Abstract
Integrin-mediated adhesion is as ancient as multicellularity, but it was not always as complex as it is in humans. Here, I examine the extent of conservation of 192 adhesome proteins across the genomes of nine model organisms spanning one and a half billion years of evolution. The work reveals that Rho GTPases, lipid- and serine/threonine-kinases, and phosphatases existed before integrins, but tyrosine phosphorylation developed concomitant with integrins. The expansion of specific functional groups such as GAPs, GEFs, adaptors, and receptors is demonstrated, along with the expansion of specific protein domains, such as SH3, PH, SH2, CH, and LIM. Expansion is due to gene duplication and creation of families of paralogues. Apparently, these paralogues share few partners and create new sets of interactions, thus increasing specificity and the repertoire of integrin-mediated signaling. Interestingly, the average number of interactions positively correlates with the evolutionary age of proteins. While shedding light on the evolution of adhesome complexity, this analysis also highlights the relevance and creates a framework for studying integrin-mediated adhesion in simpler model organisms.
Introduction
The transition of unicellular organisms into multicellular life forms relied on the appearance of proteins that can mediate cell adhesion (Harwood and Coates, 2004; King, 2004). Early metazoa developed two cell adhesion systems: cadherins mediate cell–cell adhesion by homophilic interaction, and integrins provide cell–extracellular matrix (ECM) adhesion (Hynes and Zhao, 2000).
Although integrins may have originated to facilitate physical linkage, they evolved to function in many other key processes of animal development (Gumbiner, 1996; Bökel and Brown, 2002). In both vertebrate and invertebrate development integrin function is essential for early morphogenetic processes such as gastrulation (Marsden and Burke, 1998), and convergent extension (Davidson et al., 2006), as well as for later development of complex organs such as the heart (Yang et al., 1995) and nervous system (Lallier et al., 1996; Becker et al., 2003). These additional roles rely on the connection of integrins to the cytoskeleton and their interaction with diverse cellular signaling pathways (Giancotti and Ruoslahti, 1999; Sastry and Burridge, 2000; Zamir and Geiger, 2001).
Integrin-mediated adhesion is focused at discreet structures along the cell–ECM interface, and their dynamics—coupled with the cytoskeleton's contractility—plays a vital role in cell migration across the ECM (Huttenlocher et al., 1995), the assembly of a patterned ECM (Wierzbicka-Patynowski and Schwarzbauer, 2003), and in mechanosensation (Bershadsky et al., 2003).
In accordance with their structural and bidirectional signaling functions, pivotal to so many developmental and homeostatis processes, integrin-mediated adhesion sites are overwhelmingly complex. Extensive work in hundreds of laboratories over the last 35 years has expanded the list of integrin-mediated adhesion components to over 150, and identified close to 700 interactions between them (Zaidel-Bar et al., 2007). Rigorous analysis of the adhesome network uncovered some of its functional design principles (Zaidel-Bar et al., 2007). However, a standing question, of relevance to other fields, is how did such complexity develop within cells over time?
The aim of this analysis is to shed light on the process of growing complexity in the context of integrin adhesion by examining the subset of adhesome proteins present in each of nine organisms, from yeast to mouse, spanning 1,500 million years of evolution.
Identifying human adhesome orthologues in model organisms
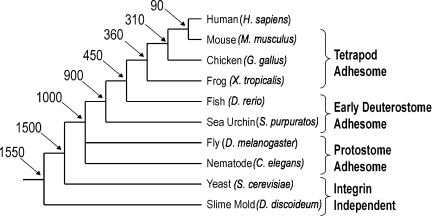
To date, a systematic parts list of integrin-mediated adhesion components has only been compiled for humans (Zaidel-Bar et al., 2007). I performed protein BLAST searches (Altschul et al., 1997) for each of the 192 human adhesome protein sequences against the full genome of each of nine organisms: mouse (Mus musculus), chicken (Galllus gallus), frog (Xenopus tropicalis), fish (Danio rerio), sea urchin (Strongylocentrotus purpuratus), fruit fly (Drosophila melanogaster), nematode (Caenorhabditis elegans), slime mold (Dictyostelium discoideum), and yeast (Saccharomyces cerevisiae). The model organisms chosen for this analysis span across 1.5 billion years of evolution from unicellular fungi and amoeba to mammals (Fig. 1).
Figure 1.
Phylogenetic tree of humans and nine model organisms. The relationships and estimated divergence times (millions of years ago) of the model organisms examined in this analysis are shown, based on recent multigene and multiprotein studies (Hedges, 2002). Branch lengths are not proportional to time.
To qualify as an orthologue, a given protein needed to be roughly the same length as the human sequence and share sequence similarity along at least 50% of the protein (bit score above 80). In addition, it had to contain all the important protein domains of the human protein, as detected by a conserved domain database (Marchler-Bauer et al., 2007), and in a reverse BLAST against the human genome it had to identify the presumed human orthologue as the first hit.
Noteworthy, certain proteins were considered orthologues based on the literature, even though they did not satisfy all the above criteria. For example, Mena and VASP orthologues share sequence similarity along less than 30% of the human proteins' length, and yet functionally they were shown to be true orthologues.
A table containing all the human adhesome proteins and the sequence IDs of their putative orthologues in each of the nine organisms is given in Table S1.
Interspecies variations in adhesome composition
Of the 192 human adhesome proteins, fewer than 50 are found in yeast or slime mold, just over 100 are found in flies or worms, and 163 are found in the fish genome. However, although the absolute number of adhesome genes has constantly grown throughout evolution, the analysis also shows many instances of genes disappearing in certain lineages. For example, worms possess 15 proteins that flies apparently do not (e.g., Vimentin, Caveolin, Plectin). Similarly, sea urchins appear to be missing 17 genes that are found in flies (e.g., Tensin, LIMK, Kindlin-1). Apparently, it is not rare for genes to be lost during evolution: there are 30 genes in fish missing from the frog genome and 14 genes in fish missing from the chicken genome (Table S1).
Estimating an evolutionary age for each gene
Although genes can appear and disappear throughout evolution, it will be useful for our analysis to appropriate an evolutionary age for each gene. For simplicity, I divided the nine model organisms into four evolutionary groups: yeast and slime mold form together the most ancient “Integrin Independent” group; nematode and fly form the “Protostome Adhesome”; sea urchin and fish form the “Early Deuterostome Adhesome”; and frog, chicken, and mouse collectively form the “Tetrapod Adhesome” (Fig. 1). Each of the 192 human adhesome proteins was assigned to one of these groups according to its most ancient orthologue. For example, if the most ancient organism with a vinculin orthologue is C. elegans, then the vinculin gene belongs to the protostome adhesome group.
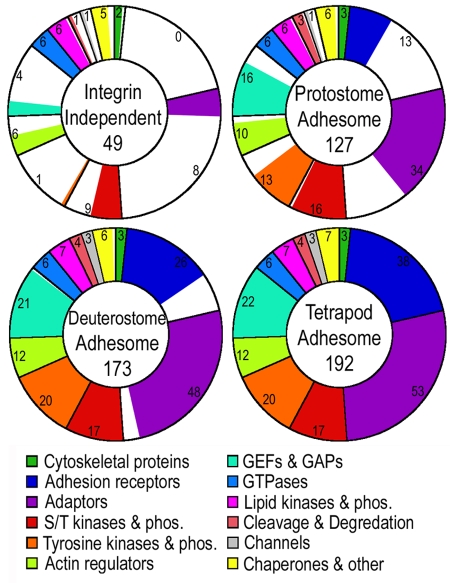
Evolution across protein functional groups
Although the total number of adhesome proteins kept growing throughout evolution, the expansion rate depended on protein function. Fig. 2 shows the composition of the adhesome at the different evolutionary stages. Remarkably, all GTPases, phospholipase enzymes, and phosphatidylinositol phosphate kinases the human adhesome utilizes existed already in yeast and slime mold. Additionaly, half of the serine/threonine kinases and phosphatases and half of the actin regulators are also found independent of integrins. Apparently, these proteins functioned in regulating the cytoskeleton before multicellularity, during processes such as cell division and cell motility. Strikingly, Dictyostelium also has homologues of the adaptor proteins talin and paxillin. These orthologues, Talin B and paxB, were reported to localize to discreet puncta at the cell–matrix interface and were shown to be necessary for force transmission crucial for cell motility (Tsujioka et al., 2004; Bukharova et al., 2005).
Figure 2.
Expansion of the adhesome through evolution. The number of proteins at each evolutionary stage is shown, broken down according to functional groups as detailed in the colored legend. All doughnuts are divided along the lines of the tetrapod adhesome protein groups and filled with color in proportion to the actual number of proteins.
Only 20% or fewer of the tetrapod adaptors, tyrosine kinases, tyrosine phosphatases, and GEF and GAP proteins existed before integrins. At the protostome stage, on the other hand, serine/threonine kinases and phosphatases are close to or reach their maximum, and over 80% of actin regulators existed. With the exception of adhesion receptors, of which there are a third, all other functional groups reach ∼70% of their tetrapod level in the protostome stage, highlighting the usefulness of studying integrin-mediated adhesion in nematode and flies. At the early deuterostome stage all but three functional groups are saturated. The only three groups still expanding in tetrapods are GEFs, adaptors, and adhesion receptors (Fig. 2).
Evolution of protein domains
Another way to examine how the adhesome expanded over evolutionary time is to look at the inventory of protein domains. Focusing on nonenzymatic protein domains, I counted the number of proteins with a given domain at each of the evolutionary stages (Table I). A dramatic expansion in the number of proteins containing SH3, PH, and SH2 occurred throughout adhesome evolution. A substantial increase in the number of proteins containing CH, LIM, and FERM domains took place as well. Indeed, these domains are significantly enriched in the adhesome compared with their general abundance in the proteome. In contrast, the number of proteins in the adhesome with PDZ, ANK, and SPEC domains did not change significantly from yeast to mouse.
Table I.
The increase in particular protein domains during adhesome evolution
| Protein domain | Number of proteins with the domain at each evolutionary stage | |||
| Integrin-independent | Protostome | Deuterostome | Tetrapod | |
| SH3 | 5 | 27 | 33 | 34 |
| PH | 5 | 16 | 21 | 23 |
| SH2 | 3 | 15 | 20 | 22 |
| CH | 2 | 7 | 9 | 10 |
| LIM | 1 | 4 | 6 | 10 |
| FERM | 1 | 7 | 9 | 9 |
| ANK | 2 | 4 | 5 | 5 |
| PDZ | 0 | 2 | 3 | 3 |
| SPEC | 2 | 4 | 4 | 4 |
PH and FERM domains target proteins to the plasma membrane, and SH3, LIM, and SH2 domains mediate protein–protein interactions that depend on specific sequence or tyrosine phosphorylation of the target protein. Thus, an increase in the number of proteins containing these domains enhanced the number and specificity of protein–protein and protein–membrane interactions within the adhesome.
Gene duplications create families of proteins in the adhesome
In theory, the number of adhesome components could increase by “adoption” of genes from other cellular pathways; by the creation of de novo genes; or by duplication of existing adhesome genes. The last method is easy to detect because duplicated genes (paralogues) display a high degree of sequence similarity. To identify families of similar proteins within the adhesome I performed an all-against-all BLAST and then used the clustering algorithm CLANS (http://toolkit.tuebingen.mpg.de/clans/) to cluster proteins connected by a P value of 10−35 or better. Over 60% of the adhesome proteins belong to a cluster (i.e., are part of a family of proteins). The largest families are α- and β-integrins, with 18 and 8 proteins, respectively. Another 62 proteins are in clusters of three or more, and there are 34 pairs of paralogues (Table S1). Tyrosine kinases and phosphatases are highly clustered, as are adaptors, suggesting multiple events of gene duplication within these functional groups. In contrast, actin regulators appear to rarely duplicate and remain mostly unrelated to each other.
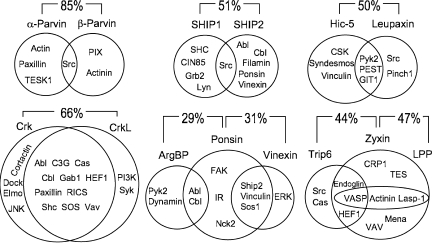
Comparing interaction repertoires of close paralogues
When proteins share high sequence similarity it is often assumed they have similar function, but the question is how similar? Do they have redundant roles or did they specialize and fit into different molecular niches? To address this question I compared the interaction partners of 15 pairs of paralogues and found evidence for both scenarios outlined above. For example, Crk and CrkL share the majority of interacting targets, whereas α-parvin and β-parvin share only one interactor and the rest of their interactors are unique (Fig. 3). It does, however, appear that the tendency of paralogues is to specialize. On average, only a third of their interactors were common to both paralogues.
Figure 3.
Common and unique interactions of paralogues. Venn diagrams show the relationship between sets of interactors of two or three paralogues. The percent similarity between the amino acid sequence of pairs of proteins is shown.
This result would be hard to explain if paralogues were the product of simple gene duplication and divergence because one would expect the duplicated protein to start off with the same set of interactors as the original protein. However, if the new protein is the result of gene duplication and translocation to another region in the genome then it may be fused to another coding region and acquire novel domains and/or its expression pattern may be altered and the new tissues in which it is expressed might not express the original interactors. Examining the location of close paralogues on the physical map of the human genome lends support to the latter option. For example: parvin-α is on chromosome 11 and parvin-β on chromosome 22, and SHIP1 is located on chromosome 2 and SHIP2 on chromosome 11.
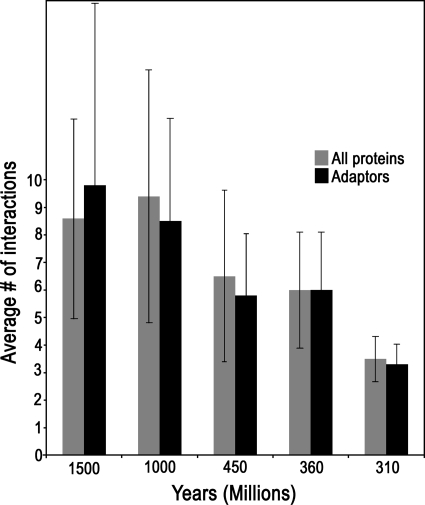
Protein interactions increase as a function of evolutionary age
Combining the recently published adhesome interaction database (Zaidel-Bar et al., 2007) with the evolutionary information obtained here, I found a positive correlation between the evolutionary age of a protein and the number of interactions it has. Because the expansion of the adhesome was not uniform across functional groups, I repeated the analysis using only adaptor proteins and got essentially the same result. Adaptors existing already in yeast or slime mold have, on average, threefold more interactions compared with adaptors first appearing in chicken (Fig. 4).
Figure 4.
Average number of interactions increases with evolutionary age. The average number of interactions (± SD) of proteins from all functional groups (gray bars) or only of adaptors (black bars) was calculated separately for proteins in each evolutionary age group.
The database of interactions was based on published papers, and therefore may be biased so that proteins receiving more attention have a larger number of known interactions. However, the evolutionary age of a protein was most likely not a factor in determining how much research attention it received. Thus, the correlation uncovered here may be a result of older genes being more essential and leading to more dramatic loss-of-function phenotypes.
The positive correlation between evolutionary age and number of interactions can be explained if proteins gradually acquire new interactors. Following this interpretation it is possible the plateau seen in Fig. 3 between 1,000 and 1,500 million years indicates that after one billion years of evolution the number of interactions per protein reaches saturation. The average number of interactions at saturation is nine.
Conclusions
A quarter of the adhesome proteins existed before integrins and were subsequently “adopted” as regulators for the newly emerging adhesion complex. Most of these “ancient” proteins continue to play multiple roles in cells and are only transiently associated with cell–matrix adhesion.
Along with the emergence of integrin receptors in metazoa, cells developed tyrosine phosphorylation as a form of regulation, and the number of tyrosine kinases and phosphatases regulating the adhesome kept growing until 450 million years ago.
Importantly, the core adhesome components are all present already in protostomia, a fact that highlights the utility of studying integrin-mediated adhesion in the fruit fly and nematode. In fact, 70% of the adhesome interaction network is accounted for by the protostome adhesome.
Between a billion and 450 million years ago, adhesome complexity grew by gene duplication and the creation of families of tyrosine kinases, GAPs, GEFs, adaptors, and receptors. These gene duplications are responsible for the enrichment of the adhesome with SH3, SH2, PH, FERM, and LIM domains. The number of GEFs, adaptors, and receptors continued growing in the last 400 million years. Although younger proteins appear to have fewer interactions relative to older family members, the new paralogues established a new set of substrates (for enzymes) and/or interactors, broadening the extent and increasing the specificity of integrin-mediated signaling and regulation.
Thus, the apparent complexity of the mammalian integrin adhesome is mainly due to the existence of multiple alternative pathways, mediated by different paralogues, which most likely are separated in time or in location, within the cell or between cell types. The challenge therefore is to tease apart these unique pathways. Such a task should be greatly assisted by studying integrin adhesion in the different model organisms described here, which present the natural advantage of lacking many of the human paralogues.
Acknowledgments
I wish to thank Morgan Kita and Cecile Ane for technical assistance, and Benny Geiger, Christoph Ballestrem, and Alon Zaslaver for comments on the manuscript.
I thank The Machiah Foundation and the National Institutes of Health (grants GM078747-01 and GM058038-09) for funding.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., McLane M.A., Becker C.G. 2003. Integrin antagonists affect growth and pathfinding of ventral motor nerves in the trunk of embryonic zebrafish.Mol. Cell. Neurosci. 23:54–68 [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D., Balaban N.Q., Geiger B. 2003. Adhesion-dependent cell mechanosensitivity.Annu. Rev. Cell Dev. Biol. 19:677–695 [DOI] [PubMed] [Google Scholar]
- Bökel C., Brown N.H. 2002. Integrins in development: moving on, responding to, and sticking to the extracellular matrix.Dev. Cell. 3:311–321 [DOI] [PubMed] [Google Scholar]
- Bukharova T., Bukharova T., Weijer G., Bosgraaf L., Dormann D., van Haastert P.J., Weijer C.J. 2005. Paxillin is required for cell-substrate adhesion, cell sorting and slug migration during Dictyostelium development.J. Cell Sci. 118:4295–4310 [DOI] [PubMed] [Google Scholar]
- Davidson L.A., Marsden M., Keller R., Desimone D.W. 2006. Integrin alpha5beta1 and fibronectin regulate polarized cell protrusions required for Xenopus convergence and extension.Curr. Biol. 16:833–844 [DOI] [PubMed] [Google Scholar]
- Giancotti F.G., Ruoslahti E. 1999. Integrin signaling.Science. 285:1028–1032 [DOI] [PubMed] [Google Scholar]
- Gumbiner B.M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis.Cell. 84:345–357 [DOI] [PubMed] [Google Scholar]
- Harwood A., Coates J.C. 2004. A prehistory of cell adhesion.Curr. Opin. Cell Biol. 16:470–476 [DOI] [PubMed] [Google Scholar]
- Hedges S.B. 2002. The origin and evolution of model organisms.Nat. Rev. Genet. 3:838–849 [DOI] [PubMed] [Google Scholar]
- Huttenlocher A., Sandborg R.R., Horwitz A.F. 1995. Adhesion in cell migration.Curr. Opin. Cell Biol. 7:697–706 [DOI] [PubMed] [Google Scholar]
- Hynes R.O., Zhao Q. 2000. The evolution of cell adhesion.J. Cell Biol. 150:F89–F96 [DOI] [PubMed] [Google Scholar]
- King N. 2004. The unicellular ancestry of animal development.Dev. Cell. 7:313–325 [DOI] [PubMed] [Google Scholar]
- Lallier T.E., Whittaker C.A., DeSimone D.W. 1996. Integrin alpha 6 expression is required for early nervous system development in Xenopus laevis.Development. 122:2539–2554 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Derbyshire M.K., DeWeese-Scott C., Gonzales N.R., Gwadz M., Hao L., He S., Hurwitz D.I., Jackson J.D., et al. 2007. CDD: a conserved domain database for interactive domain family analysis.Nucleic Acids Res. 35(Database issue):D237–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M., Burke R.D. 1998. The betaL integrin subunit is necessary for gastrulation in sea urchin embryos.Dev. Biol. 203:134–148 [DOI] [PubMed] [Google Scholar]
- Sastry S.K., Burridge K. 2000. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics.Exp. Cell Res. 261:25–36 [DOI] [PubMed] [Google Scholar]
- Tsujioka M., Yoshida K., Inouye K. 2004. Talin B is required for force transmission in morphogenesis of Dictyostelium.EMBO J. 23:2216–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I., Schwarzbauer J.E. 2003. The ins and outs of fibronectin matrix assembly.J. Cell Sci. 116:3269–3276 [DOI] [PubMed] [Google Scholar]
- Yang J.T., Rayburn H., Hynes R.O. 1995. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development.Development. 121:549–560 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., Geiger B. 2007. Functional atlas of the integrin adhesome.Nat. Cell Biol. 9:858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir E., Geiger B. 2001. Molecular complexity and dynamics of cell-matrix adhesions.J. Cell Sci. 114:3583–3590 [DOI] [PubMed] [Google Scholar]