Abstract
The end product of purine catabolism varies amongst vertebrates and is a consequence of independent gene inactivation events that have truncated the purine catabolic pathway. Mammals have traditionally been grouped into two classes based on their end product of purine catabolism: most mammals, whose end product is allantoin due to an ancient loss of allantoinase (ALLN), and the hominoids, whose end product is uric acid due to recent inactivations of urate oxidase (UOX). However little is known about purine catabolism in marsupials and monotremes. Here we report the results of a comparative genomics study designed to characterize the purine catabolic pathway in a marsupial, the South American opossum (Monodelphis domestica), and a monotreme, the platypus (Ornithorhynchus anatinus). We found that both genomes encode a more complete set of genes for purine catabolism than do eutherians and conclude that a near complete purine catabolic pathway was present in the common ancestor of all mammals, and that the loss of ALLN is specific to placental mammals. Our results therefore provide a revised history for gene loss in the purine catabolic pathway and suggest that marsupials and monotremes represent a third class of mammals with respect to their end products of purine catabolism.
Keywords: allantoinase, evolution, gene loss, mammals, purine catabolism
Introduction
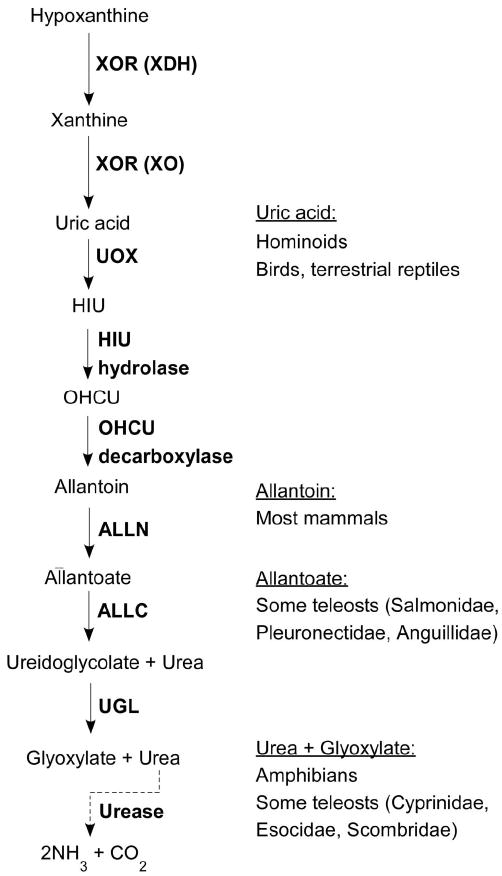
Purine metabolism is an essential biochemical pathway that is conserved across a wide-range of phyla and is considered a likely candidate for the most ancient metabolic pathway on the planet (Caetano-Anolles et al. 2007). The end products of purine catabolism, however, vary among vertebrates and have been the subject of comparative biochemical studies since the early 20th century (for example see Hunter et al. 1914b). Those studies as well as subsequent reports have led to the widely accepted view of the end products of purine catabolism in vertebrates as summarized in Fig. 1.
Figure 1.
Purine catabolism. The enzymes and catabolites of the terminal portion of the purine catabolic pathway are shown on the left. The end products of this pathway for different groups of vertebrates are shown on the right (Urich 1994; Hayashi et al. 2000).
The terminal portion of purine catabolism begins with the degradation of hypoxanthine to uric acid by xanthine dehyrogenase and xanthine oxidoreductase (XDH, EC 1.1.1.204; XOR, EC 1.2.3.2), which are encoded for by a single gene present in all vertebrates. Until recently, the next step in this pathway was reported to be the degradation of uric acid to allantoin by urate oxidase (UOX, EC 1.7.3.3), which is absent in hominoids, birds and terrestrial reptiles. However, Ramazzina et al., (2006) recently identified two enzymes, HIU hydrolase and OHCU decarboxylase, that catalyze intermediate steps in the conversion of uric acid to allantoin, which is the end product of this pathway in most mammals. Allantoinase (ALLN, EC 3.5.2.5) then degrades allantoin to allantoate, which is the final product of this pathway in some teleost fish. In other teleost fish and amphibians, allantoicase (ALLC, EC 3.5.3.4) catalyzes the hydrolysis of allantoate to ureidoglycolate and urea, which is followed by degradation of ureidoglycolate to glyoxylate and urea by ureidoglycolate lyase (UGL, EC 4.3.2.3)(Hayashi et al. 2000). Finally, urease activity (EC 3.5.1.5), whose presence has been detected in the gut of some fish but is encoded within bacteria living in the host and not the vertebrate genome, can generate the most terminal products of the pathway, ammonia and carbon dioxide (Urich 1994).
To date, comparative studies of the end products of purine catabolism in mammals have relied on the relative ratios of uric acid and allantoin in urine as a standard by which to compare this pathway between species. In placental mammals, most species primarily excrete allantoin and a small amount of uric acid (Hunter et al. 1914a; Hunter et al. 1914b), whereas hominoids primarily excrete uric acid and only trace amounts of allantoin (Wiechowski 1909; Wiechowski 1912; Wells et al. 1914). More limited information is available for the other mammals, the marsupials and monotremes. In the case of marsupials, the enzymatic activity of UOX has been directly assayed for and detected in the opossum (Caldwell et al. 1914). With the exception of human and chimpanzee (Hunter et al. 1914b), this species was shown to excrete more uric acid relative to allantoin than all placental mammals. In the monotremes, both the platypus and echidna have been shown to excrete uric acid and allantoin, but the total amount of these metabolites per a defined volume of urine was much smaller and included high proportions of uric acid than was observed in the placental mammal included in the same study (Denton et al. 1963). Purine catabolism therefore clearly proceeds beyond uric acid in both marsupials and monotremes, and the presence of significant amounts of allantoin indicates that the pathway extends at least as far in these species as in most placental mammals. Moreover, the distinct relative ratios of uric acid to allantoin in marsupials and monotremes compared to most placental mammals could be an indication that both uric acid and allantoin are intermediate metabolites in non-eutherian mammals. Thus, while purine catabolic metabolites downstream of allantoin have not been directly assayed for in marsupials and monotremes, these results are suggestive that the purine catabolic pathway in these species proceeds farther than it does in placental mammals.
Variation in the end product of purine catabolism among vertebrates is the result of truncation of the pathway by complete or partial inactivation of the genes encoding the purine catabolic enzymes (Wu et al. 1989; Wu et al. 1992; Andersen et al. 2006). Once a mutation truncates the purine catabolic pathway, the genes downstream of the mutated enzyme are presumably no longer needed and will be lost over time. Though this model is not always correct, (Fujiwara et al. 1995; Vigetti et al. 2000; Vigetti et al. 2001; Vigetti et al. 2003), a comparative genomic-based study focused strictly on the presence or absence of genes can be a good predictor of the functional content of the purine catabolic pathway (Wong et al. 2005; Ramazzina et al. 2006). Here we report the results of a comparative genomics study designed to compare the gene content in the terminal portion of the purine catabolic pathway in a marsupial, the South American opossum (Monodelphis domestica) and a monotreme, the platypus (Ornithorhynchus anatinus), versus a diverse set of other vertebrates.
Materials And Methods
Identification of orthologs encoding the purine catabolic pathway
The following strategy was used to identify orthologs of the six known genes in the terminal portion of the purine catabolic pathway. First, pre-computed orthologs and the accompanying nucleotide and protein alignments 13 species [fugu (Takifugu rubripes), pufferfish (Tetraodon nigroviridis), zebrafish (Danio rerio), stickleback (Gasterosteus aculeatu), frog (Xenopus tropicalis), chicken (Gallus gallus), cow (Bos taurus), dog (Canis familiaris), mouse (Mus musculus), rat (Rattus norvegicus), rhesus monkey (Macaca mulatta), chimpanzee (Pan troglodytes) and human (Homo sapiens)] were extracted from the TreeFam database (http://www.treefam.org/) (Li et al. 2006) for XDH (TF353036), UOX (TF323438), HIU hydrolase (TF300210), OHCU decarboxylase (TF323276), ALLN (TF300759) and ALLC (TF324677). Second, in cases where orthologs were absent in TreeFam the missing orthologs were either extracted from the most recent Ensembl (http://www.ensembl.org) (Hubbard et al. 2007) annotation, or were identified by homology searches and manual gene annotation. Homology searches and manual annotation was used to identify orthologs from the platypus genome assembly (ornAna1) (Warren et al 2008), opossum genome assembly (monDom4) (Mikkelsen et al 2007), anolis genome assembly (Anolis carolinesis) (anoCar1) generated by the Broad Institute at MIT and Harvard University, and marmoset genomic sequences generated by the Washington University Genome Sequencing Center. The complete sets of orthologs were aligned with CLUSTALX (Jeanmougin et al. 1998) and resulting alignments manually edited using the TreeFam alignments as a guide. The Ensembl gene identifiers are listed in Supplementary Table 1, and the manually annotated sequences provided in Supplementary Data File 1. The alignments of these sequences are available from the authors upon request.
Classification of orthologs
The orthologs in each species were classified as a gene, a pseudogene, or absent. Orthologs were classified as a gene if they had an open reading frame, were evolving under purifying selection, and had a conserved intron/exon structure. Orthologs were classified as a pseudogene if the protein coding sequence was truncated by early nonsense mutations and evolving neutrally. Orthologs were classified as absent from a species only if homology searches of the assembled genome (and when available EST databases) and direct analysis of the predicted syntenic genomic location failed to detect the gene of interest.
Sequence Evolution
To evaluate the rate of evolution for the purine catabolic genes, a maximum likelihood method was implemented by the “codeml” program in PAML (Yang 2003) to estimate the ratio of nonsynonymous (Ka) to synonymous (Ks) substitutions, i.e. Ka/Ks. The tree phylogeny used in the following analyses is: (((((((Marmoset,(Rhesus,(Chimp,Human))),(Rat,Mouse)),(Dog,Cow)),Opossum),Platypus), (Chicken,Anolis)),Xenopus,(((Fugu,Tetraodon),Stickleback),Zebrafish)); For each gene, Ka/Ks rate estimates were first compared between the marsupial and monotreme lineages using a likelihood ratio test (LRT) to determine if the estimated Ka/Ks values were significantly different (p < 0.01). Rate estimates for the marsupial and monotreme, either combined (p > 0.01) or individually (p < 0.01), were then compared to the Ka/Ks estimates for the genes known to have retained their function as purine catabolic enzymes in fish, amphibian, bird & reptile, and placental mammals to detect significant differences with other groups of vertebrates.
Results
The opossum and platypus genomes encode a more complete set of genes for purine catabolism than do placental mammals
The end products of purine catabolism in placental mammals are well established but have yet to be determined in marsupials and monotremes. To genetically characterize the terminal portion of the purine catabolic pathway in marsupials and monotremes we determined the presence or absence of the known genes in this pathway in the platypus (monotreme) and opossum (marsupial) genomes. In particular, the platypus and opossum genomes were searched for the presence of XOR/XDH, UOX, HIU hydrolase, OHCU decarboxylase, ALLN and ALLC as the basis for comparison to a diverse sampling of 15 other vertebrates (4 teleosts: stickleback, fugu, tetraodon and zebrafish, 1 amphibian: frog, 1 reptile: anolis lizard, 1 bird: chicken, and 8 placental mammals including 2 hominoids: human and chimpanzee and 6 other placental mammals: mouse, rat, dog, cow, rhesus monkey and marmoset) (Fig. 2). (Note that the identity of the gene encoding the UGL locus has yet to be identified in vertebrates (Takada et al. 1986), and therefore could not be included in this study.)
Figure 2.
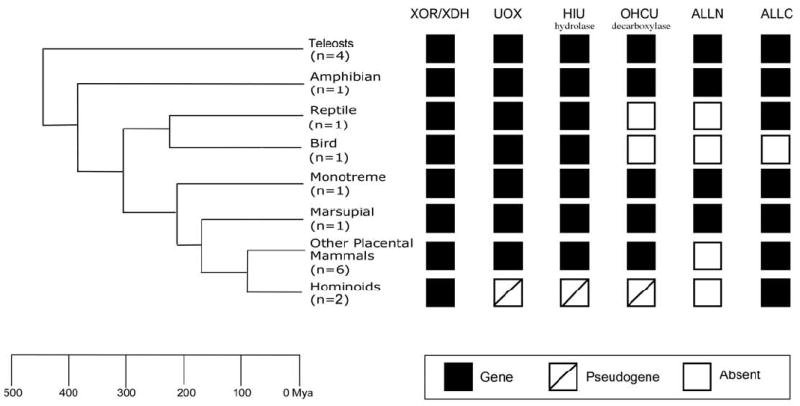
Gene content of the purine catabolic pathway in opossum and platypus. The tree on the left represents the accepted vertebrate phylogeny and divergence dates (millions of years ago, Mya) (Kumar et al. 1998). The species included in each category are teleosts (stickleback, fugu and tetraodon and zebrafish), an amphibian (frog), a reptile (anolis lizard),a bird (chicken), a monotreme (platypus), a marsupial (opossum), and placental mammals, including the hominoids (human and chimpanzee) and other placental mammals (mouse, rat, dog, cow, rhesus monkey and marmoset). The gene content of the purine catabolic pathway for each group of species is summarized on the right. Orthologs were classified as a gene if they had an open reading frame, were evolving under purifying selection, and had a conserved intron/exon structure, as pseudogenes if the protein coding sequence was truncated by early nonsense mutations and evolving neutrally, and absent when no gene or gene fragment was identified.
As expected from previous biochemical studies summarized in Fig. 1 all the known purine catabolic genes are present in the amphibian and teleost genomes (Fig. 2). Similarly, we also detected the full complement of purine catabolic genes, including ALLN which was absent in placental mammals, in both the opossum and platypus genomes (Fig. 2). (Note that the absence of ALLN in the genomes of placental mammals is consistent with allantoin being the end product of purine catabolism in those species, excluding the hominoids). Moreover, estimates of the rates at which the marsupial and monotreme purine catabolic genes have evolved were consistent with each encoding a functional protein (Table 1). In particular, the rate at which a protein has evolved can be inferred by calculating the ratio of the rates of nonsynonymous (Ka) and synonymous (Ks) nucleotide substitutions, i.e., Ka/Ks. In the case of the opossum and platypus, the genes in the purine catabolic pathway had Ka/Ks values between 0.07-0.47, consistent with all six genes evolving under purifying selection (Table 1). While statistically significant differences in the estimated Ka/Ks values were detected between the opossum, platypus and other vertebrate lineages, no obvious pattern emerged that suggested the opossum and platypus genes were evolving under different selective constraints than the orthologs that are known to encode functional purine catabolic enzymes in other vertebrates (Table 1). The presence of all the genes encoding the purine catabolic pathway in the opossum and platypus genomes and their pattern of evolution therefore suggests that marsupials and monotremes have the potential to encode a more complete purine catabolic pathway than placental mammals.
Table 1.
Estimation of Ka/Ks of the genes in the purine catabolic pathway.
| XDH | UOX | HIU hydrolase |
OHCU decarboxylase |
ALLN | ALLC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Syn 656 |
Non 1954 |
Syn 215 |
Non 625 |
Syn 78 |
Non 240 |
Syn 93 |
Non 290 |
Syn 137 |
Non 373 |
Syn 252 |
Non 686 |
| Marsupial | 0.0865a | 0.0797 | 0.0633 | 0.4613a | 0.0957 | 0.3236 | ||||||
| Monotreme | 0.1894a | 0.0786 | 0.1455 | 0.0612a | 0.0599 | 0.1401 | ||||||
| Teleosts & Amphibian | 0.1164c | 0.0350 | 0.2296 | 0.1296 | 0.0883 | 0.1102b,c | ||||||
| Other placental mammals | 0.1492b | 0.1101 | 0.1396 | 0.1916 | n/a | n/a | ||||||
Ka/Ks estimates of the genes in the purine catabolic pathway were calculated for all lineages except the hominoids, and birds and reptiles. Note that though ALLC is present in the genomes of placental mammals it was not included in this table because it is no longer required to function as a purine catabolic enzyme.
Indicates a significant difference (p<0.01) in estimated rates between marsupial and monotreme lineages.
Indicates a significant difference (p<0.01) in estimated rates compared to marsupial.
Indicates a significant difference (p<0.01) in estimated rates compared to monotreme.
‘Syn’ refers to the number of synonymous sites; ‘Non’ refers to the number of nonsynonymous sites.
Loss of ALLN is specific to placental mammals
The presence of ALLN in opossum and platypus genomes and its absence in the genomes of placental mammals suggests that ALLN was most likely present in the eutherian lineage prior to the radiation of placental mammals ~100 million years ago (Murphy et al. 2001). We therefore scrutinized the predicted syntenic location of ALLN between the flanking KIF1A and AGXT loci in the genomes of the placental mammals to see if we could detect any remnants of this gene. However, we were unable to detect ALLN, or clear evidence for any other gene, in this region of eutherian genomes, including the finished genomes of human and mouse. Homology searches using the ALLN protein sequence from other species were also unable to detect this gene elsewhere in the genome of placental mammals. Thus, these results suggest that not only was the purine catabolic function of ALLN lost in placental mammals, but the gene encoding this enzyme has also been deleted or degenerated beyond recognition in the eutherian lineage.
Discussion
Variation in the end product of purine catabolism among vertebrates has been of interest to biologists for nearly a century (Caldwell et al. 1914; Hunter et al. 1914a; Hunter et al. 1914b; Keilin 1959). In textbooks and reviews, mammals are grouped into two classes based on their end products of purine catabolism; the hominoids, whose end product is uric acid, and other mammals, whose end product is allantoin (Fig. 1). This dichotomy among mammals can be explained by the loss of ALLN activity in the common ancestor of all mammals, and subsequent recent inactivation(s) of UOX in hominoids (Wu et al. 1992; Oda et al. 2002). The results of this study provide an updated and expanded view of the sequence of genetic modifications that have shaped the evolution of this pathway in mammals.
The presence of the ALLN in opossum and platypus and absence in placental mammals strongly suggests that loss of ALLN is specific to placental mammals. The most parsimonious time frame for the loss of ALLN from the eutherian genome therefore pre-dates the most recent common ancestor of placental mammals ~100 million years ago (Murphy et al. 2001) and postdates the split of the eutherian and marsupial lineages ~173 million years ago (Kumar et al. 1998). It is possible that this genetic difference between placental mammals and marsupials and monotremes may not reflect a true biochemical difference between these groups of species. However, the presence of a full complement of the known genes in the terminal portion of the purine catabolic pathway in both the opossum and platypus demonstrates that these species have, at a minimum, retained the potential to encode a more complete pathway than is found in other mammals. As such, we hypothesize that marsupials and monotremes have end products of purine catabolism comparable to that of teleosts and/or amphibians, i.e. glyoxylate and urea (Fig. 1), and thus represent a third class of mammals with respect to the end products of purine metabolism. We further propose that the distinct ratios and amounts of uric acid and allantoin previously observed in the urine of the Virginia opossum (Didelphis virginiana) (Caldwell et al. 1914), platypus and echidna (Denton et al. 1963) compared to placental mammals is a reflection of this predicted difference in end products of purine catabolism. Direct biochemical studies will ultimately be needed to test these hypotheses and determine the specific end product of purine metabolism in marsupials and monotremes.
Purine metabolism has been repeatedly modified via differential gene loss throughout vertebrate evolution, and gene loss is increasingly being recognized as an important force in evolution (Olson 1999). Recently, clear associations have been made between gene loss and fundamental biological differences between placental mammals and monotremes (Brawand et al. 2008; Ordonez et al. 2008). Given that the loss of ALLN is specific to placental mammals, it is tempting to speculate on possible adaptive advantages the loss of this gene may have provided to our eutherian ancestors. Prior hypotheses on the adaptive advantages of modifications in nitrogen metabolism, including the urea cycle and the end products of purine catabolism, have focused on diet, water conservation and embryonic environment (Packard 1966; Campbell et al. 1987; Mommsen et al. 1989). Since the eutherian embryonic environment is distinct from that of both the marsupials and monotremes in that full-term development occurs in utero with a chorioallantoic placenta, one possibility is that loss of ALLN is somehow related to this phenotypic innovation that is common to all placental mammals. However, it should be noted that embryonic environment is not always clearly correlated with variation in the end products of purine metabolism. For example, though birds/reptiles and monotremes have a somewhat similar embryonic environment, the end product of birds/reptiles is uric acid due to the inactivation of UOX whereas our results suggest the end product in monotremes may be glyxolate and urea. One potential experimental approach to assess the biological significance of truncation in placental mammals would be to genetically alter the mouse genome in order to reconstitute the functional purine catabolic pathway inferred to have been present in the common ancestor of all mammals. In summary, the results of our modern comparative genomic survey have yielded new insights into the evolution of the purine catabolic pathway in mammals and provide a rationale to revisit and expand the comparative biochemical studies of purine catabolism in marsupials and monotremes.
Supplementary Material
Acknowledgments
The authors thank Kristin Harper, Karyn Meltz-Steinberg, Lisa McGraw and Bob Sullivan for comments on the manuscript. This research was in part supported by a grant from the National Institutes of Health (R21NS060935).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen O, Aas TS, Skugor S, Takle H, van Nes S, et al. Purine-induced expression of urate oxidase and enzyme activity in Atlantic salmon (Salmo salar). Cloning of urate oxidase liver cDNA from three teleost species and the African lungfish Protopterus annectens. Febs J. 2006;273:2839–2850. doi: 10.1111/j.1742-4658.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Brawand D, Wahli W, Kaessmann H. Loss of egg yolk genes in mammals and the origin of lactation and placentation. PLoS Biol. 2008;6:e63. doi: 10.1371/journal.pbio.0060063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Kim HS, Mittenthal JE. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A. 2007;104:9358–9363. doi: 10.1073/pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell GT, Wells HG. The purine enzymes of the opossum (Didelphus virginiana) J Biol Chem. 1914;19:279–283. [Google Scholar]
- Campbell JW, Vorhaben JE, Smith DD., Jr Uricoteley:its nature and origin during the evolution of tetrapod vertebrates. J Exp Zool. 1987;243:349–363. doi: 10.1002/jez.1402430302. [DOI] [PubMed] [Google Scholar]
- Denton DA, Reich M, Hird FJR. Ureotelism of Echidna and Platypus. Science. 1963;139:1225. doi: 10.1126/science.139.3560.1225. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Noguchi T. Degradation of purines: only ureidoglycollate lyase out of four allantoin-degrading enzymes is present in mammals. J Biol Chem. 1995;312:315–318. doi: 10.1042/bj3120315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Fujiwara S, Noguchi T. Evolution of urate-degrading enzymes in animal peroxisomes. Cell Biochem Biophys. 2000;32:123–129. doi: 10.1385/cbb:32:1-3:123. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Givens MH. Studies in the comparative biochemsitry of purine metabolism. II. The excretion of purine catabolites in the urine of ungulates. J Biol Chem. 1914a;18:403–416. [Google Scholar]
- Hunter A, Givens MH, Guion CM. Studies in the biochemistry of purine metabolism. I. The excretion of purine catabolites in the urine of marsupials, rodents and carnivora. J Biol Chem. 1914b;18:387–401. [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Keilin J. The biological significance of uric acid and guanine excretion. Biol Rev. 1959;34:265–296. [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–178. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mommsen TP, Walsh PJ. Evolution of urea synthesis in vertebrates: the piscine connection. Science. 1989;243:72–75. doi: 10.1126/science.2563172. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Oda M, Satta Y, Takenaka O, Takahata N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol Biol Evol. 2002;19:640–653. doi: 10.1093/oxfordjournals.molbev.a004123. [DOI] [PubMed] [Google Scholar]
- Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez GR, Hillier LW, Warren WC, Grutzner F, Lopez-Otin C, Puente XS. Loss of genes implicated in gastric function during platypus evolution. Genome Biol. 2008;9:R81. doi: 10.1186/gb-2008-9-5-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard GC. The influence of ambient temperature and aridity on modes of reproduction and excretion of amniote vertebrates. Am Naturalist. 1966;100:667–682. [Google Scholar]
- Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway throught phylogenetic comparison of whole genomes. Nat Chem Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- Takada Y, Noguchi T. Ureidoglycollate lyase, a new metalloenzyme of peroxisomal urate degradation in marine fish liver. J Biol Chem. 1986;235:391–397. doi: 10.1042/bj2350391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich K. Comparative Animal Biochemistry. Springer-Verlag; Berlin: 1994. [Google Scholar]
- Vigetti D, Binelli G, Monetti C, Prati M, Bernardini G, Gornati R. Selective pressure on the allantoicase gene during vertebrate evolution. J Mol Evol. 2003;57:650–658. doi: 10.1007/s00239-003-2515-5. [DOI] [PubMed] [Google Scholar]
- Vigetti D, Monetti C, Acquati F, Taramelli R, Bernardini G. Human allantoicase gene: cDNA cloning, genomic organization and chromosome localization. Gene. 2000;256:253–260. doi: 10.1016/s0378-1119(00)00342-5. [DOI] [PubMed] [Google Scholar]
- Vigetti D, Monetti C, Bernardini G. Molecular cloning of mouse allantoicase cDNA. Biochim Biophys Acta. 2001;1519:117–121. doi: 10.1016/s0167-4781(01)00207-x. [DOI] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–184. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells HG, Caldwell GT. The purine enzymes of the orang-utan (Simia satyrus) and chimpanzee (Anthropopithecus troglodytes) J Biol Chem. 1914;18:157–165. [Google Scholar]
- Wiechowski W. Das Vorhandensein von Allantoin im normalen Menschenharn und seine Bedeutung fur Beurteilung des menschlichen Harnsaurestoffwechsels. Biochem Z. 1909;19:368–383. [Google Scholar]
- Wiechowski W. Ein Beitrag zur Kenntnis des Purinstoffwechsels der Affen. Prag med Wochenschr. 1912;37:275. [Google Scholar]
- Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- Wu XW, Lee CC, Muzny DM, Caskey CT. Urate oxidase: primary structure and evolutionary implications. Proc Natl Acad Sci USA. 1989;86:9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XW, Muzny DM, Lee CC, Caskey CT. Two independent mutational events in the loss of urate oxidase during hominoid evolution. J Mol Evol. 1992;34:78–84. doi: 10.1007/BF00163854. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 2003;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.