Abstract
Maintenance of endoplasmic reticulum (ER) function is achieved in part through Ire1 (inositol-requiring enzyme 1), a transmembrane protein activated by protein misfolding in the ER. The cytoplasmic nuclease domain of Ire1 cleaves the messenger RNA (mRNA) encoding XBP-1 (X-box–binding protein 1), enabling splicing and production of this active transcription factor. We recently showed that Ire1 activation independently induces the rapid turnover of mRNAs encoding membrane and secreted proteins in Drosophila melanogaster cells through a pathway we call regulated Ire1-dependent decay (RIDD). In this study, we show that mouse fibroblasts expressing wild-type Ire1 but not an Ire1 variant lacking nuclease activity also degrade mRNAs in response to ER stress. Using a second variant of Ire1 that is activated by a small adenosine triphosphate analogue, we show that although XBP-1 splicing can be artificially induced in the absence of ER stress, RIDD appears to require both Ire1 activity and ER stress. Our data suggest that cells use a multitiered mechanism by which different conditions in the ER lead to distinct outputs from Ire1.
Introduction
The ER is responsible for folding and processing proteins entering the secretory pathway and uses a variety of mechanisms to adjust its capacity in response to changes in the folding burden. This collection of mechanisms, termed the unfolded protein response (UPR; Ron and Walter, 2007), is activated by stress caused by environmental stimuli and diseases such as viral infection (Tardif et al., 2004; Bechill et al., 2008) and multiple myeloma (Carrasco et al., 2007). In metazoans, components of the UPR are essential for developmental processes requiring high levels of secretion such as the differentiation of plasma cells (Gass et al., 2002; Iwakoshi et al., 2003) and pancreatic β cells (Harding et al., 2001; Zhang et al., 2002). The UPR restores ER function both by increasing its capacity and decreasing the load of new proteins through transcriptional induction of secretory pathway components and general translational attenuation. One of the key players in the UPR is Ire1 (inositol-requiring enzyme 1), a conserved transmembrane protein with a luminal domain that senses protein misfolding in the ER. The resulting oligomerization of Ire1 leads to activation of its cytoplasmic kinase and endoribonuclease domain. The main function of the kinase appears to be activation of the nuclease, which requires binding of ATP or ADP in the active site of the kinase (Papa et al., 2003). The nuclease in turn cleaves two specific sites in the mRNA encoding XBP-1 (X-box–binding protein 1), a conserved UPR transcription factor, which leads to XBP-1 activation and translation through removal of a regulatory intron.
We have recently shown that Ire1 in Drosophila melanogaster cells independently mediates the cleavage and degradation of mRNAs encoding proteins that traverse the secretory pathway (Hollien and Weissman, 2006). This new branch of the UPR, which we call regulated Ire1-dependent decay (RIDD), has the potential to selectively relieve the burden on the ER while clearing the translation and translocation machinery for the subsequent influx of new proteins induced by the UPR. Studies on the regulation of specific messages suggest that the RIDD pathway also operates in mammalian cells (Tirasophon et al., 2000; Iwawaki et al., 2001; Oikawa et al., 2007; Iqbal et al., 2008; Lipson et al., 2008). Mammals express two isoforms of Ire1: Ire1-α is expressed ubiquitously, and Ire1-β is expressed in intestinal epithelial cells. Overexpression of Ire1-α leads to cleavage of its own message in COS-1 cells (Tirasophon et al., 2000) and reduced levels of the message encoding CD59 (complement defense 59) in HeLa cells (Oikawa et al., 2007). Ire1-α also appears to mediate the degradation of insulin transcripts in pancreatic β cells under chronic high glucose conditions, perhaps promoting cell survival during extreme chronic stress (Lipson et al., 2008). Ire1-β appears to have alternative targets as well, mediating cleavage of the 28-S ribosomal RNA (Iwawaki et al., 2001) and the mRNA encoding microsomal triglyceride transfer protein, an ER chaperone important for the assembly of lipid transport vesicles (Iqbal et al., 2008). Together, these examples of Ire1 function in mRNA decay may explain observations that Ire1 in metazoans has a broader range of physiological outputs than XBP-1 splicing (Zhang et al., 2005).
In this study, we take advantage of mouse fibroblasts lacking Ire1 activity, both to confirm that RIDD is conserved in mammalian cells and to investigate the functional requirements of RIDD. By expressing wild-type and mutant variants of Ire1-α, we find that the nuclease activity of Ire1 is required for both splicing and RIDD. However, these two outputs can be differentially triggered, revealing an unexpected complexity in Ire1 activation.
Results and discussion
Ire1 degrades mRNAs in response to ER stress in mouse fibroblasts
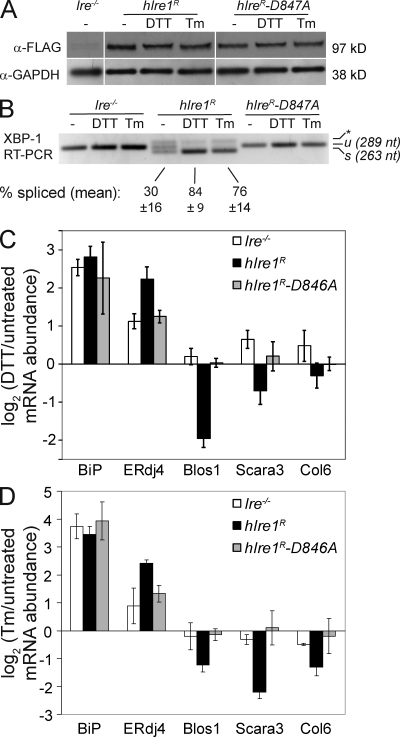
To determine whether the RIDD pathway functions in mammals as well as in Drosophila, we used a mouse embryonic fibroblast (MEF) cell line established by Lee et al. (2002) in which the Ire1-α gene has been disrupted. We stably introduced human Ire1-α (hIre1) into these cells using flippase-mediated, site-directed recombination (Cohen and Panning, 2007), which allowed us to insert the wild-type and hIre1 variants discussed in the next sections into the same sites within the genome. In response to various forms of chemically induced ER stress, the reconstituted cells (referred to here as hIre1R) but not the Ire1 knockout (Ire1−/−) cells induced the splicing of XBP-1 (see Fig. 3 B). Both cell lines strongly induced immunoglobin-binding protein (BiP), an abundant ER chaperone, in response to ER stress (Fig. 1), which is in agreement with previous observations that BiP induction is Ire1 independent in mouse fibroblasts (Lee et al., 2002). As expected, induction of ERdj4, a transcriptional target of the Ire1/XBP-1 branch of the UPR (Lee et al., 2003), was stronger in the hIre1R cells compared with Ire1−/− cells, especially in response to DTT, a reducing agent which disrupts disulfide bond formation in the ER (Fig. 1 A).
Figure 3.
The nuclease activity of Ire1 is required for RIDD. (A) Western blot of Flag-tagged hIre1 proteins. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (B) Ethidium bromide–stained agarose gel showing XBP-1 reverse transcribed from total RNA and PCR amplified using primers surrounding the splice site. The asterisk denotes a hybrid band amplified from spliced and unspliced (u) products (Shang and Lehrman, 2004). s, spliced product after the removal of the 26 nucleotide intron. Bands were quantified in ImageJ (National Institutes of Health) to determine the percentage of splicing. (C and D) RNA levels of RIDD targets and controls in DTT (C)- and Tm (D)-treated cells measured by qPCR and normalized to signals from Rpl19. (B–D) The means and SDs for three independent experiments are shown.
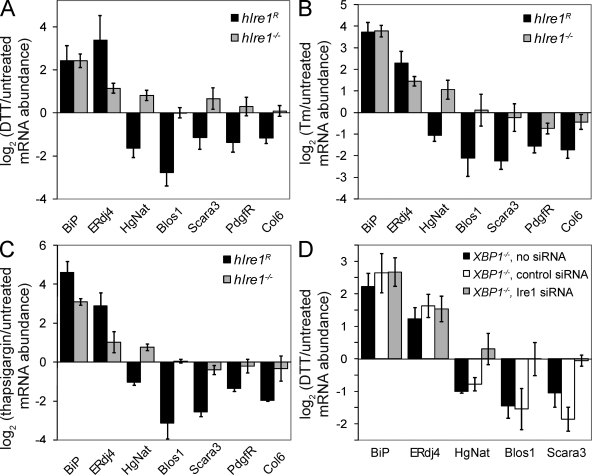
Figure 1.
mRNAs are down-regulated in response to multiple stressors in an Ire1-dependent manner. (A–C) mRNA abundance for targets of ER stress in hIre1R and Ire1−/− cells treated with 2 mM DTT (A), 3 µg/ml Tm (B), or 500 nM thapsigargin (C) for 5 h relative to untreated cells. (D) Relative mRNA abundance in cells lacking functional XBP-1. Cells were untransfected or transfected with negative control siRNA (QIAGEN) or siRNAs targeting Ire1, followed by DTT treatment as in A. (A–D) The means and SDs for three to four independent experiments were measured by qPCR; each sample was normalized using the mRNA levels of Rpl19.
To investigate the RIDD pathway, we took an unbiased, microarray-based approach. We induced ER stress in the Ire1−/− and hIre1R cells with DTT, purified total RNA from treated and untreated cells, amplified and labeled these samples, and hybridized them to whole-genome MEEBOChip arrays. As expected, treatment of our hIre1R cells with DTT led to the induction of classical UPR targets to levels comparable with that seen by others in wild-type cells (Fig. S1; Lee et al., 2003).
Consistent with the RIDD pathway functioning in these cells, we observed that several RNAs were down-regulated in response to ER stress in the hIre1R but not the Ire1−/− cells (Fig. 1 and Fig. S1). Although ∼120 mRNAs fell into this category in the array data, the magnitudes of the changes in expression were generally small (many were twofold or less) compared with those seen in Drosophila cells, where many RNAs were down-regulated by 5–10-fold. Despite the relatively small changes for individual messages, the effect of down-regulating mRNAs at the ER surface may profoundly impact the folding burden of the ER. For example, the redistribution of certain nascent proteins from the ER to the cytosol during ER stress significantly impacts cell survival, although the effect on translocation is similar in magnitude to RIDD (Kang et al., 2006).
To limit false positives, we applied a series of strict criteria for identifying the most likely RIDD targets (see Materials and methods); the results are shown in Table I. We confirmed the regulation of several of these targets by reverse transcription followed by quantitative real-time PCR (qPCR; Fig. 1 and Fig. S1). To determine whether the down-regulation was a general response to ER stress or was restricted to DTT, we also treated cells with tunicamycin (Tm), which inhibits N-linked glycosylation or thapsigargin, which disrupts calcium homeostasis. All candidate RIDD targets tested were down-regulated in response to these three forms of ER stress in an Ire1-dependent manner (Fig. 1, A–C).
Table I.
Candidate targets of the RIDD pathway in mouse cells
| Target name | UniGene ID | Localization | Mean log2 (DTT/untreated) | Decreased stability in DTT | |
| hIre1R | Ire1−/− | ||||
| Heparan-α-glucosaminide N-acetyltransferase (Hgsnat) | Mm.28326 | Membrane (lysos.) | −1.34 | 0.45 | Yes |
| Biogenesis of lysosome-related organelles complex-1, subunit 1 (Blos1) | Mm.30118 | Cytosol | −1.22 | −0.10 | Yes |
| Scavenger receptor class A, member 3 (Scara3) | Mm.344095 | Membrane (ER/Golgi) | −1.11 | 0.17 | Yes |
| PDGF receptor (Pdgfrb) | Mm.4146 | Membrane (PM) | −0.89 | 0.12 | Yes |
| Peripheral myelin protein (Pmp22) | Mm.1237 | ECM | −0.88 | −0.07 | Yes |
| Collagen, type VI, α1 (Col6) | Mm.2509 | ECM | −0.85 | −0.15 | Yes |
| Ephrin B2 | Mm.209813 | Membrane (PM) | −1.06 | −0.14 | No |
| Microtubule-associated protein 7 domain containing 1 (MAP7d1) | Mm.266716 | NA | −0.69 | 0.10 | No |
| Splicing factor, Arg/Ser rich 3 (SRp20) | Mm.6787 | Spliceosome | −0.89 | −0.14 | No |
| Tripartite motif protein 16 (Trim16) | Mm.117087 | Cytosol | −0.73 | 0.12 | No |
| Galnt10 | Mm.271670 | Membrane (Golgi) | −0.59 | 0.14 | Yes |
| Laminin B1 subunit 1 | Mm.172674 | ECM | −1.40 | 0.08 | ND |
| Mannose receptor C type 2 | Mm.235616 | Membrane (PM) | −1.14 | −0.17 | ND |
| Leu-rich repeat containing 8C | Mm.319847 | Membrane | −0.99 | −0.17 | ND |
| Neural cell adhesion molecule 1 | Mm.4974 | Membrane (PM) | −0.88 | 0.14 | ND |
| PDGFA-associated protein 1 | Mm.188851 | ECM | −0.88 | −0.21 | ND |
| Syndecan 4 | Mm.3815 | Membrane (PM) | −0.85 | −0.18 | ND |
| Homeo box B4 | Mm.3546 | Nucleus | −0.84 | −0.03 | ND |
| Expressed sequence C79267 | Mm.30464 | NA | −0.77 | −0.12 | ND |
| 3-oxoacid CoA transferase 1 | Mm.13445 | ECM | −0.77 | −0.02 | ND |
| Tripeptidyl peptidase I | Mm.20837 | ECM, mitoch., lysos. | −0.74 | −0.12 | ND |
| RIKEN cDND 4932417H02 | Mm.209933 | NA | −0.71 | 0.21 | ND |
| Pre–B cell leukemia transcription factor–interacting protein 1 | Mm.65906 | NA | −0.69 | 0.33 | ND |
| Chondroitin sulfate proteoglycan 4 (CSP4) | Mm.41329 | Membrane (PM) | −0.68 | 0.28 | ND |
| SHANK-associated RH domain–interacting protein | Mm.41463 | Cytosol | −0.67 | 0.41 | ND |
| ST3 β-galactoside α-2,3-sialyltransferase 5s | Mm.38248 | Membrane (Golgi) | −0.62 | 0.17 | ND |
lysos., lysosomal; mitoch., mitochondria; NA, not applicable (no annotation available); PM, plasma membrane. Localization data are based on Gene Ontology annotations of the mouse genome. Log2 (DTT/untreated) RNA levels were determined by microarray analysis and represent the mean of three independent experiments. Stability data are shown in Fig. S2.
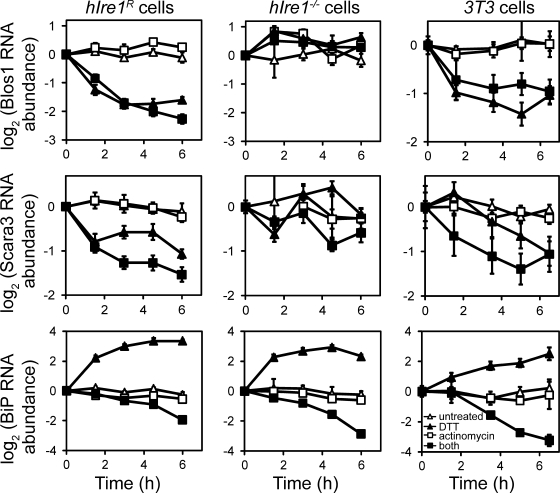
Several observations indicate that the RIDD pathway, if not the specific targets, is conserved in mammalian cells and Drosophila. First, the down-regulation of mouse target mRNAs was independent of XBP-1 (Fig. 1 D). Using RNAi to deplete Ire1 from cells lacking a functional XBP-1 (Lee et al., 2003), we found that Ire1-dependent down-regulation of several target mRNAs occurred in the absence of XBP-1. As a control, in cells lacking XBP-1, ERdj4 was induced only to the level seen in Ire1−/− cells, and this induction was insensitive to Ire1 depletion (Fig. 1 D). Second, the down-regulation of many mouse targets was achieved through an increase in their decay rates. We inhibited transcription with actinomycin D and monitored mRNA levels over time in the absence and presence of ER stress (Fig. 2 and Fig. S2). For 7 of the 11 targets tested, the decreases in mRNA abundance were dependent not on transcription but on increased rates of mRNA degradation. Third, as in Drosophila cells, the set of targets in mouse cells is highly enriched for mRNAs encoding membrane proteins. 17 of the 22 targets in Table I that were assigned localization annotations in the mouse genome encode membrane or ECM proteins, as do 6 of the 7 confirmed targets that displayed increased decay rates. The seventh confirmed target, Blos1, encodes a protein that does not appear to contain a membrane-spanning domain itself but is part of a complex detected in both cytosolic and peripheral membrane fractions (Starcevic and Dell'Angelica, 2004).
Figure 2.
mRNAs down-regulated by Ire1 are degraded faster in the presence of ER stress. hIre1R, Ire1−/−, or NIH-3T3 cells were treated with 1 µg/ml actinomycin D and/or 2 mM DTT, and relative mRNA abundance was monitored over time by qPCR. RNA levels were normalized to those of Rpl19 and to untreated controls. Error bars represent the SDs in qPCR replicates; representative time course data from two independent experiments are shown.
We consistently observed partial splicing of XBP-1 in our hIre1R cells in the absence of ER stress, which is likely caused by overexpression of hIre1 in these cells. To confirm that the mRNA decay we observed was not an artificial product of additional stress or Ire1 activity caused by overexpression, we measured the mRNA decay rates of two confirmed RIDD targets in NIH-3T3 cells (Fig. 2). As in our reconstituted cells, NIH-3T3 cells displayed increased decay rates and lower abundance for RIDD targets in the presence of DTT. Collectively, our data indicate that RIDD is a general, conserved pathway associated with folding stress in the ER.
The nuclease activity of Ire1 is required for RIDD
Because the parent Ire1−/− cell line lacks Ire1 activity, we could probe specific functions of Ire1 by inserting variants of hIre1 containing point mutations into these cells in an isogenic manner. Using this approach, we expressed an hIre1 variant containing a single point mutation (D847A) that has been previously shown to compromise the nuclease activity of hIre1 while leaving the kinase activity intact (Tirasophon et al., 2000). For these experiments, we used C-terminally Flag-tagged versions of both the wild-type and mutant Ire1 and confirmed by Western blotting that the hIre-D847A variant was expressed at similar levels to the wild type (Fig. 3 A). Consistent with a lack of nuclease activity, cells expressing hIre1-D847A did not support XBP-1 splicing in the absence or presence of ER stress (Fig. 3 B) and induced ERdj4 only to the level seen in Ire1−/− cells (Fig. 3, C and D). All cell lines induced BiP by similar amounts, indicating that they are experiencing a similar level of ER stress. However, unlike the Ire1R cells, cells expressing hIre1-D847A displayed no change in the abundance of RIDD targets upon induction of ER stress (Fig. 3, C and D). These results indicate that the nuclease activity of Ire1 is required for degradation of RIDD targets, which is consistent with a mechanism in which Ire1 directly cleaves these RNAs in response to ER stress.
Bypassing the kinase activity of Ire1
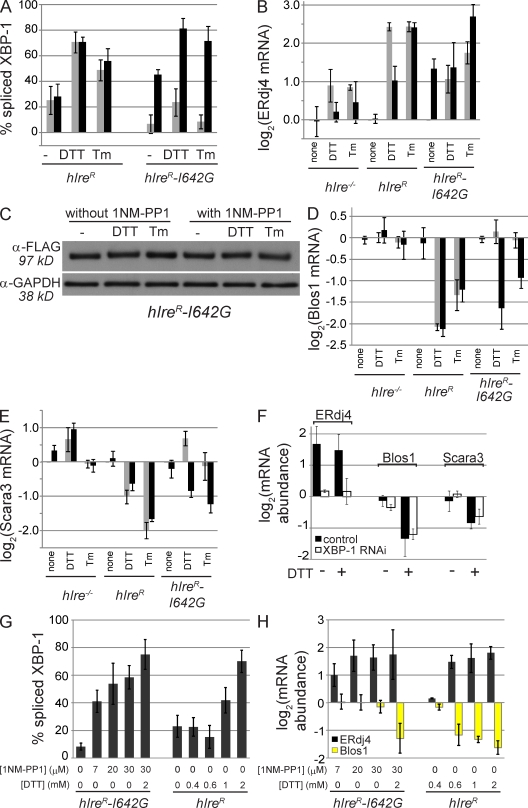
Recently, it has been shown that for yeast and human Ire1, the cytoplasmic kinase activity can be bypassed using a small molecule that binds to the enlarged ATP pocket of an engineered Ire1 variant (Papa et al., 2003; Lin et al., 2007). This variant, I642G in hIre1, has very little activity in the absence of the ATP analogue 1NM-PP1 (4-amino-1-tert-butyl-3-[1'-naphthylmethyl]pyrazolo[3,4-d] pyrimidine). Binding allows activation of the nuclease while simultaneously inhibiting the kinase activity of Ire1-I642G. In HEK293 cells (Lin et al., 2007) and MEF cells (Han et al., 2008), hIre1-I642G splices XBP-1 and induces the UPR upon binding 1NM-PP1 alone, even in the absence of ER stress. To test whether activation of Ire1 is sufficient for degradation of RIDD substrates, we introduced hIre1-I642G into Ire1−/− cells and tested its activity in the absence and presence of 1NM-PP1 and ER stress (Fig. 4). Treatment of mouse cells expressing hIre1-I642G with 1NM-PP1 was sufficient to induce splicing of XBP-1 (Fig. 4 A) and transcriptional induction of the Ire1/XBP-1 target ERdj4 (Fig. 4 B). This Ire1 activity was not caused by a general induction of ER stress, as cells lacking Ire1 or expressing the wild-type hIre1 were insensitive to 1NM-PP1 (Fig. 4), although the drug did suppress the Ire1-independent induction of ERdj4 by DTT in all strains through an unknown mechanism (Fig. 4 B). The 1NM-PP1–induced activation of hIre1-I642G was not caused by increased levels of the protein, as assayed by Western blotting (Fig. 4 C). Thus, our data are consistent with a 1NM-PP1–induced conformational change in the Ire1 variant itself, as proposed previously (Papa et al., 2003).
Figure 4.
Activation of Ire1 is not sufficient for RIDD. (A) Percentage of spliced XBP-1 in the absence (gray bars) and presence (black bars) of 7 µM 1NM-PP1 in the indicated cell lines. Splicing was measured as in Fig. 3 B. (B) Relative RNA abundance of ERdj4, measured by qPCR, in the absence (gray bars) and presence (black bars) of 1NM-PP1. RNA abundance was normalized to that of Rpl19, and ERdj4/Rpl19 ratios for experimental samples were divided by those for untreated cells from the same experiment. (C) Western blot of hIre-I642G–Flag. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (D and E) Relative RNA abundance of two RIDD targets, Blos1 (D) and Scara3 (E), measured as in B. Gray bars, no 1NM-PP1; black bars, 7 µM 1NM-PP1. (F) Relative RNA abundance in hIre1R-I642G cells measured as in B. Control cells (shaded bars) and cells depleted of XBP-1 using RNAi (open bars) were treated with 1NM-PP1 only or 1NM-PP1 and DTT. (G and H) Percentage of spliced XBP-1 (G) and relative mRNA abundance for ERdj4 and Blos1 (H) in various concentrations of 1NM-PP1 and/or DTT. Measurements are as described for A and B. (A–H) Cells were treated with 2 mM DTT, 3 µg/ml Tm, and/or 7 µM 1NM-PP1 for 5 h unless otherwise indicated. The means and SD for three to five independent experiments are shown.
In contrast to XBP-1 splicing and ERdj4 induction, no degradation of RIDD substrates was observed with 1NM-PP1 alone, indicating that XBP-1 splicing and RIDD are separable functions of Ire1 and have distinct requirements for activation (Fig. 4, D and E). Treatment of cells expressing Ire1-I642G with DTT or Tm alone (in the absence of 1NM-PP1) had very little effect on splicing or RIDD targets. However, cells treated with both 1NM-PP1 and ER stress degraded RIDD targets to similar levels as those expressing the wild-type Ire1 (Fig. 4, D and E). As a further control, we confirmed the XBP-1 independence of RIDD in these cells. XBP-1 is transcriptionally induced in our cells in a largely Ire1-independent manner and therefore does not occur to a significant extent in cells treated with 1NM-PP1 alone. Therefore, we depleted XBP-1 from cells expressing Ire1-I642G using RNAi, and although this blocked induction of ERdj4 by DTT, RIDD remained intact when cells were treated with both 1NM-PP1 and DTT (Fig. 4 F).
To determine whether the lack of RIDD in our 1NM-PP1–induced cells is merely a threshold effect, we treated cells with higher concentrations of 1NM-PP1, such that no further XBP-1 splicing or ERdj4 activation could be achieved with 1NM-PP1 alone (Fig. 4, G and H). The levels of XBP-1 splicing in cells expressing hIre1-I642G treated with 30 µM 1NM-PP1 were as high or higher than those seen in wild-type cells treated with low concentrations of DTT (Fig. 4 G) or with Tm (Fig. 4 A), where RIDD was clearly functioning. Note that although XBP-1 splicing appears insensitive to low concentrations of DTT in Fig. 4 G, we observed partial splicing shortly after the addition of DTT followed by rapid recovery before changes in gene expression could be reliably measured. For cells expressing wild-type hIre1, ERdj4 and RIDD appeared to be activated in parallel by low DTT concentrations, suggesting that RIDD is not limited to extreme ER stress (Fig. 4 H). In contrast to splicing and activation of ERdj4, no change in mRNA abundance for RIDD targets was detected in cells expressing hIre1-I642G and treated with 1NM-PP1 alone, even at high concentrations, although the cells were still capable of inducing RIDD when treated with 1NM-PP1 and DTT together (Fig. 4 H). In summary, although XBP-1 splicing and downstream transcriptional induction can be artificially induced with 1NM-PP1, RIDD cannot.
Implications for the mechanism of RIDD
Our results indicate that in mammalian cells, RIDD requires an active Ire1 nuclease domain but does not, per se, depend on Ire1's kinase activity. ER stress–dependent degradation of RNAs by hIre1-I642G was seen at concentrations of 1NM-PP1 ∼1.5-fold higher than those that inhibited the kinase activity of the protein in vitro (Papa et al., 2003) and sixfold higher than those that inhibited kinase activity in HEK293 cells (Lin et al., 2007). These data, together with the observation that RIDD does not require new transcription, are consistent with a direct mechanism of mRNA target cleavage by Ire1, although we cannot rule out an alternative role for Ire1's nuclease activity and/or the involvement of other nucleases.
However, our data also indicate that activation of Ire1's nuclease is not sufficient for RIDD. This observation represents a mechanistic divergence between RIDD and Ire's well-established role in XBP-1 splicing, which is induced by 1NM-PP1 activation of hIre1-I642G. There are several potential explanations for this. It may be that Ire1 assumes a distinct conformation or oligomerization state when activated by 1NM-PP1 versus ER stress and that although the former is sufficient for splicing XBP-1, the latter is required for RIDD. For example, it was recently found that yeast Ire1 forms higher order oligomers that lead to higher levels of RNase activity (Aragon et al., 2009; Korennykh et al., 2009). Such oligomers may form only in the presence of misfolded proteins or, conversely, in the absence of binding to BiP, an ER chaperone which binds Ire1 and is titrated off by misfolded proteins during ER stress. A second possibility is that other factors activated by ER stress are necessary for RIDD. Although new transcription of such a factor is not required (Fig. 2), there may be proteins recruited or activated by ER stress that are important for mRNA decay but not for splicing. Finally, the complement of mRNAs available for targeting to this decay pathway may change upon subjecting cells to ER stress. Certain signal sequences are sensitive to the ER folding environment and influence how associated ribosomes partition between the ER and cytoplasm (Kang et al., 2006); thus, a substantial change in the pool of mRNAs associated with the ER membrane under stress conditions could affect the RIDD pathway.
The fact that the two outputs of Ire1's nuclease activity, RIDD and XBP-1 splicing, can be differentially activated reveals an unanticipated complexity in the UPR. Growing evidence suggests that various sensors associated with the UPR are activated under different conditions, allowing for specific responses to different forms of ER stress in different cell types (Gass et al., 2002; DuRose et al., 2006). This customization of responses may extend to activation within Ire1 itself. Activation of XBP-1 leads to a protective remodeling and expansion of the secretory pathway, whereas RIDD reduces the load of incoming proteins. Under certain physiological situations, e.g., during plasma cell development, activating the XBP-1–dependent remodeling pathway without inducing RIDD may be more beneficial; thus, cells may induce an active state of Ire1 that is similar to the 1NM-PP1–mediated activation described in this study. In contrast, conditions such as viral infection may call for a more destructive response that limits the load of incoming proteins, which is analogous to the effects of reduced translation mediated by the PERK (PKR-like ER kinase) branch of the UPR (Harding et al., 2000). Intriguingly, both hepatitis C virus and human cytomegalovirus induce Ire1 but appear to block downstream effects of XBP-1 (Tardif et al., 2004; Isler et al., 2005). However, hepatitis C virus protein production is increased in the absence of Ire1 (Tardif et al., 2004), suggesting that an alternate activity of Ire1 such as RIDD may be attenuating viral protein synthesis. It remains to be determined what role RIDD may play in viral infection and other physiological stress conditions, but the ability of this pathway to function in mammalian cells and the potential to decouple it from XBP-1 splicing could allow for a more specific and effective response to changing conditions within the ER.
Materials and methods
Establishment of Ire1-expressing cell lines and RNAi
Ire1-α−/− MEFs were obtained from R. Kaufman (University of Michigan Medical Center, Ann Arbor, MI), and XBP1−/− MEFs were obtained from L. Glimcher (Harvard School of Public Health, Boston, MA). All cells were maintained at 37°C and 5% CO2 in DME supplemented with 10% fetal calf serum, Gln, and antibiotics. We generated Ire1-α−/− MEFs containing ferritin-like protein recombination target (FRT) sites for site-specific transgene expression by transfection with pFRTlacZeo (Invitrogen), and stable clonal integrants were selected with Zeocin (Cohen and Panning, 2007). We then transfected the Ire1-α−/− FRT cells with the pOG44 ferritin-like protein recombinase vector (Invitrogen) and FRT vectors containing hIre1 under the control of the human ER-1α promoter (Lin et al., 2007). QuikChange mutagenesis (Agilent Technologies) was used to make the point mutations in the hIre1 coding sequence. Multiple independent isogenic clones were analyzed for transgene mRNA and protein expression with identical findings. The ATP analogue 1NM-PP1 was a gift from C. Zhang and K. Shokat (University of California, San Francisco, San Francisco, CA; Bishop et al., 2000).
To deplete cells of Ire1 or XBP-1 by RNAi, we transfected ∼3 × 104 cells with 150 ng total of a mixture of four different siRNAs (QIAGEN). After 48 h, we treated cells with DTT and/or 1NM-PP1, and purified RNA as described in the next section. We typically attained 90% knockdown as measured by qPCR.
Microarray experiments and analysis
We passaged and plated Ire1−/− and hIre1R cells into 150-cm2 flasks for collection of microarray samples. After a 3-d growth (at ∼70% confluency), we either left cells untreated or induced ER stress by adding 2 mM DTT for 6 h. We then harvested the cells and purified total RNA using TRIZOL (Invitrogen). To generate labeled samples for array hybridization, we amplified 0.5 µg RNA in a single round using the amino allyl MessageAmp II aRNA kit (Applied Biosystems). As a reference for the array hybridizations, we also amplified and labeled a pool of RNA samples from NIH-3T3, Ire1−/−, and hIre1R cells (both untreated and DTT treated). In parallel, we synthesized unlabeled cDNA from 2 µg of total RNA for qPCR measurements to confirm the array data. We repeated the entire experiment for a total of three times.
We hybridized 5 µg each of a Cy5-labeled experimental sample and the Cy3-labeled reference pool to whole-genome mouse arrays at 65°C for 48 h. The arrays were produced in-house using the MEEBOChip platform. We extracted and processed image data using GenePix 6 (MDS Analytical Technologies), normalized each array to achieve a mean Cy5/Cy3 ratio of 1.0, and removed spots containing low signal or poor signal uniformity. To simplify the analysis, we disregarded spots for which one or more samples did not pass these quality control measures and for which the SD of the three measurements was >30% of the mean. For the remaining spots (representing ∼25% of the total spots on the array), we calculated the log2 ratio of DTT treated/untreated for each of the three replicates in the two cells lines. We then performed hierarchical clustering of the averaged data (Fig. S1).
To select robust RIDD candidates, we applied the following criteria to the 122 spots in clusters displaying Ire1-dependent down-regulation. First, we required the candidate RNA to be down-regulated by 1.5-fold (log2 [DTT/untreated signal] ≤ −0.58) in at least two of the three replicates and in the mean of the three replicates. Second, to select those RNAs whose down-regulation was truly Ire1 dependent, we required that the targets be down-regulated 1.5-fold more in the hIre1R cells compared with the Ire1−/− cells and display no more than a 25% decrease in signal in response to ER stress in the Ire1−/− cells. Lastly, to rule out artifacts caused by higher expression of the candidate RNAs in the hIre1R cells in the absence of ER stress, we also required that the mean signal intensity in the presence of ER stress was lower (by 15%) in the hIre1R cells compared with the Ire1−/− cells. 26 RIDD candidates fit these criteria; these are listed in Table I.
qPCR and XBP-1–splicing assays
We purified RNA samples for all experiments using TRIZOL and synthesized cDNA from total RNA samples using Superscript II (Invitrogen). We then performed qPCR measurements using the primers shown in Table S1. We measured each sample in triplicate using the Opticon (Bio-Rad Laboratories) or Realplex (Eppendorf) qPCR machines and normalized them using the signal from Rpl19, which did not change significantly relative to total RNA input concentration in any of the treatments used. We used mock cDNA samples containing no reverse transcription to ensure that the qPCR signals arose from cDNA and not from contaminating genomic DNA or other sources. To quantify the amount of XBP-1 splicing in each experiment, we amplified cDNA using primers surrounding the splice site (Table S1) and ran products on 2% agarose gels.
Western blotting
We washed cells in PBS and lysed them in radioimmunoprecipitation assay buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% NP-40, 1% Na-deoxycholate, and 0.1% SDS) with added protease inhibitors. We resolved proteins on NuPage Bis-Tris 4–12% acrylamide gels (Invitrogen), transferred them to nitrocellulose membranes, and probed blots using primary polyclonal anti-Flag antibodies (Sigma-Aldrich) at a 1:2,000 dilution followed by secondary peroxidase-conjugated anti–rabbit antibodies (Jackson ImmunoResearch Laboratories) at a 1:5,000 dilution. As a loading control, we also probed blots with polyclonal anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase; ProSci, Inc.). We visualized the immunoblots using a chemiluminescent assay (ECL; GE Healthcare).
Online supplemental material
Fig. S1 shows the array data for hIre1−/− and hIre1R cell lines, a comparison with published data for UPR targets, and confirmation of RIDD target regulation measured by qPCR. Fig. S2 shows decay rate measurements for 11 RIDD candidates in the presence and absence of DTT. Table S1 displays the sequences for primers used in qPCR measurements. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903014/DC1.
Acknowledgments
We thank Samantha Cooper and Charlie Kim for technical advice regarding the MEEBO arrays, Chao Zhang and Kevan Shokat for providing 1NM-PP1, Randal Kaufman for the Ire1−/− cells, and Laurie Glimcher for the XBP-1−/− cells. We also thank Dale Cameron, Vladimir Denic, Mike Bassik, and members of the Weissman and Walter laboratories for discussion.
This work was supported by funds from the National Institutes of Health and Howard Hughes Medical Institute.
Note added in proof. Ire1 but not Ire1-I642G activated by 1NM-PP1 was also recently found to degrade ER-localized mRNAs and promote apoptosis in INS-1 cells (Han et al. 2009. Cell. doi:10.1016/j.cell.2009.07.017).
Footnotes
Abbreviations used in this paper: BiP, immunoglobin-binding protein; FRT, ferritin-like protein recombination target; hIre1, human Ire1-α; MEF, mouse embryonic fibroblast; qPCR, quantitative real-time PCR; RIDD, regulated Ire1-dependent decay; Tm, tunicamycin; UPR, unfolded protein response.
References
- Aragon T., van Anken E., Pincus D., Serafimova I.M., Korennykh A.V., Rubio C.A., Walter P. 2009. Messenger RNA targeting to endoplasmic reticulum stress signalling sites.Nature. 457:736–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill J., Chen Z., Brewer J.W., Baker S.C. 2008. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression.J. Virol. 82:4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase.Nature. 407:395–401 [DOI] [PubMed] [Google Scholar]
- Carrasco D.R., Sukhdeo K., Protopopova M., Sinha R., Enos M., Carrasco D.E., Zheng M., Mani M., Henderson J., Pinkus G.S., et al. 2007. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis.Cancer Cell. 11:349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H.R., Panning B. 2007. XIST RNA exhibits nuclear retention and exhibits reduced association with the export factor TAP/NXF1.Chromosoma. 116:373–383 [DOI] [PubMed] [Google Scholar]
- DuRose J.B., Tam A.B., Niwa M. 2006. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress.Mol. Biol. Cell. 17:3095–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J.N., Gifford N.M., Brewer J.W. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells.J. Biol. Chem. 277:49047–49054 [DOI] [PubMed] [Google Scholar]
- Han D., Upton J.-P., Hagen A., Callahan J., Oakes S.A., Papa F.R. 2008. A kinase inhibitor activates the IRE1alpha RNase to confer cytoprotection against ER stress.Biochem. Biophys. Res. Commun. 365:777–783 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response.Mol. Cell. 5:897–904 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D.D., Ron D. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival.Mol. Cell. 7:1153–1163 [DOI] [PubMed] [Google Scholar]
- Hollien J., Weissman J.S. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response.Science. 313:104–107 [DOI] [PubMed] [Google Scholar]
- Iqbal J., Dai K., Seimon T., Jungreis R., Oyadomari M., Kuriakose G., Ron D., Tabas I., Hussain M.M. 2008. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA.Cell Metab. 7:445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler J.A., Skalet A.H., Alwine J.C. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response.J. Virol. 79:6890–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi N.N., Lee A.H., Vallabhajosyula P., Otipoby K.L., Rajewsky K., Glimcher L.H. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1.Nat. Immunol. 4:321–329 [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Hosoda A., Okuda T., Kamigori Y., Nomura-Furuwatari C., Kimata Y., Tsuru A., Kohno K. 2001. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress.Nat. Cell Biol. 3:158–164 [DOI] [PubMed] [Google Scholar]
- Kang S.W., Rane N.S., Kim S.J., Garrison J.L., Taunton J., Hegde R.S. 2006. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway.Cell. 127:999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., Stroud R.M., Walter P. 2009. The unfolded protein response signals through high-order assembly of Ire1.Nature. 457:687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Glimcher L.H. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response.Mol. Cell. Biol. 23:7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R.J. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response.Genes Dev. 16:452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response.Science. 318:944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson K.L., Ghosh R., Urano F. 2008. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells.PLoS ONE. 3:e1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D., Tokuda M., Iwawaki T. 2007. Site-specific cleavage of CD59 mRNA by endoplasmic reticulum-localized ribonuclease, IRE1.Biochem. Biophys. Res. Commun. 360:122–127 [DOI] [PubMed] [Google Scholar]
- Papa F.R., Zhang C., Shokat K., Walter P. 2003. Bypassing a kinase activity with an ATP-competitive drug.Science. 302:1533–1537 [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response.Nat. Rev. Mol. Cell Biol. 8:519–529 [DOI] [PubMed] [Google Scholar]
- Shang J., Lehrman M.A. 2004. Discordance of UPR signaling by ATF6 and Ire1p-XBP1 with levels of target transcripts.Biochem. Biophys. Res. Commun. 317:390–396 [DOI] [PubMed] [Google Scholar]
- Starcevic M., Dell'Angelica E.C. 2004. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1).J. Biol. Chem. 279:28393–28401 [DOI] [PubMed] [Google Scholar]
- Tardif K.D., Mori K., Kaufman R.J., Siddiqui A. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response.J. Biol. Chem. 279:17158–17164 [DOI] [PubMed] [Google Scholar]
- Tirasophon W., Lee K., Callaghan B., Welihinda A., Kaufman R.J. 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response.Genes Dev. 14:2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Wong H.N., Song B., Miller C.N., Scheuner D., Kaufman R.J. 2005. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis.J. Clin. Invest. 115:268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., McGrath B., Li S., Frank A., Zambito F., Reinert J., Gannon M., Ma K., McNaughton K., Cavener D.R. 2002. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas.Mol. Cell. Biol. 22:3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]