Abstract
Expression of the p16Ink4a tumor suppressor gene, a sensor of oncogenic stress, is up-regulated by a variety of potentially oncogenic stimuli in cultured primary cells. However, because p16Ink4a expression is also induced by tissue culture stress, physiological mechanisms regulating p16Ink4a expression remain unclear. To eliminate any potential problems arising from tissue culture–imposed stress, we used bioluminescence imaging for noninvasive and real-time analysis of p16Ink4a expression under various physiological conditions in living mice. In this study, we show that oncogenic insults such as ras activation provoke epigenetic derepression of p16Ink4a expression through reduction of DNMT1 (DNA methyl transferase 1) levels as a DNA damage response in vivo. This pathway is accelerated in the absence of p53, indicating that p53 normally holds the p16Ink4a response in check. These results unveil a backup tumor suppressor role for p16Ink4a in the event of p53 inactivation, expanding our understanding of how p16Ink4a expression is regulated in vivo.
Introduction
Oncogenic proliferative signals are coupled to a variety of growth inhibitory processes (Lowe et al., 2004; Campisi, 2005; Sharpless and DePinho, 2005). For example, in cultured primary human fibroblasts, ectopic expression of oncogenic Ras or its downstream mediator initiates cellular senescence, the state of irreversible cell cycle arrest, through up-regulation of Cdk inhibitors such as p16Ink4a (Serrano et al., 1997; Lin et al., 1998; Zhu et al., 1998; Ohtani et al., 2001). Some contribution to cellular senescence presumably underlies the importance of p16Ink4a as a tumor suppressor, but the roles and mechanisms regulating its expression in vivo remain elusive (Michaloglou et al., 2005; Gil and Peters, 2006; Kim and Sharpless, 2006; Takahashi et al., 2006). To date, much of our current knowledge of how human p16Ink4a gene expression is induced by oncogenic stimuli derives from studies undertaken in cultured primary cells (Serrano and Blasco, 2001; Gil and Peters, 2006; Kim and Sharpless, 2006). However, because human p16Ink4a gene expression is also induced by tissue culture–imposed stress (Ramirez et al., 2001; Benanti and Galloway, 2004; Ince et al., 2007; Shay and Wright, 2007), it remains unclear whether the induction of human p16Ink4a gene expression in tissue-cultured cells truly reflects an anticancer process or an artifact of tissue culture–imposed stress.
We believe that p16Ink4a knockout mice are powerful tools to elucidate the physiological roles of p16Ink4a gene expression in vivo. However, one limitation of this approach is the developmental or somatic compensation by up-regulation of remaining p16Ink4a family genes in p16Ink4a knockout mice (Krimpenfort et al., 2007; Ramsey et al., 2007; Wiedemeyer et al., 2008). Moreover, the possibility of cross-species differences between human p16Ink4a expression and mouse p16Ink4a expression also complicates the interpretation of p16Ink4a knockout mouse data (Gil and Peters, 2006). Therefore, alternative approaches are also needed to supplement the tissue culture studies and to assist in understanding the roles and mechanisms regulating human p16Ink4a gene expression in vivo.
In this study, we developed a new transgenic mouse model that carries the entire human p16Ink4a gene locus tagged with firefly luciferase. In this mouse model, luciferase activity allows expression of the human p16Ink4a gene to be monitored under various physiological conditions in the context of living animals. Interestingly, the human p16Ink4a gene behaved in the same way as the mouse p16Ink4a gene in living mice, indicating that this mouse model is an ideal tool for studying physiological response of p16Ink4a gene expression in vivo. Using this system, in conjunction with various tissue culture experiments, we show in this study that oncogenic Ras signaling provokes p16Ink4a gene expression through a reduction of DNMT1 (DNA methyl transferase 1) levels as a DNA damage response (DDR) in vivo. Furthermore, this pathway is accelerated in the setting of p53 deletion, indicating that there is a regulatory feedback circuit between the p53 and p16Ink4a tumor suppressors. This unexpected regulatory circuit is likely to elicit a backup tumor suppressor role for p16Ink4a after p53 inactivation, facilitating our understanding of how these critical tumor suppressor genes are networked in vivo.
Results
Visualization of human p16Ink4a gene expression in living animals
To monitor human p16Ink4a gene expression as accurately as possible, we used a large genomic DNA segment of the human chromosome that contains the entire Ink4a/Arf gene locus (Fig. 1 A). Furthermore, this chromosome segment was engineered to express a fusion protein of human p16Ink4a and firefly luciferase (p16-luc) without deleting any genomic DNA sequences at the Ink4a/Arf gene locus (Fig. 1, A and B). This is very important because BMI-1, which is a negative regulator of p16Ink4a gene expression (Jacobs et al., 1999), is known to bind not only to the promoter region but also to the intron region of the p16Ink4a locus (Bracken et al., 2007; Kotake et al., 2007). Moreover, the expression of p16-luc fusion protein enables us to specify p16Ink4a gene expression but not Arf gene expression from this overlapping gene locus. It is noteworthy that overexpression of p16-luc did not have any significant impact on phosphorylation of retinoblastoma tumor suppressor protein (pRb), although similar levels of p16Ink4a expression efficiently blocked pRb phosphorylation in U2OS cells, suggesting that p16-luc does not function as a Cdk inhibitor (Fig. S1 A). However, importantly, a significant level of luciferase activity was observed and was reduced when this recombinant human chromosome segment (p16-luc DNA) was cointroduced with a BMI-1 expression vector into tissue culture cells (Fig. 1 C). Together, these results indicate that p16-luc DNA would be an ideal tool to monitor human p16Ink4a gene expression in vivo, especially in the context of living animals.
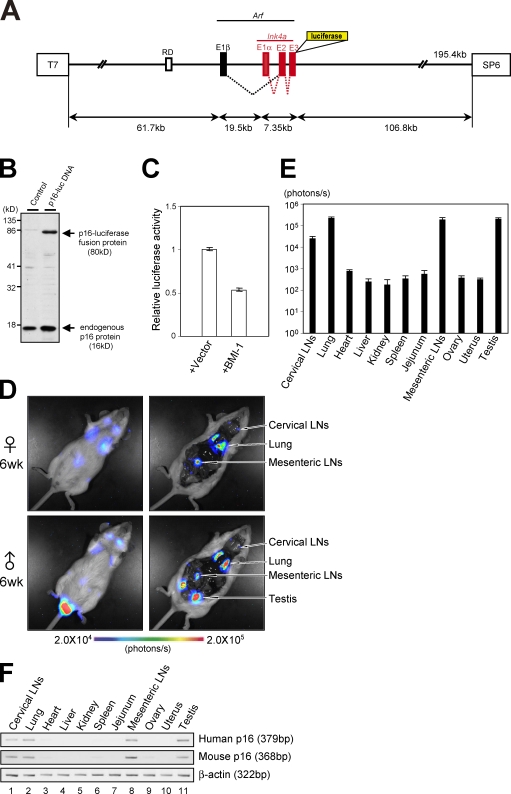
Figure 1.
Generation of human p16Ink4a reporter mice. (A) A large genomic DNA segment (195.4 kb) of human chromosome that contains the entire INK4a/ARF gene locus and surrounding sequences, including a putative DNA replication origin (RD) known to regulate p16Ink4a gene expression (Gonzalez et al., 2006), was engineered to express luciferase-tagged p16Ink4a. (B) BAC vector containing p16-luc DNA or empty BAC vector was transfected into 293T cells. Expression of the p16Ink4a-luciferase fusion protein was analyzed by Western blotting after selection with antibiotics. (C) BAC vector containing p16-luc DNA was introduced into 293T cells with or without BMI-1 expression plasmid along with 0.2 mg of MMLV (Moloney murine leukaemia virus)-lacZ plasmid. Luciferase activities were normalized by lacZ activities. Error bars indicate SD. (D) The 6-wk-old p16-luc mice were subjected to noninvasive BLI. Representative images of five different experiments are shown (left). The same mice were incised through the mouth and anus under anesthesia. Representative BLI data of five different experiments are shown. The color bar indicates photons with minimum and maximum threshold values. (E) Bioluminescence intensity emitted from the organs was graphed (log10 scale). The mean ± SD of five independent experiments is shown. (F) The levels of exogenous (human) p16Ink4a gene expression and endogenous (mouse) p16Ink4a gene expression in p16-luc mice were analyzed by semiquantitative RT-PCR. β-Actin was used as a loading control. Representative data of five different experiments are shown.
Therefore, a transgenic mouse line that carries p16-luc DNA (p16-luc mouse) was established that had incorporated the entire p16-luc DNA segment into its genome as judged by FISH, Southern blotting, and PCR analysis (Fig. S1 B and not depicted). To examine the copy number of the transgene, genomic DNA was prepared from early passage mouse embryonic fibroblasts (MEFs) derived from the p16-luc mice. The DNA sequence specific to this human chromosome segment was amplified and quantified by real-time PCR analysis (Ballester et al., 2004; Chandler et al., 2007) using genomic DNA prepared from the same number of early passage human diploid fibroblasts (HDFs) as a control. The signals obtained from the p16-luc MEFs were almost half compared with those from HDFs, which have two copies of the human Ink4a/Arf gene (Fig. S1 C). These results, together with the FISH analysis data (Fig. S1 B), suggest that the p16-luc mouse strain contains a single copy of the p16-luc DNA fragment. The p16-luc mice were then subjected to noninvasive bioluminescence imaging (BLI) under anesthesia as described previously (Ohtani et al., 2007). Although previous studies reported that p16Ink4a protein expression is virtually undetectable in the normal tissues of young humans and rodents (Zindy et al., 1997; Nielsen et al., 1999), BLI was sufficiently sensitive to detect p16-luc expression in the cervix, chest, central abdomen, and testis of 6-wk-old mice (Fig. 1 D, left). To define the organs expressing a high level of bioluminescence signal, the same mice were again subjected to BLI after incision through the mouth and anus under anesthesia (Fig. 1 D, right). As expected from noninvasive BLI data (Fig. 1 D, left), significant levels of bioluminescence signals were observed in cervical lymph nodes (LNs), lung, mesenteric LNs, and testis (Fig. 1, D [right] and E). Notably, these levels were well correlated with not only exogenous (human) but also endogenous (mouse) p16Ink4a mRNA expression (Fig. 1, E and F; and Fig. S1 D), indicating that the tissue-specific gene expression of the human p16Ink4a gene is very similar to that of the mouse p16Ink4a gene.
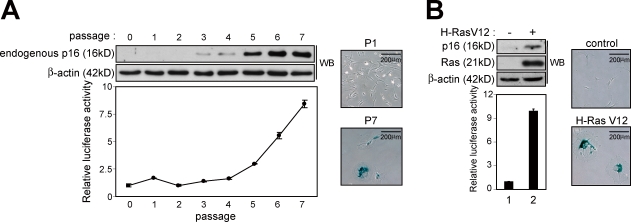
Induction of p16Ink4a expression in cellular senescence and organismal aging
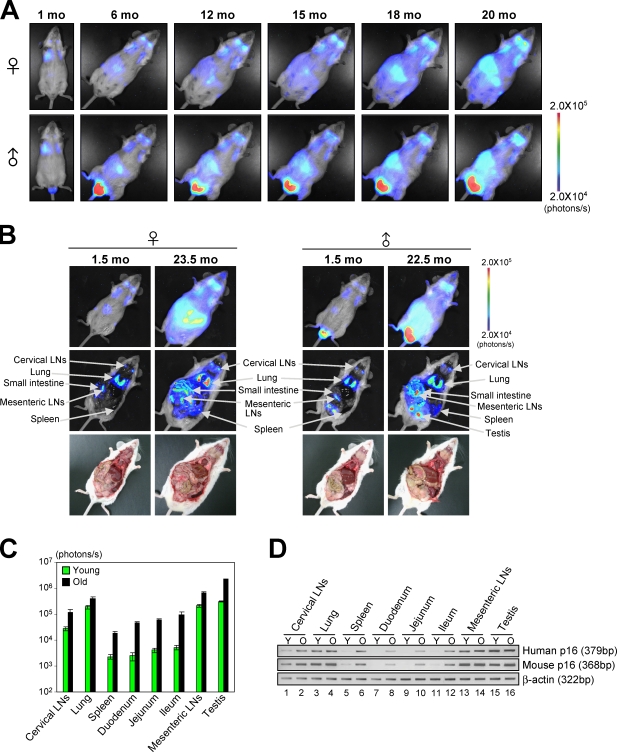
Because induction of p16Ink4a expression is a hallmark of cellular senescence (Hara et al., 1996; Serrano et al., 1997), we next asked whether the human p16Ink4a gene responds to senescence stimuli in murine cells. To this end, MEFs prepared from p16-luc mice were rendered senescent by either serial passage or ectopic expression of oncogenic Ras in culture. Concurrent with the level of endogenous p16Ink4a expression, luciferase activity was significantly increased upon induction of cellular senescence in both settings (Fig. 2), indicating that the human p16Ink4a gene also responds to senescence stimuli in murine cells. Moreover, as the levels of mouse p16Ink4a gene expression are increased in many different tissues with age (Zindy et al., 1997; Krishnamurthy et al., 2004), the levels of bioluminescent signals in p16-luc mice were dramatically increased during the aging process throughout the body (Fig. 3, A and B). However, the most striking increase was observed in the central abdomen and was found to be localized to the small intestine and spleen (Fig. 3, A and B; and Fig. S2). These levels were well correlated with both exogenous (human) and endogenous (mouse) p16Ink4a mRNA expression (Fig. 3, C and D). Collectively, these results indicate that overall regulation of human p16Ink4a gene expression is very similar to that of mouse p16Ink4a gene expression, at least in mouse cells, illustrating the potential of p16-luc mice for analysis of p16Ink4a gene expression against oncogenic stimuli in vivo.
Figure 2.
Induction of human p16Ink4a gene expression during the onset of cellular senescence in cultured MEFs. (A and B) Primary MEFs derived from the p16-luc mice were rendered senescent by either serial passage (A) or oncogenic Ras expression (retrovirus infection; B). The levels of endogenous p16Ink4a expression and luciferase activity were measured. Representative data of three independent experiments are shown. As confirmation of senescence, representative photographs of the cells stained for SA β-gal activity are shown. β-Actin was used as a loading control. The means ± SD of three independent experiments are shown. WB, Western blot.
Figure 3.
The dynamics of p16Ink4a gene expression during the aging process in vivo. (A) The same p16-luc mice were subjected to noninvasive BLI every 2 wk throughout their entire life span. Representative images of 10 independent mice are shown. (B) The p16-luc mice (young and old) were subjected to noninvasive BLI. The same mice were then incised through mouth and anus under anesthesia. (A and B) The color bars indicate photons with minimum and maximum threshold values. (C) Bioluminescence intensity emitted from the organs from young (1.5 mo) and old (22.5 mo) mice were graphed (log10 scale). The mean ± SD of five independent experiments is shown. (D) The levels of exogenous (human) p16Ink4a gene expression and endogenous (mouse) p16Ink4a gene expression from young (Y; 1.5 mo) and old (O; 22.5 mo) mice were analyzed by semiquantitative RT-PCR. β-Actin was used as a loading control. Representative data of five independent experiments are shown.
Delayed response of p16Ink4a expression against oncogenic Ras signaling in vivo
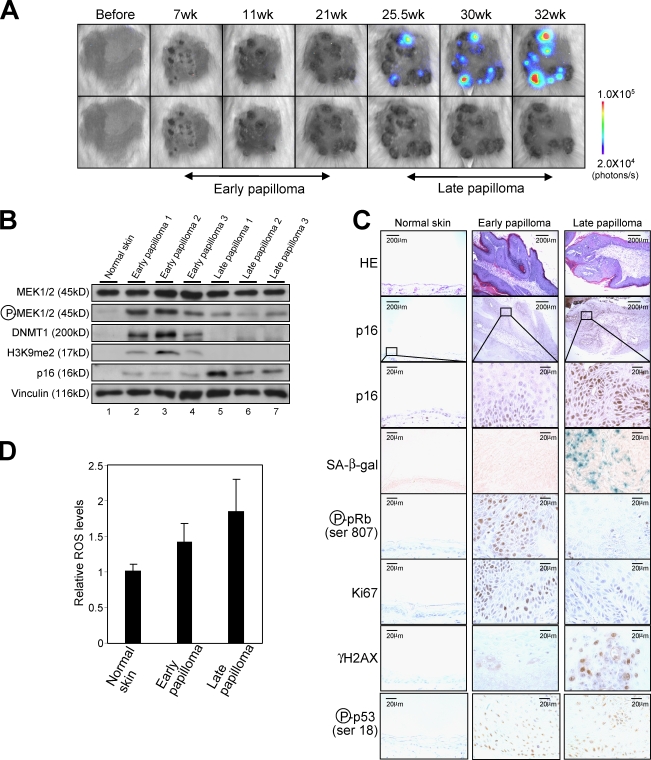
Although oncogenic Ras–induced senescence is widely observed in cultured normal HDFs (Serrano et al., 1997; Serrano and Blasco, 2001; Campisi, 2005; Gil and Peters, 2006; Kim and Sharpless, 2006), freshly isolated HDFs are resistant to oncogenic Ras–induced senescence because of low level p16Ink4a gene expression (Benanti and Galloway, 2004). To explore this notion in a more physiological setting rather than using ectopic expression of oncogenic Ras in cultured cells, the p16-luc mice were subjected to a conventional chemically induced skin papilloma protocol with a single dose of DMBA followed by multiple treatments with TPA. Because this protocol causes an oncogenic mutation in the H-ras gene (Quintanilla et al., 1986), it appeared to be ideal for studying physiological responses of p16Ink4a gene expression against oncogenic Ras signaling in living animals. In agreement with previous studies (Quintanilla et al., 1986; Kemp, 2005), benign skin papillomas began to appear after 7 wk of treatment and continued to grow to a larger size for a further 18 wk (Fig. 4 A, bottom, early papilloma). Although bioluminescent signals were hardly detectable during this time, a significant level of bioluminescent signal was induced as papillomas stopped growing (Fig. 4 A, top, late papilloma). The levels of bioluminescent signals were well correlated with those of endogenous p16Ink4a expression as well as other senescence markers such as senescence-associated (SA) β-galactosidase (β-gal) activity (Dimri et al., 1995) and dephosphorylation of pRb (Fig. 4, A–C; Campisi, 2005), indicating that oncogenic Ras signaling derived from the endogenous H-ras gene indeed provokes p16Ink4a expression accompanied by senescence cell cycle arrest in vivo.
Figure 4.
The dynamics of p16Ink4a gene expression during skin papilloma formation. (A) The p16-luc mice treated with DMBA/TPA were subjected to noninvasive BLI at the indicated time points after initiation of TPA treatment. Representative images of 10 independent experiments are shown (top). These papillomas were photographed in dimmed light (bottom). The color bar indicates photons with minimum and maximum threshold values. (B) Representative Western blots of biopsy samples of skin papillomas or control normal skin are shown using the antibodies indicated on the left. Vinculin was used as a loading control. (C) Hematoxylin and eosin (HE) staining, SA β-gal staining, and immunohistochemistry for endogenous p16Ink4a expression, phosphorylation of pRb at Ser807, Ki67 expression, phosphorylation of histone H2AX (γ-H2AX), and phosphorylation of p53 at Ser18 were performed by using biopsy samples of skin papillomas or control normal skin. The boxes denote regions shown below at higher magnification (p16 staining). (D) The levels of ROS were measured by using biopsy samples of skin papillomas or control normal skin. The means ± SD of three independent experiments are shown.
Note that the level of p16Ink4a expression in early papillomas was slightly but consistently higher than those seen in normal skin (Fig. 4, B and C). However, because significant levels of pRb phosphorylation and Ki67 expression, which are markers for cell proliferation, were observed in early papillomas (Fig. 4 C), this level of p16Ink4a expression does not appear to be high enough to induce senescence cell cycle arrest. Thus, it is tempting to speculate that p16Ink4a may play a more important role or roles in late papillomas, presumably preventing malignant conversion of benign tumors. Indeed, by 30 wk after DMBA/TPA treatment, 33% of p16Ink4a knockout mice (C57BL/6 background) had at least one carcinoma compared with 5% of the wild-type mice (unpublished data), indicating that p16Ink4a plays an important role or roles in preventing malignant conversion of benign tumors. These results are somewhat consistent with a previous study showing that the tumor-free survival of DMBA-treated mice was substantially reduced in p16Ink4a knockout mice (Sharpless et al., 2004).
The global level of histone 3 Lys9 (H3K9) dimethylation (H3K9me2) is reduced in late papillomas
Given that oncogenic mutation in the H-ras gene is known to occur immediately after DMBA treatment (Quintanilla et al., 1986) and was indeed observed in early papillomas (Fig. S3), it was puzzling that p16Ink4a gene expression was fully induced in late but not early papillomas (Fig. 4, A–C). We first reasoned that the long latency period of p16Ink4a gene expression may be caused by a spontaneous up-regulation of the intensity of Ras signaling because high levels but not low levels of oncogenic Ras expression have been shown to induce p16Ink4a gene expression in transgenic mice carrying a tetracycline-inducible ras oncogene (Sarkisian et al., 2007). However, unexpectedly, the levels of phosphorylated MEK (MAPK/ERK kinase), which is an indicator of Ras signaling, in early papillomas were even higher than those seen in late papillomas (Fig. 4 B), implying that the effect or effects of oncogenic Ras signaling on the p16Ink4a promoter (Serrano et al., 1997; Ohtani et al., 2001) may be counteracted in early papillomas.
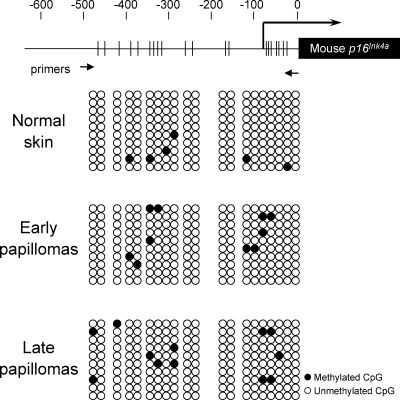
To substantiate this idea, we next sought negative regulators of p16Ink4a expression whose activities are increased in early papillomas. In the course of our study, we found that the levels of DNMT1, which is known to repress p16Ink4a expression (Robert et al., 2003), were significantly increased in early papillomas and were subsequently reduced in late papillomas (Fig. 4 B). These results led us to hypothesize that increased levels of DNMT1 may counteract the effects of oncogenic Ras signaling on the p16Ink4a promoter in early papillomas and that the subsequent reduction of DNMT1 levels thereby allows derepression of p16Ink4a gene promoter, presumably through altering the DNA methylation status of the p16Ink4a gene promoter in late papillomas. However, unexpectedly, the status of the CpG methylation around the p16Ink4a promoter was not substantially different between early and late papillomas (Fig. 5), indicating that DNMT1 may regulate p16Ink4a gene expression either indirectly or through a different activity of DNMT1 in this setting.
Figure 5.
Bisulfite sequence analysis of the mouse p16Ink4a gene promoter. A schematic illustration of the mouse p16Ink4a gene promoter is shown at the top, with the sequence numbered backwards from immediately before the initiating Met codon (the positions of the numbers are indicated by vertical dashes). The transcriptional start site is indicated by a thick vertical line, with the direction of transcription shown by the associated arrow. The coding region of the mouse p16Ink4a gene is annotated and indicated with a closed box. The clustering of CpG dinucleotides is shown with thin vertical lines through the promoter region. The positions of the primers for methyl-specific PCR are indicated below with two small arrows. Bisulfite-treated DNA was prepared from normal skin or early or late papillomas (Fig. 4). Methylation-specific PCR products were subcloned into the pGEM-T vector, and 10 clones for each sample were sequenced. Representative results of three different experiments are shown.
It has recently been shown that DNMT1 also possesses an activity to enhance H3K9 methylation through interacting with G9a, a major H3K9 mono- and dimethyltransferase (Estève et al., 2006). Thus, we next examined the levels of H3K9 methylation during skin papilloma development. Interestingly, global levels of H3K9me2 were remarkably increased in early papillomas and were reduced in late papillomas, which is consistent with the levels of DNMT1 during skin papilloma development (Fig. 4 B). These results, in conjunction with previous studies showing that the silencing of p16Ink4a gene expression was associated with H3K9 methylation in several human cancer cell lines (Nguyen et al., 2002; Bachman et al., 2003), led us to the hypothesis that the levels of DNMT1 may regulate p16Ink4a expression, at least partly, through altering the levels of H3K9me2 around the p16Ink4a gene promoter region during DMBA/TPA-induced skin papilloma development.
The balance between the intensity of Ras signaling and the levels of DNMT1 expression regulates p16Ink4a expression
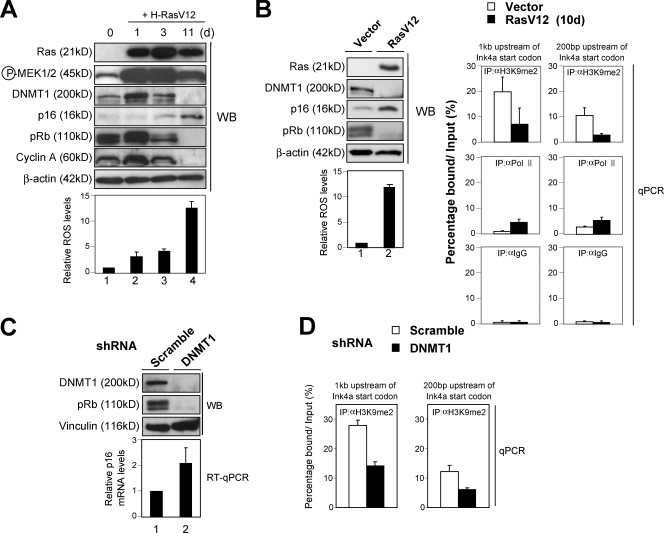
To test this hypothesis and to delineate the molecular mechanisms involved, we next used primary HDFs because mice lacking the DNMT1 gene are embryonic lethal (Li et al., 1992) and the p16Ink4a gene is exquisitely sensitive to tissue culture stress in mouse primary cells (Zindy et al., 1997; Parrinello et al., 2003). Of note, the levels of DNMT1 were initially increased by oncogenic Ras expression and were subsequently reduced as cells reached to the senescence stage in cultured HDFs (Fig. 6 A). This is reminiscent of our results in DMBA/TPA-induced skin papillomas (Fig. 4 B). Note that the levels of DNMT1 bound to the p16Ink4a gene promoter were significantly reduced in oncogenic Ras–induced senescence in cultured HDFs (Fig. S4 A). Moreover, the levels of H3K9me2 around the p16Ink4a gene promoter were also substantially reduced in the same setting (Fig. 6 B), suggesting that a similar mechanism is likely to be involved in the regulation of p16Ink4a gene expression by oncogenic Ras signaling both in vitro (cultured HDFs) and in vivo (mouse skin).
Figure 6.
Correlation between DNMT1 level and H3K9me2 level around the p16Ink4a gene promoter. (A) Early passage TIG-3 cells were infected with retrovirus encoding oncogenic Ras (+H-RasV12). Cells were then subjected to Western blot (WB) analysis at the indicated times with the antibodies shown on the left and to analysis of intracellular levels of ROS. (B) Early passage TIG-3 cells were infected with retrovirus encoding oncogenic Ras (RasV12) or control empty vector for 10 d and were subjected to Western blotting with the antibodies shown, analysis of intracellular levels of ROS, and to ChIP analysis using the antibodies indicated (IP). The precipitated DNA was amplified by real-time quantitative PCR (qPCR) using primers specific for the p16Ink4a gene promoter described previously (Bracken et al., 2007). (C and D) Early passage TIG-3 cells were infected with retrovirus encoding shRNA against DNMT1 or control scramble shRNA. Cell extracts were prepared from cells at 7 d after selection with puromycin and were subjected to Western blotting and quantitative real-time RT-PCR (RT-qPCR) analysis for p16Ink4a gene expression (C) and subjected to ChIP analysis using antibodies against H3K9me2 (D). (D) The precipitated DNA was amplified by real-time quantitative PCR using primers specific for the p16Ink4a gene promoter described previously (Bracken et al., 2007). (A–D) The means ± SD of three independent experiments are shown.
It is noteworthy that substantial levels of both phosphorylated MEK, an indicator of Ras signaling (positive factor), and DNMT1 (negative factor) were observed in proliferating early passage HDFs (Fig. 6 A), and depletion of DNMT1 level by RNAi caused a substantial increase of p16Ink4a gene expression accompanied by a senescence-like cell cycle arrest in early passage HDFs (Fig. 6 C and Fig. S4, B and C). Moreover, the levels of DNMT1 bound to the p16Ink4a gene promoter and H3K9me2 around the p16Ink4a gene promoter were also substantially reduced in DNMT1 knockdown cells (Fig. 6 D and Fig. S4 D). Therefore, it is likely that the level of the p16Ink4a gene is regulated, at least partly, through the balance between the intensity of Ras signaling (positive factor) and the levels of DNMT1 expression (negative factor). Note that the intensity of the Ras signaling was very low in normal skin (Fig. 4 B). This may explain why the level of p16Ink4a expression is very low despite the low level of DNMT1 expression in normal skin (Fig. 4 B). Together, these results indicate that the increased level of DNMT1 is likely to counteract the effects of oncogenic Ras signaling on the p16Ink4a gene promoter, presumably through facilitating H3K9me2 around the p16Ink4a gene promoter in early papillomas. It is also interesting to note that oncogenic Ras signaling is shown to activate the DNMT1 gene promoter (MacLeod et al., 1995). Thus, the induction of DNMT1 expression appears to be caused by a direct effect of oncogenic Ras expression. However, it remains unclear why DNMT1 levels are subsequently reduced in late papillomas and in oncogenic Ras–induced senescence in cultured HDFs (Figs. 4 B and 6 A).
DDR provokes p16Ink4a expression through reduction of DNMT1 levels
Because several lines of evidence suggest that DDRs triggered by hyper–cell proliferation play a key role or roles in the onset of oncogene-induced senescence (Bartkova et al., 2006; Di Micco et al., 2006; Mallette et al., 2007), we next asked whether DDR triggered by hyper–cell proliferation might be involved in the reduction of DNMT1 level in late papillomas. Indeed, a significant increase of phosphorylation of pRb and expression of Ki67, which are markers for hyper–cell proliferation, were observed before detection of γ-H2AX foci and phosphorylation of proteins containing the ATM (ataxia telangiectasia mutated)/ATR (ATM and Rad3 related) substrate motif, which are signs of DDR, in skin papillomas (Fig. 4 C and Fig. S4 E). Moreover, treatment with doxorubicin (DXR), a DNA-damaging agent, caused significant reductions of DNMT1 and the H3K9me2 modification around the p16Ink4a gene promoter, accompanied by induction of p16Ink4a gene expression in cultured HDFs (Fig. S5, A and B). Together, these results suggest that the accumulation of DNA damage triggered by hyper–cell proliferation is likely to cause a reduction of DNMT1 levels in late papillomas.
To understand how DNA damage causes a reduction of DNMT1 levels, we next focused on the intracellular levels of reactive oxygen species (ROS) because ROS levels are known to be increased by both oncogenic Ras expression and DNA damage (Fig. S5 A; Finkel and Holbrook, 2000; Macleod, 2008), and increased levels of ROS are shown to be essential for the onset of oncogenic Ras–induced senescence in cultured HDFs (Lee et al., 1999). The levels of ROS were substantially increased in late papillomas and in oncogenic Ras–induced senescence in cultured HDFs (Figs. 4 D and 6 A). Moreover, reduction of DNMT1 levels by oncogenic Ras expression was somewhat diminished when ROS production was attenuated by the addition of catalase in cultured HDFs (Fig. S5 C). Conversely, treatment with H2O2 to increase intracellular levels of ROS significantly reduced the levels of DNMT1 gene expression in HDFs (Fig. S5 D), presumably through blocking the activity of E2F transcription factor because E2F activity is known to activate DNMT1 gene expression (McCabe et al., 2005) and is reduced by treatment with H2O2 (not depicted). Together, these results provide compelling evidence that DNA damage provokes p16Ink4a gene expression, at least partly, through blocking the levels of DNMT1 gene expression by elevated levels of ROS.
Inactivation of p53 accelerates the DDR pathway activating p16Ink4a gene expression
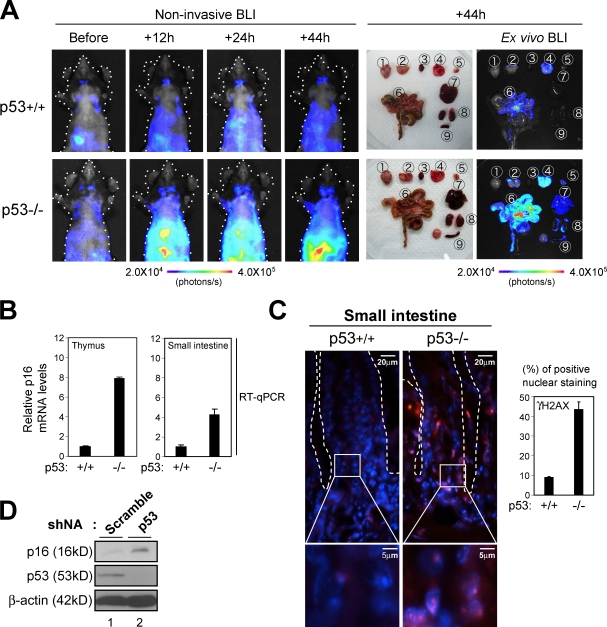
Because the p53 tumor suppressor is known to be activated immediately after detection of DNA damage, preventing proliferation of damaged cells (Vousden and Lane, 2007; Riley et al., 2008), we wondered whether p53 might have some influence over the DDR pathway activating p16Ink4a gene expression in vivo. Indeed, phosphorylation of p53 at Ser18, which is a marker of p53 activation, was observed in early papillomas (Fig. 4 C), implying that p53 may prevent accumulation of DDR and thereby counteract p16Ink4a gene expression in vivo. To explore this idea, we again took advantage of using p16-luc mice. The p16-luc mice were crossed into a p53 homozygous-null genetic background, and p16-luc mice lacking the p53 gene were subject to noninvasive BLI at the indicated times after treatment with DXR. Intriguingly, although bioluminescent signals were only slightly induced after treatment with DXR in p16-luc mice, this effect was dramatically enhanced by p53 deletion, especially in highly proliferative tissues such as thymus or small intestine (Fig. 7 A). Note that the levels of bioluminescence signals were well correlated with those of endogenous p16Ink4a gene expression as well as the number of DNA damage foci in p16-luc mice (Fig. 7, B and C). Moreover, RNAi-mediated depletion of p53 also increased the levels of p16Ink4a expression in cultured HDFs (Fig. 7 D), precluding the possibility that the enhanced expression of the p16Ink4a gene in p53 knockout mice was an artifact of BLI and/or the generation of p53 knockout mice. Therefore, these results indicate that loss of p53 function indeed accelerates the DDR pathway activating p16Ink4a gene expression, unveiling a regulatory circuit between the p53 and p16Ink4a tumor suppressors.
Figure 7.
Accelerated induction of p16Ink4a gene expression by DNA damage in mice lacking p53. (A) 8-wk-old p16-luc mice lacking the p53 gene (p53−/−) or their wild-type controls (p53+/+) were injected i.p. with 20 µg/g DXR and subjected to noninvasive BLI at the indicated time points. The same mice were then sacrificed, tissues were rapidly removed and placed in culture dishes, and ex vivo tissue BLI was performed. The tissues examined were as follows: 1, brain; 2, cervical LNs; 3, heart; 4, lung; 5, thymus; 6, small intestine; 7, liver; 8, kidney; 9, spleen. The color bars indicate photons with minimum and maximum threshold values. Representative images of five different experiments are shown. Schematic drawings illustrating the mouse bodies are shown (dotted lines). (B and C) Isolated tissues were subjected to analysis of quantitative real-time RT-PCR (RT-qPCR) for p16Ink4a gene expression (B) or to immunofluorescence analysis using antibody against γ-H2AX (red; C). DNA was stained with DAPI (blue). The histogram indicates the percentage of nuclei that were positive for γ-H2AX staining. The means ± SD of three independent experiments are shown. (C) Enlarged images of the boxed areas are shown below. Small intestinal crypts are marked by dashed lines. (D) Early passage TIG-3 cells were infected with retrovirus encoding shRNA against p53 or control scramble shRNA. Cell extracts were prepared after selection with puromycin and subjected to Western blotting using the antibodies shown on the left. β-Actin was used as a loading control.
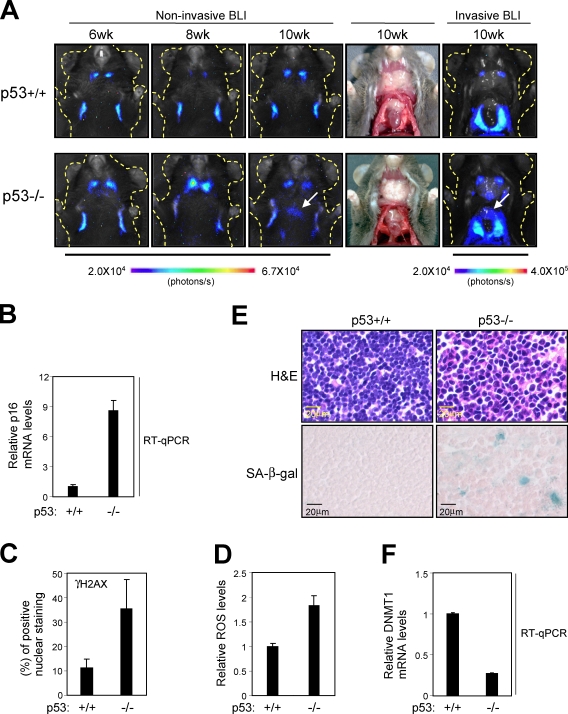
To further verify the biological significance of the regulatory circuit between p53 and p16Ink4a, we next asked whether p16Ink4a gene expression is induced naturally in the setting of p53 deletion. Interestingly, a substantial increase of bioluminescence signals over controls was indeed observed in the thymus of nearly all mice lacking p53 around 10–20 wk after birth (Fig. 8 A). Note that p53 knockout mice are shown to be highly susceptible to thymic lymphoma (Donehower et al., 1992). Therefore, it is very likely that deletion of the p53 gene causes a strong oncogenic stimulus that provokes p16Ink4a expression in the thymus. Indeed, the levels of bioluminescent signals were well correlated with those of endogenous p16Ink4a expression as well as γ-H2AX foci, ROS, and SA β-gal activity in thymic tissues of p53 knockout mice (Fig. 8, B–E). Moreover, a substantial reduction of DNMT1 gene expression was also observed in thymic tissues of p53 knockout mice (Fig. 8 F). Altogether, although additional mechanisms may be at play here, these data strongly suggest that inactivation of p53 potentiates the DDR pathway activating p16Ink4a gene expression and thereby causing senescent cell cycle arrest in highly proliferating tissues such as thymus. However, because p53 knockout mice are highly susceptible to thymic lymphoma (Donehower et al., 1992), induction of p16Ink4a appears to be insufficient for long-term stability of senescence cell cycle arrest in the absence of p53, which is the guardian of the genome.
Figure 8.
Induction of p16Ink4a gene expression in p53 knockout mice. (A) p16-luc mice lacking the p53 gene (p53−/−) or their wild-type controls (p53+/+) were subjected to noninvasive BLI at the indicated age. The same mice were then incised under anesthesia (photographed under regular light) and subjected to BLI again (invasive BLI). Representative images of five different experiments are shown. The color bars indicate photons with minimum and maximum threshold values. Schematic drawings illustrating the mouse bodies are shown (dotted lines). The arrows show bioluminescence signals derived from thymus. (B–F) Thymus tissue was isolated from both genotypes of 10-wk-old mice (A) and subjected to analysis of quantitative real-time RT-PCR (RT-qPCR) for p16Ink4a gene expression (B) or DNMT1 gene expression (F) or histochemistry (hematoxylin and eosin [H&E] staining and SA β-gal staining; E), γ-H2AX foci (C), or intracellular levels of ROS (D). The means ± SD of three independent experiments are shown.
Discussion
The regulation of human p16Ink4a gene expression has remained incompletely defined, at least in part, because p16Ink4a gene expression is rapidly induced by tissue culture stress in nontransformed cells (Shay and Wright, 2007). To circumvent this problem, we developed a new transgenic mouse model that carries the entire human p16Ink4a gene locus tagged with luciferase. In this mouse model, luciferase activity allows expression of the human p16Ink4a gene to be monitored under various physiological conditions in living mice. We did not observe any phenotypic differences between p16-luc mice and control nontransgenic mice (unpublished data), although Matheu et al. (2009) reported that one or two additional mouse Ink4a/Arf gene loci without a reporter gene caused an impairment of male germ cell production as well as reduced incidence of aging-associated cancer and extended longevity. However, because the p16-luc fusion protein does not function as a Cdk inhibitor (Fig. S1 A), it is most likely that these seemingly contradictory data are, at least partly, caused by the difference of p16Ink4a activity between our mice and the mice of Matheu et al. (2009). Interestingly, the human p16Ink4a gene and mouse p16Ink4a gene are regulated similarly in mouse cells, as judged by several well-accepted criteria (Figs. 1–4). Collectively, these results indicate that the p16-luc transgenic mouse line could be an ideal tool for studying physiological response of p16Ink4a gene expression against oncogenic stress in vivo.
By monitoring and quantifying p16Ink4a gene expression repeatedly in the same mouse throughout DMBA/TPA-induced skin papilloma development, we unveiled the dynamics of p16Ink4a gene expression against oncogenic Ras signaling in vivo (Fig. 4 A). Curiously, although the intensity of Ras signaling in early papillomas is higher than that in late papillomas (Fig. 4 B), a significant induction of p16Ink4a expression was observed in late but not in early papillomas (Fig. 4, A–C), suggesting that the effect or effects of oncogenic Ras signaling on the p16Ink4a gene promoter (Serrano et al., 1997; Ohtani et al., 2001) may be counteracted by negative regulators of p16Ink4a gene expression in early papillomas. Interestingly, the levels of DNMT1, which is a negative regulator of p16Ink4a gene expression (Robert et al., 2003), were substantially increased in early papillomas and were subsequently reduced in late papillomas (Fig. 4 B). Moreover, similar kinetics of DNMT1 expression were also observed in the setting of oncogenic Ras–induced senescence in cultured HDFs (Fig. 6 A), and depletion of DNMT1 by RNAi caused an increase of p16Ink4a gene expression in proliferating HDFs (Fig. 6 C). Thus, it is most likely that increased levels of DNMT1 expression counterbalance the levels of p16Ink4a gene expression in early papillomas, with the subsequent reduction of DNMT1 levels resulting in the induction of p16Ink4a expression in late papillomas.
Because Ras signaling is known to activate the DNMT1 gene promoter (MacLeod et al., 1995), it is most likely that the induction of DNMT1 in early papillomas is a direct effect of oncogenic Ras signaling (Fig. 4 B). However, it was unclear how DNMT1 is reduced in late papillomas (Fig. 4 B). Our results strongly suggest that DDR triggered by hyper–cell proliferation plays a critical role or roles in blocking the level of DNMT1 gene expression, at least partly, through the elevation of ROS level (Figs. 4 D and 6 A and Fig. S5). Note that DNMT1 gene expression is regulated by E2F (McCabe et al., 2005) and that E2F activity is reduced by H2O2 treatment (unpublished data). Therefore, it is possible that ROS regulates DNMT1 expression, at least in part, through E2F. These results, together with the observation that depletion of DNMT1 causes up-regulation of p16Ink4a gene expression in cultured HDFs (Fig. 6 C), indicate that DDR plays a key role in the induction of p16Ink4a gene expression through blocking the levels of DNMT1 expression in the setting of Ras-induced senescence both in vitro and in vivo.
Despite significant differences in the levels of DNMT1 expression, we were unable to see substantial differences in the status of the CpG methylation around the p16Ink4a gene promoter between early and late papillomas (Fig. 5). However, interestingly, global levels of H3K9me2 were strikingly increased in early papillomas and were reduced in late papillomas, which is consistent with the changing levels of DNMT1 in these settings (Fig. 4 B). Moreover, local levels of H3K9me2 around the p16Ink4a promoter were also reduced in oncogenic Ras–induced senescence and in DNMT1 depletion in cultured HDFs (Fig. 6, B and D). These results, together with a recent observation that DNMT1 possesses an activity to enhance H3K9me2 through interacting with G9a (Estève et al., 2006), suggest that DNMT1 regulates p16Ink4a gene expression through altering the H3K9me2 status around the p16Ink4a gene promoter during oncogenic Ras–induced senescence. It is noteworthy that Narita et al. (2003) have reported an increase in H3K9 methylation specifically at E2F target promoters in senescent HDFs. Collectively, with our observations at the p16Ink4a gene promoter, it appears that widespread redistribution of H3K9 methylation may occur during senescence onset.
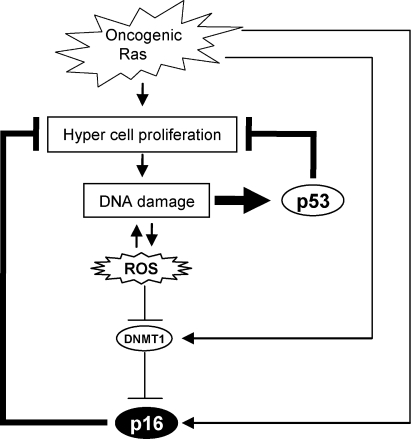
Therefore, we propose a model in which although oncogenic Ras signaling has a potential to activate p16Ink4a gene expression (Serrano et al., 1997; Serrano and Blasco, 2001; Ohtani et al., 2001), this effect is initially counteracted by the elevation of DNMT1 levels and thereby causes hyper–cell proliferation. However, because hyper–cell proliferation tends to cause DNA damage and elevation of ROS, DNMT1 gene expression is eventually reduced by this ROS, leading to epigenetic derepression of p16Ink4a gene expression and thus senescence cell cycle arrest (Fig. 9). Importantly, moreover, this pathway is more potentiated in the setting of p53 deletion because p53 tends to prevent proliferation of damaged cells that cause further accumulation of DNA damage (Fig. 9; Vousden and Lane, 2007; Riley et al., 2008). Indeed, the DDR pathway activating p16Ink4a gene expression was provoked in the thymus of nearly all of the mice lacking p53 around 10–20 wk after birth (Fig. 8). Therefore, it is possible that p16Ink4a may play a backup tumor suppressor role in case p53 is accidentally inactivated, especially in highly proliferative tissue such as thymus. Indeed, it has previously been shown that overexpression of Aurora A resulted in a significant induction of p16Ink4a expression in the mammary glands of p53 knockout mice (Zhang et al., 2008). Together, our finding may help to explain why mice doubly deficient for p53 and p16Ink4a showed an increased rate of tumor formation (Sharpless et al., 2002; Terzian et al., 2008) and why the combination of p53 and p16Ink4a loss frequently occurred in human cancer cells (Malumbres and Barbacid, 2001).
Figure 9.
Cross talk between the p53 and p16 pathways through DDR. Although oncogenic Ras signaling has a potential to activate p16Ink4a gene expression, this effect is initially counteracted by an elevation of DNMT1 level and thereby causes a strong proliferative burst, resulting in the accumulation of DNA damage. The accumulation of DNA damage activates ROS production, which in turn blocks DNMT1 gene expression, thereby causing epigenetic derepression of p16Ink4a gene expression and thus senescence cell cycle arrest. This pathway is counterbalanced by the p53 pathway because p53 is immediately activated by DNA damage and blocks proliferation of damaged cells that cause further accumulation of DNA damage. Thus, the DDR pathway activating p16Ink4a expression is accelerated in the event of p53 inactivation.
However, it is clear that all aspects of p16Ink4a regulation cannot be explained by factors described in this study and that the p16Ink4a gene is subject to multiple levels of control (Jacobs et al., 1999; Passegué and Wagner, 2000; Ohtani et al., 2001, 2003; Gonzalez et al., 2006; Bracken et al., 2007; Kotake et al., 2007; Baker et al., 2008; Kia et al., 2008; Agger et al., 2009; Barradas et al., 2009; Tzatsos et al., 2009; Witcher and Emerson, 2009). Nonetheless, our results uncover an unexpected link between p53 and p16Ink4a gene expression, expanding our understanding of how p16Ink4a gene expression is induced by oncogenic stimuli in vivo and open up new possibilities for its control.
Materials and methods
Generation of p16-luc mice
A large human genomic DNA segment (195.4 kb) containing the human p16Ink4a gene was obtained from BACPAC Resources Center. The terminal sequences were determined with primers complementary to the insert-flanking elements of the bacterial artificial chromosome (BAC) vector (pBACe3.6). The cDNA fragment encoding firefly luciferase was inserted to the 3′ end of the p16Ink4a coding sequences using a Counter Selection BAC Modification kit (Gene Bridges). The DNA was digested with NotI and then used for microinjection. The transgenic mouse strain was generated by pronuclear microinjection of the reporter transgene into fertilized ICR oocytes. One transgenic line, p16-luc (line number CDB0416T-53), was selected because transgene integrity was complete. For the generation of compound mice, p16-luc mice were crossed with C57BL/6 mice for eight generations and then crossed with p53 knockout mice (C57BL/6; Ohtani et al., 2007). p16Ink4a knockout mice (C57BL/6) were provided by N.E. Sharpless (University of North Carolina Lineberger Comprehensive Cancer Center, Chapel Hill, NC; Sharpless et al., 2004). All animals were cared for by using protocols approved by the Committee for the Use and Care of Experimental Animals of the Japanese Foundation for Cancer Research and/or the University of Tokushima.
BLI and image acquisition
For the detection of luciferase expression, mice were anesthetized and injected i.p. with 75 mg/kg d-luciferin sodium salt 5 min before beginning photon recording. Photons emitted from living mice or from isolated organs were acquired as described previously (Ohtani et al., 2007). In brief, mice were placed in the light-tight chamber, and a grayscale image of the mice was first recorded with dimmed light followed by acquisition of the luminescence image using a cooled charge-coupled device (CCD) camera (PIXIS 1024B; Princeton Instruments). The signal to noise ratio was increased by 2 × 2 binning and 15-min exposure. For colocalization of the luminescent photon emission on the animal body, grayscale and pseudocolor images were merged by using Image-Pro Plus (Media Cybernetics).
Tumor induction experiments
10 mice of the p16-luc line in the resting phase of the hair cycle (8 wk old) were shaved and treated with 100 µg DMBA (7,12-dimethylbenz[a]anthracene) in 100 µl acetone. 1 wk after DMBA treatment, mice were subsequently treated twice a week with 12.5 µg TPA (12-o-tetradecanoylphorbol 13-acetate) in 100 µl acetone for 15 wk. Control mice were treated with acetone instead of DMBA/TPA.
H-ras sequencing
Total RNA was isolated from skin papilloma and control normal tissues using TRIZOL reagent (Invitrogen). RNA was converted to cDNA by using oligo (dT) primer, and a 330-bp PCR fragment containing exon 2 of the H-ras gene was amplified with the 5′-TGGGGCAGGAGCTCCTGGAT-3′ and 5′-CTGTACTGATGGATGTCCTC-3′ primers. PCR fragments were subcloned using pGEM-T Easy vector system (Promega) and were sequenced by using Dye-Terminator and Big-Dye cycle sequencing systems (Applied Biosystems).
Semiquantitative RT-PCR
Total RNA was isolated using TRIZOL reagent, and 2 µg of total RNA was used for the reverse transcription reaction. The PCR was performed using Blend Taq polymerase (TOYOBO) with primers specific for the mouse p16Ink4a gene, human p16Ink4a gene, and mouse β-actin gene as shown below. Human p16Ink4a, 5′-ACCAGAGGCAGTAACCATGC-3′ (forward) and 5′-TGTCGTTCGCGGGCGCAACTG-3′ (reverse); mouse p16Ink4a, 5′-GAACTCTTTCGGTCGTACCC-3′ (forward) and 5′-TGGGCGTGCTTGAGCTGA-3′ (reverse); and β-actin, 5′-GTATGGAATCCTGTGGCATC-3′ (forward) and 5′-AAGCACTTGCGGTGCACGAT-3′ (reverse).
Real-time RT-PCR
Quantitative real-time RT-PCR was performed using SYBER Premix EX Taq system (TAKARA) and a Prism 7900HT (ABI). Amplified signals were confirmed to be single bands by gel electrophoresis and were normalized to the levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data were analyzed using SDS2.1 software (ABI). The PCR primer sequences used are as follows: GAPDH, 5′-CAACTACATGGTTTACATGTTC-3′ (forward) and 5′-GCCAGTGGACTCCACGAC-3′ (reverse); human p16Ink4a, 5′-CCCAACGCACCGAATAGTTA-3′ (forward) and 5′-ACCAGCGTGTCCAGGAAG-3′ (reverse); mouse p16Ink4a, 5′-GAACTCTTTCGGTCGTACCC-3′ (forward) and 5′-CGAATCTGCACCGTAGTTGA-3′ (reverse); human DNMT1, 5′-TACCTGGACGACCCTGACCTC-3′ (forward) and 5′-CGTTGGCATCAAAGATGGACA-3′ (reverse); mouse DNMT1, 5′-GAGGAAGGCTACCTGGCTAA-3′ (forward) and 5′-AGTGAGAGTGTGTGTTCCGT-3′ (reverse); 5′ end of the transgene, 5′-GCAGAAAGCCAGGAGAGGTG-3′ (forward) and 5′-GCCGCATGGACACAGGTGAC-3′ (reverse); 3′ end of the transgene, 5′-TCCCTTATCCTTACCCCACT-3′ (forward) and 5′-TGATGATGATCCTATGGTGT-3′ (reverse); and GAPDH (human/mouse common sequence), 5′-AGACCACAGTCCATGCCATC-3′ (forward) and 5′-TTGCCCACAGCCTTGGCAG-3′ (reverse).
Histology and immunohistochemistry
Immunohistochemistry was performed as previously described (Ohtani et al., 2007). The primary antibodies used were mouse p16 (Santa Cruz Biotechnology, Inc.), γ-H2AX (Millipore), phospho-pRb (Ser807/811; Cell Signaling Technology), Ki67 (RTU-Ki67-MM1; Novocastra), and phospho (Ser/Thr)-ATM/ATR substrate (Cell Signaling Technology). The fluorochromes used were DAPI (Dojindo) and Alexa Fluor 546 (Invitrogen). Microscopic images were obtained at RT by using a fluorescence microscope (AxioImager.A1; Carl Zeiss, Inc.) connected with a CCD camera (AxioCamMRc5; Carl Zeiss, Inc.) through an EC Plan-Neofluar 10× NA 0.3 Ph1, 40× NA 0.75 Ph2, or 63× NA 1.25 oil Ph3 objective using AxioVision software (Carl Zeiss, Inc.).
Cells and cell culture experiments
Early passage (45 PDLs) HDFs (TIG-3 cells), MEFs, 293T cells, and U20S cells were cultured in DME supplemented with 10% FBS at 37°C. Retroviral infection was performed as previously described (Takahashi et al., 2006). For RNAi experiments, HDFs were infected with retrovirus encoding short hairpin RNA (shRNA) as described previously (Maehara et al., 2005). RNAi sequences used were as follows: scramble, 5′-CATTGCTATAGAGGCAGAT-3′; human DNMT1, 5′-TGGTCCGCATGGGCTATCAGT-3′; and human p53, 5′-GACTCCAGTGGTAATCTAC-3′.
SA β-gal staining was performed as described previously (Dimri et al., 1995). Microscopic images were obtained at RT by using an inverted microscope (AxioVert 135; Carl Zeiss, Inc.) connected with an AxioCamMRc5 CCD camera through an Achrostigmat 10× NA 0.25 Ph1 objective using AxioVision software. SA heterochromatic foci analysis was performed using DAPI as described previously (Narita et al., 2003). Microscopic images were obtained at RT by using an AxioImager.A1 fluorescence microscope connected with an AxioCamMRc5 CCD camera through an EC Plan-Neofluar 63× NA 1.25 oil Ph3 objective using AxioVision software.
Western blot analysis
Immunoblotting was performed as previously described (Takahashi et al., 2006). Primary antibodies used were human p16 (EMD), Ras (EMD), pRb (BD), vinculin (hVIN-1; Sigma-Aldrich), mouse p16 (Santa Cruz Biotechnology, Inc.), DNMT1 (Santa Cruz Biotechnology, Inc.), H3K9me2 (Abcam), phospho-MEK1/2 (Ser217/221; Cell Signaling Technology), MEK1/2 (Cell Signaling Technology), β-actin (AC-74; Sigma-Aldrich), and phospho-p53 (Cell Signaling Technology).
Bisulfite sequencing analysis
Bisulfite sequencing analysis was performed as described previously (Suzuki et al., 2006).
Chromatin immunoprecipitation (ChIP) analysis
ChIP analysis was performed using EZ-ChIP (Millipore). The antibodies used were anti-H3K9me2 (Abcam), anti–RNA polymerase II (Millipore), and anti-IgG (Millipore) as a negative control. The immunoprecipitated DNA was quantified by real-time quantitative PCR as described in Real-time RT-PCR. The sequences of the PCR primers were described previously (Bracken et al., 2007).
Online supplemental material
Fig. S1 shows the analysis of transgene integrity and expression of the endogenous p16Ink4a gene in nontransgenic mice. Fig. S2 shows ex vivo images of luciferase expression in tissues of p16-luc mice. Fig. S3 shows detection of the H-ras mutation at codon 61 (A→T). Fig. S4 shows the effects of oncogenic Ras or DNMT1 knockdown in HDFs. Fig. S5 shows that DDR causes a reduction of DNMT1 level. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904105/DC1.
Acknowledgments
We thank Drs. N.E. Sharpless for p16Ink4a knockout mice, R. Agami and R. Bernards (Netherland Cancer Institute, Amsterdam, Netherlands) for RNAi vector, and M. Serrano (Spanish National Cancer Research Center, Madrid, Spain) for retrovirus vector encoding H-RasV12. We also thank Dr. Y. Shinkai (Kyoto University, Kyoto, Japan) for valuable suggestions for the analysis of H3K9me2 and to Ms. C. Sugita (Japanese Foundation for Cancer Research, Koto-ku, Tokyo, Japan) and Ms. S. Chiba (University of Tokushima, Kuramoto-cho, Tokushima, Japan) for their assistance in mouse experiments.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Cell Science Research Foundation, the Mitsubishi Foundation, the Naito Foundation, the Princess Takamatsu Cancer Research Fund, the Takeda Science Foundation, and the Vehicle Racing Commemorative Foundation.
Footnotes
Abbreviations used in this paper: BAC, bacterial artificial chromosome; β-gal, β-galactosidase; BLI, bioluminescence imaging; CCD, charge-coupled device; ChIP, chromatin immunoprecipitation; DDR, DNA damage response; DXR, doxorubicin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; H3K9, histone 3 Lys9; H3K9me2, H3K9 dimethylation; HDF, human diploid fibroblast; LN, lymph node; MEF, mouse embryonic fibroblast; pRb, retinoblastoma tumor suppressor protein; ROS, reactive oxygen species; SA, senescence associated; shRNA, short hairpin RNA.
References
- Agger K., Cloos P.A., Rudkjaer L., Williams K., Andersen G., Christensen J., Helin K. 2009. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence.Genes Dev. 23:1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman K.E., Park B.H., Rhee I., Rajagopalan H., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene.Cancer Cell. 3:89–95 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Perez-Terzic C., Jin F., Pitel K., Niederländer N.J., Jeganathan K., Yamada S., Reyes S., Rowe L., Hiddinga H.J., et al. 2008. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency.Nat. Cell Biol. 10:825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester M., Castelló A., Ibáñez E., Sánchez A., Folch J.M. 2004. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals.Biotechniques. 37:610–613 [DOI] [PubMed] [Google Scholar]
- Barradas M., Anderton E., Acosta J.C., Li S., Banito A., Rodriguez-Niedenführ M., Maertens G., Banck M., Zhou M.M., Walsh M.J., et al. 2009. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS.Genes Dev. 23:1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints.Nature. 444:633–637 [DOI] [PubMed] [Google Scholar]
- Benanti J.A., Galloway D.A. 2004. Normal human fibroblasts are resistant to RAS-induced senescence.Mol. Cell. Biol. 24:2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Mönch K., Minucci S., Porse B.T., Marine J.C., et al. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells.Genes Dev. 21:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors.Cell. 120:513–522 [DOI] [PubMed] [Google Scholar]
- Chandler K.J., Chandler R.L., Broeckelmann E.M., Hou Y., Southard-Smith E.M., Mortlock D.P. 2007. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression.Mamm. Genome. 18:693–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre' M., Nuciforo P.G., Bensimon A., et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication.Nature. 444:638–642 [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo.Proc. Natl. Acad. Sci. USA. 92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr., Butel J.S., Bradley A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours.Nature. 356:215–221 [DOI] [PubMed] [Google Scholar]
- Estève P.O., Chin H.G., Smallwood A., Feehery G.R., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S. 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication.Genes Dev. 20:3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T., Holbrook N.J. 2000. Oxidants, oxidative stress and the biology of ageing.Nature. 408:239–247 [DOI] [PubMed] [Google Scholar]
- Gil J., Peters G. 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all.Nat. Rev. Mol. Cell Biol. 7:667–677 [DOI] [PubMed] [Google Scholar]
- Gonzalez S., Klatt P., Delgado S., Conde E., Lopez-Rios F., Sanchez-Cespedes M., Mendez J., Antequera F., Serrano M. 2006. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus.Nature. 440:702–706 [DOI] [PubMed] [Google Scholar]
- Hara E., Smith R., Parry D., Tahara H., Stone S., Peters G. 1996. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence.Mol. Cell. Biol. 16:859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince T.A., Richardson A.L., Bell G.W., Saitoh M., Godar S., Karnoub A.E., Iglehart J.D., Weinberg R.A. 2007. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes.Cancer Cell. 12:160–170 [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom K., Marino S., DePinho R.A., van Lohuizen M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus.Nature. 397:164–168 [DOI] [PubMed] [Google Scholar]
- Kemp C.J. 2005. Multistep skin cancer in mice as a model to study the evolution of cancer cells.Semin. Cancer Biol. 15:460–473 [DOI] [PubMed] [Google Scholar]
- Kia S.K., Gorski M.M., Giannakopoulos S., Verrijzer C.P. 2008. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus.Mol. Cell. Biol. 28:3457–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Sharpless N.E. 2006. The regulation of INK4/ARF in cancer and aging.Cell. 127:265–275 [DOI] [PubMed] [Google Scholar]
- Kotake Y., Cao R., Viatour P., Sage J., Zhang Y., Xiong Y. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene.Genes Dev. 21:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P., Ijpenberg A., Song J.Y., van der Valk M., Nawijn M., Zevenhoven J., Berns A. 2007. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a.Nature. 448:943–946 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J., Torrice C., Ramsey M.R., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E. 2004. Ink4a/Arf expression is a biomarker of aging.J. Clin. Invest. 114:1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.C., Fenster B.E., Ito H., Takeda K., Bae N.S., Hirai T., Yu Z.X., Ferrans V.J., Howard B.H., Finkel T. 1999. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species.J. Biol. Chem. 274:7936–7940 [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality.Cell. 69:915–926 [DOI] [PubMed] [Google Scholar]
- Lin A.W., Barradas M., Stone J.C., van Aelst L., Serrano M., Lowe S.W. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling.Genes Dev. 12:3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W., Cepero E., Evan G. 2004. Intrinsic tumour suppression.Nature. 432:307–315 [DOI] [PubMed] [Google Scholar]
- MacLeod A.R., Rouleau J., Szyf M. 1995. Regulation of DNA methylation by the Ras signaling pathway.J. Biol. Chem. 270:11327–11337 [DOI] [PubMed] [Google Scholar]
- Macleod K.F. 2008. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system.Nat. Rev. Cancer. 8:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara K., Yamakoshi K., Ohtani N., Kubo Y., Takahashi A., Arase S., Jones N., Hara E. 2005. Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional pRB and p53.J. Cell Biol. 168:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette F.A., Gaumont-Leclerc M.F., Ferbeyre G. 2007. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence.Genes Dev. 21:43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. 2001. To cycle or not to cycle: a critical decision in cancer.Nat. Rev. Cancer. 1:222–231 [DOI] [PubMed] [Google Scholar]
- Matheu A., Maraver A., Collado M., Garcia-Cao I., Cañamero M., Borras C., Flores J.M., Klatt P., Viña J., Serrano M. 2009. Anti-aging activity of the Ink4/Arf locus.Aging Cell. 8:152–161 [DOI] [PubMed] [Google Scholar]
- McCabe M.T., Davis J.N., Day M.L. 2005. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway.Cancer Res. 65:3624–3632 [DOI] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi.Nature. 436:720–724 [DOI] [PubMed] [Google Scholar]
- Narita M., Nũnez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence.Cell. 113:703–716 [DOI] [PubMed] [Google Scholar]
- Nguyen C.T., Weisenberger D.J., Velicescu M., Gonzales F.A., Lin J.C., Liang G., Jones P.A. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine.Cancer Res. 62:6456–6461 [PubMed] [Google Scholar]
- Nielsen G.P., Stemmer-Rachamimov A.O., Shaw J., Roy J.E., Koh J., Louis D.N. 1999. Immunohistochemical survey of p16INK4A expression in normal human adult and infant tissues.Lab. Invest. 79:1137–1143 [PubMed] [Google Scholar]
- Ohtani N., Zebedee Z., Huot T.J., Stinson J.A., Sugimoto M., Ohashi Y., Sharrocks A.D., Peters G., Hara E. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence.Nature. 409:1067–1070 [DOI] [PubMed] [Google Scholar]
- Ohtani N., Brennan P., Gaubatz S., Sanij E., Hertzog P., Wolvetang E., Ghysdael J., Rowe M., Hara E. 2003. Epstein-Barr virus LMP1 blocks p16INK4a–RB pathway by promoting nuclear export of E2F4/5.J. Cell Biol. 162:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N., Imamura Y., Yamakoshi K., Hirota F., Nakayama R., Kubo Y., Ishimaru N., Takahashi A., Hirao A., Shimizu T., et al. 2007. Visualizing the dynamics of p21(Waf1/Cip1) cyclin-dependent kinase inhibitor expression in living animals.Proc. Natl. Acad. Sci. USA. 104:15034–15039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts.Nat. Cell Biol. 5:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Wagner E.F. 2000. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression.EMBO J. 19:2969–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla M., Brown K., Ramsden M., Balmain A. 1986. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis.Nature. 322:78–80 [DOI] [PubMed] [Google Scholar]
- Ramirez R.D., Morales C.P., Herbert B.S., Rohde J.M., Passons C., Shay J.W., Wright W.E. 2001. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions.Genes Dev. 15:398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey M.R., Krishnamurthy J., Pei X.H., Torrice C., Lin W., Carrasco D.R., Ligon K.L., Xiong Y., Sharpless N.E. 2007. Expression of p16Ink4a compensates for p18Ink4c loss in cyclin-dependent kinase 4/6-dependent tumors and tissues.Cancer Res. 67:4732–4741 [DOI] [PubMed] [Google Scholar]
- Riley T., Sontag E., Chen P., Levine A. 2008. Transcriptional control of human p53-regulated genes.Nat. Rev. Mol. Cell Biol. 9:402–412 [DOI] [PubMed] [Google Scholar]
- Robert M.F., Morin S., Beaulieu N., Gauthier F., Chute I.C., Barsalou A., MacLeod A.R. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells.Nat. Genet. 33:61–65 [DOI] [PubMed] [Google Scholar]
- Sarkisian C.J., Keister B.A., Stairs D.B., Boxer R.B., Moody S.E., Chodosh L.A. 2007. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis.Nat. Cell Biol. 9:493–505 [DOI] [PubMed] [Google Scholar]
- Serrano M., Blasco M.A. 2001. Putting the stress on senescence.Curr. Opin. Cell Biol. 13:748–753 [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a.Cell. 88:593–602 [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., DePinho R.A. 2005. Cancer: crime and punishment.Nature. 436:636–637 [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Alson S., Chan S., Silver D.P., Castrillon D.H., DePinho R.A. 2002. p16(INK4a) and p53 deficiency cooperate in tumorigenesis.Cancer Res. 62:2761–2765 [PubMed] [Google Scholar]
- Sharpless N.E., Ramsey M.R., Balasubramanian P., Castrillon D.H., DePinho R.A. 2004. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis.Oncogene. 23:379–385 [DOI] [PubMed] [Google Scholar]
- Shay J.W., Wright W.E. 2007. Tissue culture as a hostile environment: identifying conditions for breast cancer progression studies.Cancer Cell. 12:100–101 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Yamada T., Kihara-Negishi F., Sakurai T., Hara E., Tenen D.G., Hozumi N., Oikawa T. 2006. Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b.Oncogene. 25:2477–2488 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Ohtani N., Yamakoshi K., Iida S., Tahara H., Nakayama K., Nakayama K.I., Ide T., Saya H., Hara E. 2006. Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence.Nat. Cell Biol. 8:1291–1297 [DOI] [PubMed] [Google Scholar]
- Terzian T., Suh Y.A., Iwakuma T., Post S.M., Neumann M., Lang G.A., Van Pelt C.S., Lozano G. 2008. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss.Genes Dev. 22:1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatsos A., Pfau R., Kampranis S.C., Tsichlis P.N. 2009. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus.Proc. Natl. Acad. Sci. USA. 106:2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K.H., Lane D.P. 2007. p53 in health and disease.Nat. Rev. Mol. Cell Biol. 8:275–283 [DOI] [PubMed] [Google Scholar]
- Wiedemeyer R., Brennan C., Heffernan T.P., Xiao Y., Mahoney J., Protopopov A., Zheng H., Bignell G., Furnari F., Cavenee W.K., et al. 2008. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development.Cancer Cell. 13:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witcher M., Emerson B.M. 2009. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary.Mol. Cell. 34:271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Shimizu T., Araki N., Hirota T., Yoshie M., Ogawa K., Nakagata N., Takeya M., Saya H. 2008. Aurora A overexpression induces cellular senescence in mammary gland hyperplastic tumors developed in p53-deficient mice.Oncogene. 27:4305–4314 [DOI] [PubMed] [Google Scholar]
- Zhu J., Woods D., McMahon M., Bishop J.M. 1998. Senescence of human fibroblasts induced by oncogenic Raf.Genes Dev. 12:2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Quelle D.E., Roussel M.F., Sherr C.J. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging.Oncogene. 15:203–211 [DOI] [PubMed] [Google Scholar]