3A) and up to 67% of atypical lesions (Figure 3B). In cancerous lesions, up to 61% of cells showed low and 20% high mEGFR staining (Figure 4).
cEGFR staining in the control group was mainly basal and significantly lower than mEGFR staining (2.4%). Basal and suprabasal cEGFR expression was lowest in the control group, increased in abnormal and atypical hyperplastic lesions, and was highest in cancerous lesions group.
In simple hyperplasia and dysplastic lesions group, no superficial cytoplasmic staining was noticed; it appeared only in cancerous lesions group (1.3%).
In cancerous lesions group, an increase in mEGFR expression in the superficial layer was found with increased tumor grade. The number of negative cells in the suprabasal layer was significantly higher in gradus 1 than in gradus 2 (P = 0.023 Mann-Whitney U-test) and in gradus 1 than in gradus 3 (P = 0.031 Mann-Whitney U-test). The differences between cEGFR staining and grade were not observed (Kruskal-Wallis test). There was a borderline significant difference of mEGFR positive cells of the superficial layer between preinvasive and invasive tumors (P = 0.051; Mann-Whitney U-test). A significant difference between advanced and early staged tumors was found in highly positive basal cells (P = 0.044; Mann-Whitney U-test) and weakly positive superficial cells (P = 0.050; Mann-Whitney U-test). There were no differences in cEGFR staining between early and advanced tumor types (Mann-Whitney U-test).
EGFR protein expression did not show significant differences between non-smokers and smokers in precancerous and cancerous lesions. Smokers in the control group showed an increase in high mEGFR positive cells in the suprabasal transforming zone (P = 0.023; Mann-Whitney U-test).
FISH evaluation of EGFR gene amplification
FISH results were successfully obtained for 118 tissue samples. Failure to obtain results in 27 samples was mainly due to the loss of tissue materials during the TMA sectioning and hybridization procedure. EGFR gene amplification was found in 10 patients: 2 patients with precancerous lesions and 8 patients with laryngeal cancer. A significant difference in EGFR gene amplification was found between the analyzed groups. First, we evaluated 3 main groups (P = 0.031 between controls, hyperplastic lesions, and cancerous lesions; χ2 test). For better insight in hyperplastic lesions, this group was divided into abnormal and atypical lesions subgroups (P = 0.045 between controls, abnormal hyperplastic lesions, atypical hyperplastic lesions, and cancerous lesions; χ2 test, Table 1). All amplified lesions displayed 5 or more signals or clusters of EGFR gene (Figure 5A and B).
Table 1.
Number (%) of biopsies and amplification status of epidermal growth factor receptor (EGRF) by fluorescent in situ hybridization (FISH) in different glottic lesions
| Group | Amplification of EGFR gene (%) |
Total | |
|---|---|---|---|
| no | yes | ||
| Control | 31 (100) | 0 (0) | 31 (100) |
| Abnormal hyperplastic lesions | 16 (100) | 0 (0) | 16 (100) |
| Atypical hyperplastic lesions | 19 (90.5) | 2 (9.5) | 21 (100) |
| Squamous carcinoma | 42 (84.0) | 8 (16.0) | 50 (100) |
| Total number of biopsies* | 108 (91.5) | 10 (8.5) | 118 (100) |
*P = 0.045 for controls, abnormal hyperplastic lesions, atypical hyperplastic lesions and squamous carcinomas (χ2 test).
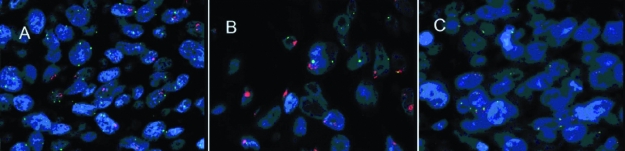
Figure 5.
Fluorescence in situ hybridization (FISH) for epidermal growth factor receptor (EGFR) gene in glottic lesions. FISH was performed by applying DNA probes: Spectrum Orange-labeled for EGFR gene and Spectrum Green-labeled centromeric probe for chromosome 7. (A) Increased EGFR gene copy number in glottic cancer: the score between EGFR gene (orange) and centromere (green) signals in tumor cell nuclei over 2.2. (B) EGFR gene amplification in form of clusters. (C) No amplification of EGFR gene in control tissue (magnification ×100).
There was no amplification in the control group (Figure 5C). In hyperplastic lesions group, amplification was detected in 2 cases of atypical hyperplasia (9.5%), one in atypical hyperplasia that developed into cancer within one month of follow-up, and the other in highly atypical dysplasia. The amplification was not detected in any case of abnormal hyperplasia.
In cancerous lesions group, EGFR gene amplification was found in 16% of cases. Some patients had previous biopsies. One of them presented with EGFR gene amplification in highly atypical hyperplasia prior to malignant transformation. In another patient, the amplification of the EGFR gene appeared in preinvasive cancer. Gene amplification was not connected either with tumor stage and grade or smoking habits (χ2 test).
Comparison between EGFR protein expression and gene amplification
The 10 samples with EGFR gene amplification had higher IHC scores of cEGFR expression throughout all layers of squamous epithelium (basal layer P = 0.003; suprabasal layer P = 0.001; superficial layer P = 0.042; Mann-Whitney U-test). Increased values of high mEGFR staining were detected in suprabasal (P = 0.069; Mann-Whitney U-test) and superficial (P = 0.217; Mann-Whitney U-test) layers of 10 amplified samples, but they were not significantly increased.
In the group of precancerous lesion samples with and without gene amplification, there was no statistical difference in IHC EGFR protein expression.
In cancerous lesions, patients with and without EGFR gene amplification showed a significant difference in cEGFR staining in the basal (P = 0.003; Mann-Whitney U-test) and suprabasal layers (P = 0.002; Mann-Whitney U-test). Significant results are shown in Table 2. Amplified cancerous lesions also showed increased high mEGFR staining in the suprabasal (P = 0.491; Mann-Whitney U-test) and superficial (P = 0.502; Mann-Whitney U-test) layers, although without significance.
Table 2.
Cytoplasmic epidermal growth factor receptor (cEGFR) expression in comparison with EGFR gene amplification status*
| cEGFR-expression | Amplification of EGFR gene (mean†±standard deviation) |
P | |
|---|---|---|---|
| no (n=42) | yes (n=8) | ||
| Basal zone | 3.25 ± 4.10 | 5.98 ± 5.06 | 0.003 |
| Suprabasal zone | 2.61 ± 2.31 | 7.34 ± 4.21 | 0.002 |
*Mann-Whitney U-test.
†Mean values of the number of cells on 0.01 mm2 in defined zones of squamous cell carcinoma with cytoplasmic epidermal growth factor receptor expression.
‡Number of analyzed tissue samples.
Discussion
Reports about EGFR gene amplification in head and neck tumors are still lacking, especially in laryngeal cancerogenesis. Chromosome 7 aberrations have been found using FISH analysis of dysplastic and cancerous laryngeal tissue and connected with prognosis (19,20), but there has been no gene analysis. The significance of EGFR gene amplification as a possible mechanism for protein overexpression still remains unclear in glottic cancer because of the heterogeneous results of different studies (21-23). The EGFR gene was discovered more than 20 years ago, but research has been stimulated only recently due to new anti-EGFR targeted therapeutic strategies (2,7-14).
In this study, TMA technology enabled us to perform a uniform analysis of protein expression and gene amplification on the same study material (21,22,24) in order to obtain better insight into glottic tumor progression and the relation of these parameters with standard clinicopathological data.
Most studies on EGFR protein expression use semiquantitative IHC analysis, which is a relatively available and affordable technique, but subjective to the pathologist’s experience. There is still a lack of technique standardization, staining intensity interpretation, and result evaluation, which sometimes causes difficulty in comparisons of results from different laboratories and authors. Some researchers consider only membranous staining as relevant, while others also take cytoplasmic staining into account and find it even more important for evaluation of tumor aggressiveness and prognosis (25-29). Setting cut-off values is also difficult in evaluation of overexpression in a multilayer squamous epithelium like the glottic region, where changes occur in layers and cannot be analyzed as, for example, glandular tissue in well-researched breast cancers and pancreatic adenocarcinomas (3,28).
In this study, a more objective IHC evaluation of EGFR protein expression has been offered. The method is accurate, precise, and reproducible, and is more detailed and objective compared with the semiquantitative analysis, even though still dependent on the experience of the pathologist.
In our study, mEGFR expression showed a gradual change from negative to weak positive and strong positive throughout all layers, from control samples and hyperplastic lesions to cancerous tissue, with the changes being most striking in the suprabasal-transforming zone. These results indicate that high and low mEGFR staining can facilitate the differentiation between dysplastic lesions and simple hyperplasia and cancerous tissue, as well as between different degrees of atypia among the hyperplastic lesions. A gradual increase in cEGFR expression with an increase in dysplasia was also observed. Moreover, the study showed a significant correlation between protein expression and different levels of atypia in hyperplastic lesions, as well as aggressiveness (gradus) and stage (early vs advanced) in glottic cancer tissue, confirming EGFR as a possible predictor of clinical behavior and prognosis in glottic cancer.
In some reports, high levels of EGFR protein expression have been reported even in simple and abnormal hyperplastic lesions (25,26,29,30). In our study, high EGFR protein expression was also found in some control patients and patients with abnormal hyperplastic lesions; none of them showed gene amplification, which seems to be connected to the progression of highly atypical lesions into cancer tissue. Based on our results and available literature data (25,26,30-33), routine IHC analysis of EGFR in follow-up protocols of glottic lesions is suggested. IHC methodology is relatively simple and affordable, but needs standardization. This is why a more objective quantitative IHC assessment of EGFR protein levels was offered in this research. In cases where IHC of EGFR and classical pathohistological analysis are inconclusive, we suggest the detection of gene amplification by FISH, which could improve the identification of lesions at high risk of cancer development. In this way, already genetically altered tissue, with high probabilities of cancer formation, could be diagnosed before it develops into cancer.
To the best of our knowledge and available literature data, we are the first to report results about EGFR gene amplification in precancerous lesions. EGFR gene amplification was found in high-risk atypical hyperplastic lesions in 2 cases. There was no EGFR gene amplification in simple or abnormal hyperplastic lesions. This indicates that EGFR gene amplification can facilitate differentiation of precancerous lesions with high malignant potential before they develop into glottic cancer. We find these results important and believe they could improve follow-up of hyperplastic lesions, prevention, and early detection of laryngeal cancer.
In cancerous lesions, EGFR gene amplification was found in 16% of the cases, which is in accordance with the few published studies on laryngeal cancer (21,29,34). Some of these patients had previous biopsies. One patient displayed EGFR gene amplification in highly atypical hyperplasia prior to malignant transformation. In another patient, who showed no EGFR gene amplification in atypical hyperplasia, the amplification of the EGFR gene appeared in preinvasive cancer. Other patients showing gene amplification were diagnosed with glottic cancer on their first visit and information on EGFR gene status of their hyperplastic lesions is lacking. In this research, in already developed carcinomas, EGFR gene amplification was not connected with stage or grade. The results are similar to those of other authors, who found no difference in EGFR gene amplification and tumor stage (21) and grade (29,34). This indicates that gene amplification seems to be a genotype change that happens even before phenotype changes at levels of atypical hyperplastic and preinvasive cancerous lesions in some cancer types, acting more in pathogenesis than in progression of glottic cancer. Even though only a small percentage of EGFR protein overexpression could be explained by gene amplification, both were found to happen early in glottic carcinogenesis.
Comparing IHC and FISH results in our tumor samples, an increase in cytoplasmic, basal, and suprabasal EGFR protein levels in tumors showing EGFR gene amplification was noticed. The only previous study comparing EGFR at protein and gene level by IHC and chromogenic in situ hybridization on laryngeal tumors was performed by Tsiambas et al (29). In contrast to our study, these authors did not observe any significant correlation between EGFR protein expression and gene amplification. Our findings in glottic cancer have yet to be confirmed in future trials. Further research is needed to evaluate the impact of cEGFR expression and gene amplification in glottic cancer, since some authors have found that cytoplasmic expression is connected with poor prognosis in some tumor types (ie, renal carcinoma and pancreatic adenocarcinoma) (27,28). Our research was limited to the evaluation of the glottic region and the strict relation between EGFR gene and protein expression, along with their correlation with classic pathohistological findings. EGRF gene mutations in laryngeal carcinogenesis and the analysis of nuclear EGFR are planned for future research.
In conclusion, our study demonstrated gene amplification as an early event in glottic cancerogenesis at the levels of high-risk atypical hyperplastic lesions and preinvasive carcinomas. This finding could improve glottic cancer prevention and early detection before genotype changes undergo tumor phenotype formation. An association was confirmed between mEGFR expression and gene amplification in glottic lesions, as well as a significant correlation between cEGFR staining and gene amplification in cancerous tissue. The implication of these findings in laryngeal carcinogenesis and the impact on prognosis and treatment have yet to be discovered.
Acknowledgments
The authors thank Miss Tanja Kovačević and Mr Ozren Štanfel for their excellent technical support in FISH and IHC procedures, and Mrs Bosa Licul and Mrs Smiljana Mavrinac for their assistance. The research was done as part of project “EGFR Molecular Marker in Stage Evaluation and Therapy Protocol of Glottic Carcinoma” and program “Important Morphological and Molecular Factors of Tumor Progression” of the Croatian Ministry of Science, Education, and Sports (Research grant No: 062-0620095-0083).
References
- 1.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–43. doi: 10.1016/S1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 2.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 3.Dowsett M, Hanna WM, Kockx M, Penault-Llorca F, Ruschoff J, Gutjahr T, et al. Standardization of HER2 testing: results of an international proficiency-testing ring study. Mod Pathol. 2007;20:584–91. doi: 10.1038/modpathol.3800774. [DOI] [PubMed] [Google Scholar]
- 4.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Khademi B, Shirazi FM, Vasei M, Doroudchi M, Gandomi B, Modjtahedi H, et al. The expression of p53, c-erbB-1 and c-erbB-2 molecules and their correlation with prognostic markers in patients with head and neck tumors. Cancer Lett. 2002;184:223–30. doi: 10.1016/S0304-3835(02)00242-2. [DOI] [PubMed] [Google Scholar]
- 6.Pivot X, Magne N, Guardiola E, Poissonnet G, Dassonville O, Francoual M, et al. Prognostic impact of the epidermal growth factor receptor levels for patients with larynx and hypopharynx cancer. Oral Oncol. 2005;41:320–7. doi: 10.1016/j.oraloncology.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol. 2003;21:2787–99. doi: 10.1200/JCO.2003.01.504. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–68. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 9.van Krieken JH, Jung A, Kirchner T, Carneiro F, Seruca R, Bosman FT, et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–31. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 10.Bonomi PD, Buckingham L, Coon J. Selecting patients for treatment with epidermal growth factor tyrosine kinase inhibitors. Clin Cancer Res. 2007;13:s4606–12. doi: 10.1158/1078-0432.CCR-07-0332. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–45. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 12.Pinter F, Papay J, Almasi A, Sapi Z, Szabo E, Kanya M, et al. Epidermal growth factor receptor (EGFR) high gene copy number and activating mutations in lung adenocarcinomas are not consistently accompanied by positivity for EGFR protein by standard immunohistochemistry. J Mol Diagn. 2008;10:160–8. doi: 10.2353/jmoldx.2008.070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 14.Italiano A, Follana P, Caroli FX, Badetti JL, Benchimol D, Garnier G, et al. Cetuximab shows activity in colorectal cancer patients with tumors for which FISH analysis does not detect an increase in EGFR gene copy number. Ann Surg Oncol. 2008;15:649–54. doi: 10.1245/s10434-007-9667-2. [DOI] [PubMed] [Google Scholar]
- 15.Znaor A. Cancer incidence in Croatia [in Croatian]. Registar za rak. Biliten. 2006;31:23. [Google Scholar]
- 16.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ.Cancer statistics 2009 CA Cancer J Clin 2009[Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Hellquist H, Cardesa A, Gale N, Kambic V, Michaels L. Criteria for grading in the Ljubljana classification of epithelial hyperplastic laryngeal lesions. A study by members of the Working Group on Epithelial Hyperplastic Laryngeal Lesions of the European Society of Pathology. Histopathology. 1999;34:226–34. doi: 10.1046/j.1365-2559.1999.00581.x. [DOI] [PubMed] [Google Scholar]
- 18.Patel SG, Shah JP. TNM staging of cancers of the head and neck: striving for uniformity among diversity. CA Cancer J Clin. 2005;55:242–58. doi: 10.3322/canjclin.55.4.242. [DOI] [PubMed] [Google Scholar]
- 19.Morrison LE, Jacobson KK, Friedman M, Schroeder JW, Coon JS. Aberrant EGFR and chromosome 7 associate with outcome in laryngeal cancer. . Laryngoscope. 2005;115:1212–8. doi: 10.1097/01.MLG.0000163755.21035.8F. [DOI] [PubMed] [Google Scholar]
- 20.Veltman JA, van Weert I, Aubele M, Bot FJ, Ramaekers FC, Manni JJ, et al. Specific steps in aneuploidization correlate with loss of heterozygosity of 9p21, 17p13 and 18q21 in the progression of pre-malignant laryngeal lesions. Int J Cancer. 2001;91:193–9. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1029>3.3.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Freier K, Joos S, Flechtenmacher C, Devens F, Benner A, Bosch FX, et al. Tissue microarray reveals prevalence of oncogene amplification in head and neck squamous cell carcinoma (HNSCC). Correlation with localization, stage and clinical outcome. Cancer Res. 2003;63:1179–82. [PubMed] [Google Scholar]
- 22.Chen B, van den Brekel MW, Buschers W, Balm AJ, van Velthuysen ML. Validation of tissue array technology in head and neck squamous cell carcinoma. Head Neck. 2003;25:922–30. doi: 10.1002/hed.10308. [DOI] [PubMed] [Google Scholar]
- 23.Lee JW, Soung YH, Kim SY, Nam HK, Park WS, Nam SW. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:2879–82. doi: 10.1158/1078-0432.CCR-04-2029. [DOI] [PubMed] [Google Scholar]
- 24.Skacel M, Skilton B, Pettay JD, Tubbs RR. Tissue microarrays: a powerful tool for high-throughput analysis of clinical specimens: a review of the method with validation data. Appl Immunohistochem Mol Morphol. 2002;10:1–6. doi: 10.1097/00022744-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Krecicki T, Jelen M, Zalesska-Krecicka M, Rak J, Szkudlarek T, Jelen-Krzeszewska J. Epidermal growth factor receptor (EGFR), proliferating cell nuclear antigen (PCNA) and Ki-67 antigen in laryngeal epithelial lesions. Oral Oncol. 1999;35:180–6. doi: 10.1016/S1368-8375(98)00100-6. [DOI] [PubMed] [Google Scholar]
- 26.Gale N, Kambic V, Poljak M, Cor A, Velkavrh D, Mlacak B. Chromosomes 7,17 polysomies and overexpression of epidermal growth factor receptor and p53 protein in epithelial hyperplastic laryngeal lesions. Oncology. 2000;58:117–25. doi: 10.1159/000012088. [DOI] [PubMed] [Google Scholar]
- 27.Langner C, Ratschek M, Rehak P, Schips L, Zigeuner R. Are heterogenous results of EGFR immunoreactivity in renal cell carcinoma related to non-standardised criteria for staining evaluation? J Clin Pathol. 2004;57:773–5. doi: 10.1136/jcp.2003.015743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda S, Ogata S, Tsuda H, Kawarabayashi N, Kimura M, Sugiura Y, et al. The correlation between cytoplasmic overexpression of epidermal growth factor receptor and tumor aggressiveness: poor prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas. 2004;29:e1–8. doi: 10.1097/00006676-200407000-00061. [DOI] [PubMed] [Google Scholar]
- 29.Tsiambas E, Stavrakis I, Lazaris AC, Karameris A, Patsouris E. Evaluation of epidermal growth factor receptor gene and chromosome 7 alterations in squamous cell carcinoma of the larynx, using chromogenic in situ hybridization on tissue microarrays. J Laryngol Otol. 2007;121:563–70. doi: 10.1017/S0022215106002374. [DOI] [PubMed] [Google Scholar]
- 30.Rubin Grandis J, Tweardy DJ, Melhem MF. Asynchronous modulation of transforming growth factor alpha and epidermal growth factor receptor protein expression in progression of premalignant lesions to head and neck squamous cell carcinoma. Clin Cancer Res. 1998;4:13–20. [PubMed] [Google Scholar]
- 31.Putti TC, To KF, Hsu HC, Chan AT, Lai GM, Tse G, et al. Expression of epidermal growth factor receptor in head and neck cancers correlates with clinical progression: a multicentre immunohistochemical study in the Asia-Pacific region. Histopathology. 2002;41:144–51. doi: 10.1046/j.1365-2559.2002.01436.x. [DOI] [PubMed] [Google Scholar]
- 32.Miyaguchi M, Olofsson J, Hellquist HB. Expression of epidermal growth factor receptor in laryngeal dysplasia and carcinoma. Acta Otolaryngol. 1990;110:309–13. doi: 10.3109/00016489009122553. [DOI] [PubMed] [Google Scholar]
- 33.Ranelletti FO, Almadori G, Rocca B, Ferrandina G, Ciabattoni G, Habib A, et al. Prognostic significance of cyclooxygenase-2 in laryngeal squamous cell carcinoma. Int J Cancer. 2001;95:343–9. doi: 10.1002/1097-0215(20011120)95:6<343::AID-IJC1060>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Koynova DK, Tsenova VS, Jankova RS, Gurov PB, Toncheva DI. Tissue microarray analysis of EGFR and HER2 oncogene copy number alterations in squamous cell carcinoma of the larynx. J Cancer Res Clin Oncol. 2005;131:199–203. doi: 10.1007/s00432-004-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]