Abstract
Although the consequences of genotoxic injury include cell cycle arrest and apoptosis, cell survival responses after genotoxic injury can produce intrinsic death-resistance and contribute to the development of a transformed phenotype. Protein tyrosine phosphatases (PTPs) are integral components of key survival pathways, and are responsible for their inactivation, while PTP inhibition is are often associated with enhanced cell proliferation. Our aim was to elucidate signaling events that modulate cell survival after genotoxin exposure. Diploid human lung fibroblasts (HLF) were treated with Cr(VI) (as Na2CrO4), the soluble oxyanionic dissolution product of certain particulate chromates, which are well-documented human respiratory carcinogens. In vitro soluble Cr(VI) induces a wide spectrum of DNA damage, in both the presence and absence of a broad-range PTP inhibitor, sodium orthovanadate (SOV). Notably, SOV abrogated Cr(VI)-induced clonogenic lethality. The enhanced survival of Cr(VI)-exposed cells after SOV treatment was predominantly due to a bypass of cell cycle arrest, as there was no effect of the PTP inhibitor on Cr-induced apoptosis. Moreover, the SOV effect was not due to decreased Cr uptake as evidenced by unchanged Cr-DNA adduct burden. Additionally, the bypass of Cr-induced growth arrest by SOV was accompanied by a decrease in Cr(VI)-induced expression of cell cycle inhibiting genes, and an increase in Cr(VI)-induced expression of cell cycle promoting genes. Importantly, SOV resulted in an increase in forward mutations at the HPRT locus, supporting the hypothesis that PTP inhibition in the presence of certain types of DNA damage may lead to increased genomic instability, via bypass of cell cycle checkpoints.
Keywords: Protein tyrosine phosphatase, hexavalent chromium, cell survival, genomic instability
1. Introduction
Deregulated cell proliferation and resistance to apoptosis are thought to be at the foundation of neoplastic evolution. Tightly orchestrated signaling pathways govern both cell proliferation and apoptosis. Thus, inappropriate activation/inactivation of key signals that control cell survival can contribute to autonomous growth and neoplastic transformation. There is considerable evidence that protein tyrosine phosphorylation is responsible for the maintenance of proliferative signals and is involved in the early stages of neoplasia (for review see [1]). Protein tyrosine phosphatases (PTPs), such as PTEN (phosphatase and tensin homolog deleted on chromosome ten) and MKP (MAP kinase phosphatase) are integral components of survival pathways, and are responsible for their respective inactivation [2;3]. Indeed, certain of these PTPs have been described as tumor suppressors since their overall effect is to decrease cell proliferation (for review see [4;5]).
Dysregulated cell proliferation underlies carcinogenesis and can be caused by genetic/epigenetic alterations induced by endogenous and environmental genotoxins. The initial consequence of genotoxic injury is usually cell cycle checkpoint arrest but may also activate apoptotic or terminal growth arresting pathways. Cellular survival in the face of genotoxic insult may produce an intrinsically death-resistant phenotype; such a selective growth advantage may allow for the emergence of cells that are more prone to neoplastic evolution.
Certain forms of hexavalent chromium [(Cr(VI)] are known human respiratory carcinogens that can be employed as useful genotoxic tools with relevant toxicological importance [6]. The intracellular metabolic reduction of Cr(VI) to its toxic metabolites is well documented and there is an extensive background on the mechanisms of Cr(VI)-induced macromolecular damage. The structural and functional aspects of Cr(VI)-induced DNA damage are summarized in several recent review articles [7;8]. Epidemiological studies carried out in the U.K., Europe, Japan and the U.S. have consistently shown that workers in the chromate production industry have an elevated risk of respiratory disease, fibrosis, perforation of the nasal septum, development of nasal polyps, and lung cancer [9;10]. Indeed, environmental and occupational exposure to chromate continues to loom large as a major public health issue and a source of continuous high-profile litigation.
The overall objective of our laboratory is to elucidate the coordinate signaling events that mediate cell fate determination and survival, and consequently mutagenesis, after genotoxic insult. The present study tested the hypothesis that maintenance of protein tyrosine phosphorylation by SOV modulates survival after Cr(VI)-induced genotoxic insult. The data show that SOV reversed Cr(VI)-induced clonogenic lethality. The enhanced survival of Cr(VI)-exposed cells after SOV treatment was predominantly due to a non-lethal bypass of Cr-induced growth arrest and was not due to decreased Cr-DNA adduct burden. This was accompanied by a decreased induction of negative cell cycle regulatory genes by Cr(VI) and an increased induction of positive cell cycle regulatory genes. Notably, co-treatment with SOV resulted in an increase in forward mutations at the hypoxanthine-guanine phosphoribosyltransferase (HPRT) locus, which underscored the potential for genomic instability as a result of loss of checkpoint control. Taken together, this work suggests that regulators of tyrosine phosphorylation may govern cell survival as an initial event after Cr(VI) genotoxic insult and potentially facilitate the earliest stages of neoplastic evolution.
2. Material and methods
2.1 Materials
Sodium chromate (Na2CrO4.4H2O; [Cr(VI)]) was purchased from J.T. Baker Chemical Company, Phillipsburg, NJ. Sodium orthovanadate (Na3O4V; SOV) was purchased from Aldrich Chemical Company, St. Louis, MO. Cell culture reagents were from Invitrogen Life Technologies, Gaithersburg, MD, and fetal bovine serum (FBS) was from Hyclone, Logan, Utah. Other chemicals were either from Sigma (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA), and were of the highest purity available.
2.2 Cell Culture
The normal diploid human lung fibroblast (HLF) cell line, LL24, was originally isolated from normal autopsy tissue of an 11-year-old male, and was obtained from the ATCC (Manassas, VA). HLF cells were maintained in Ham's F-12 nutrient mixture, supplemented with 15% FBS, and containing 50 units/ml penicillin and 50 μg/ml streptomycin. Chinese hamster ovary cells (CHO, AA8) were maintained in minimal Eagles medium (MEM), supplemented with 10% FBS, 1% penicillin-streptomycin and 2 mM glutamine. All cells were maintained at 37°C in a humidified incubator under an atmosphere containing 5% CO2.
2.3 Experimental Treatment
In all experiments, cells were seeded 24 h prior to exposure with Cr(VI) and/or SOV. Sodium chromate and SOV were respectively dissolved in deionized H2O and sterilized by passage through a 0.2 micron filter before each use. Cells were treated with the indicated final concentration of Cr(VI) for 24 h in complete medium. In experiments in which SOV was co-incubated with Cr(VI), the SOV was added 30 min prior to Cr(VI) addition. The cells were either collected for analysis after treatment or rinsed with phosphate buffered saline (PBS) and allowed to recover in complete medium for an additional 24 h before analysis.
2.4 Clonogenicity Analysis
Cells were seeded at 1 × 105/60 mm dish, and incubated with 0, 1 or 2 μM Cr(VI), in the absence or presence of 10 and 25 μM SOV. Following 24 h treatment, the cells were collected by trypsinization, washed and reseeded at 2 × 103/60 mm dish (in the absence of any agent), as previously described [11]. Cells were incubated for 14 days, and were fixed and stained with crystal violet (80% methanol, 2% formaldehyde, 2.5g/L crystal violet). Colonies of 20 cells or greater were counted.
2.5 Cell Growth Analysis
Cells were seeded at 1 × 105/100 mm dish and incubated with 0, 1, or 2 μM Cr(VI) in the absence or presence of 10 and 25 μM SOV. A group of replicate dishes were seeded for each dose tested and all of the replicates within the group received the same treatment. One replicate was taken at day 2, 4, 5, 6, 8, 10, 12, and 14 after treatment and counted to determine total cell number at each dose and time. Cells were gently harvested with trypsin, centrifuged at 600 × g for 5 min, and the cell pellets were resuspended in 1 ml PBS. Total cell number was determined using a hemacytometer (Hausser Scientific, Horsham, PA). Data were normalized to the same day non-treated control.
2.6 mRNA expression analysis
Cells were seeded at 2 × 106/150 mm dish and incubated without or with 1 μM Cr(VI) in the absence or presence of 10 μM SOV. Following 24 h treatment, RNA was isolated using RNA-Bee (Tel-Test, Friendswood, TX) followed by a secondary purification on Qiagen RNAeasy Mini columns (Qiagen, Valencia, CA). RNA quality was assessed by Agilent bioanalyzer (Agilent Technologies, Santa Clara, CA) and RNA Integrity Numbers (RIN) for all samples were >9.9. Labeled cRNA was hybridized to GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA) and arrays were scanned with an Agilent laser scanner. The raw expression data were analyzed by GeneSpring (Agilent Technologies) for differentially regulated genes after respective treatment. Data set was filtered by 1-way ANOVA with 5% FDR and 1,880 genes out of total 54,675 transcripts were chosen. Transcripts associated with cell cycle and cell proliferation (according to GO Biological Process) were selected and further filtered by expression fold cutoff at 1.5. All original microarray data has been deposited to the GEO web site http://www.ncbi.nlm.nih.gov/geo/, accession number GSE12205.
2.7 Mutation Frequency
HLF cells were seeded at 5×105/100 mm dish, and incubated for 24 h with either 0−3 μM Cr(VI) or 1 h with 0, 250, and 250 μM methyl methanesulfonate (MMS, as positive control) in the presence or absence of 10 μM SOV. CHO cells were seeded at 1×105/60 mm dish, and incubated for 24h with either 0−12 μM Cr(VI) or 1 h with 0 and 500 μM MMS in the presence or absence of 10 μM SOV. After washing, HLF and CHO cells were grown exponentially for 13 and 7 days, respectively, following treatment. For mutant selection, HLF cells were plated at 2×105 in 8 replicate 100 mm dishes and CHO cells were plated at 1×106 in 2 replicate 150 mm dishes. Cells were maintained in hypoxanthine-free EMEM containing 15% FBS and 30−60 μM 6-TG, which selectively inhibits the growth of HPRT wild-type cells [12]. In parallel with mutant selection, 300−800 cells were plated in nonselective medium to determine cloning efficiency. After 21 and 8 days, respective HLF and CHO colonies were stained with crystal violet, and colonies with ≥50 cells were counted and HPRT mutation frequency was calculated per million clonable cells.
2.8 Statistical analysis
To assess significant differences among experimental groups, statistical analyses were performed using GraphPad Prism version 4.00 for Windows (San Diego, CA). A two tailed, unpaired t test and one-way analysis of variance (ANOVA) with either a Tukey or Dunnetts post test were used with an α level of 0.05 when comparing two experimental groups and for multiple sample comparisons, respectively.
3. Results
3.1 SOV reverses Cr(VI)-induced clonogenic lethality
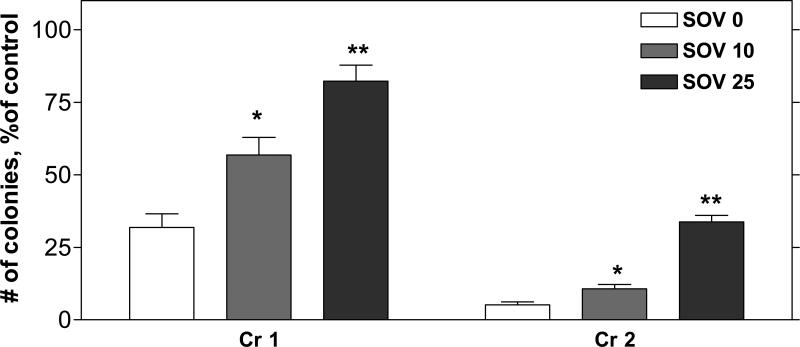
PTP inhibition by SOV increased the abundance of proteins containing phosphotyrosine moieties (Supplemental Figure 1). As shown in Figure 1, a 24 h exposure to Cr(VI) induced a concentration-dependent decrease in clonogenic survival, which was almost 100% lethal at 3 μM (data not shown), consistent with our previous studies [11;13;14]. SOV resulted in a significant increase in percent clonogenic survival, which ranged from 2 to 3-fold and 2 to 7-fold after 1 and 2 μM Cr(VI), respectively. The PTP inhibitor alone had no effect on clonogenic survival at a concentration of 10 μM, however at 25 μM, it caused a 57% decrease in clonogenic survival (data not shown). At a concentration of 50 μM, SOV alone was over 75% clonogenically lethal, and was not used for further studies (data not shown).
Figure 1. Effect of tyrosine phosphatase inhibition on clonogenic survival after exposure to Cr(VI).
HLF cells were incubated without (control) or with 1 or 2 μM Cr(VI) for 24 h (Cr 1, Cr 2), in the absence (SOV 0) or presence of 10 or 25 μM SOV (SOV 10, SOV 25), and were washed and reseeded to allow for colony formation. Colonies were stained with crystal violet, and counted 12 days later. The data are expressed as percentage of respective control, in the absence of Cr(VI) and are the means ± SE of 4−5 independent experiments. *, **: Statistically significant difference from SOV 0 at p<0.05, p<0.01.
3.2 SOV attenuates Cr(VI)-induced growth arrest
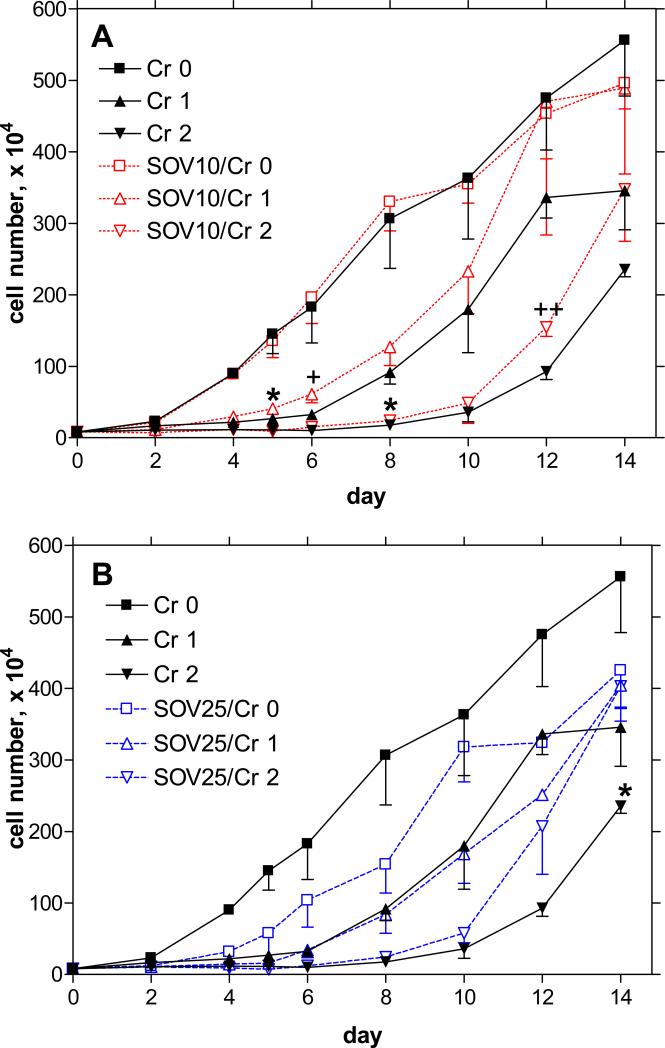
Clonogenicity is an indicator of long-term cell survival and replicative potential after exposure to a genotoxic agent. We have previously shown that cell populations exposed to Cr(VI) have a concentration-dependent spectrum of responses, presumably correlated to the extent of DNA damage, ranging from transient cell cycle checkpoint arrest to terminal growth arrest and apoptosis [14]. In order to understand the mechanism of enhanced clonogenic survival in the presence of Cr(VI) after SOV treatment, we monitored HLF cell growth over a 14-day period following a 24 h exposure to Cr(VI) (1−2 μM), in the presence and absence of SOV (10 and 25 μM). As shown in Figure 2A, 10 μM SOV alone had no effect on cell growth, however it attenuated Cr-induced growth arrest for the period of the study, which was significant at days 5/6 and 8/12 for 1 and 2 μM Cr(VI), respectively. Moreover, this concentration of SOV completely abrogated Cr-induced growth arrest 12 days after incubation with 1 μM Cr(VI), however these data were not statistically significant. In keeping with the clonogenic survival data, 25 μM SOV was associated with a persistent growth delay, averaging at around 40−50% less than that of the control growth rate (Figure 2B). Nevertheless, there was a clear trend for this concentration of SOV to attenuate Cr-induced growth arrest starting 8 days after incubation with 2 μM Cr(VI), which was statistically significant at day 14. The attenuation of Cr(VI)-induced growth arrest by SOV was concomitant with both G1/S and G2/M checkpoint bypass (data not shown).
Figure 2. Effect of tyrosine phosphatase inhibition on Cr(VI)-induced growth arrest.
HLF cells were incubated without or with 1 or 2 μM Cr(VI) for 24 h in the absence or presence of A) 10 or B) 25 μM SOV, and were washed, and cell numbers were counted for 14 days after Cr(VI) removal. Data are expressed as number of cells counted, and are the means ± SE of 3 independent experiments. *: Statistically significant difference from SOV 0 at p<0.05; +: p=0.079; ++: p=0.067.
We also studied the sensitivity of HLF cells to Cr(VI)-induced apoptosis, in the absence and presence of SOV. The basal apoptotic level in the untreated cells was around 5% of the total, as measured by phosphatidylserine (PS) translocation to the outer leaflet of the plasma membrane. Treatment with 6 μM Cr(VI) induced apoptosis in around 18% of the cells, regardless of SOV treatment. Moreover, neither 10 nor 25 μM SOV alone had any effect on PS translocation after 24 h exposure (Supplemental Figure 2).
Notably, co-incubation with 10 μM SOV had no effect on Cr-DNA adduct levels which were approximately 5 and 10 pmol/ug DNA at 1 and 2 μM Cr(VI), respectively (Supplemental Table 1), consistent with our previous report [15].
3.3 Microarray screening of cell cycle regulatory genes after co-treatment with Cr(VI) and SOV
We used microarray analyses to characterize the effect of SOV on changes in gene expression after Cr(VI) exposure. Due to the observed effect of this PTP inhibitor on the attenuation of Cr-induced growth arrest, we focused on transcripts associated with cell cycle and cell proliferation (according to GO Biological Process). Seventy-one transcripts were identified that exhibited at least a 1.5-fold change in expression when compared to the control after 24h exposure to Cr(VI), either in the presence or absence of SOV (Supplemental Table 2).
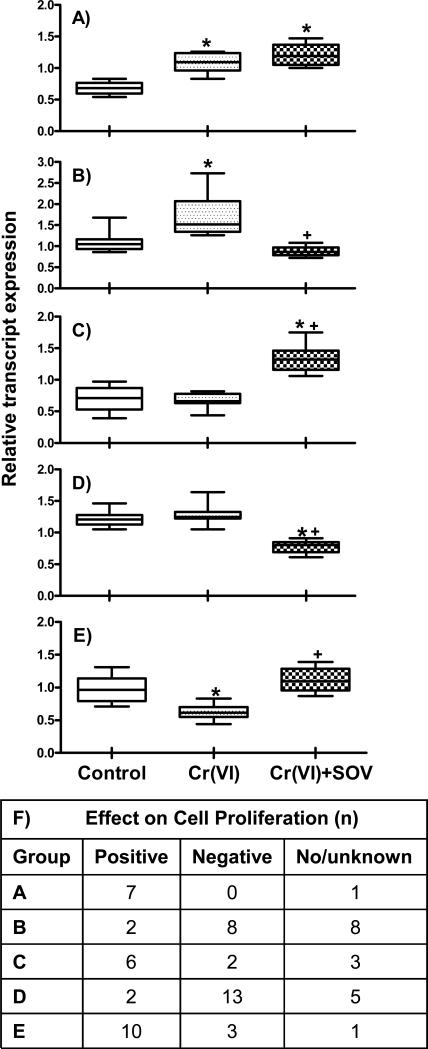
When the data were analyzed for differential gene expression profiles among untreated (control), Cr(VI), and Cr(VI) + SOV-treated groups, distinct patterns of expression emerged. The expression data were combined accordingly into 5 groups, as shown in Figure 3A-E. In group A, 8 genes were found to be upregulated by Cr(VI) treatment both in the absence and presence of SOV, as compared to control. In contrast, in group B, SOV co-treatment abrogated the upregulation of 18 genes induced by Cr(VI) treatment. Group C depicts 11 genes that were upregulated by Cr(VI) + SOV co-treatment, as compared to both control and Cr(VI) treatment alone. Conversely, in Group D, 20 genes were found to be downregulated by Cr(VI) + SOV co-treatment, when compared to both control and Cr(VI) treatment alone. Finally, the 14 genes comprising Group E, were downregulated by Cr(VI) treatment as compared to control, however SOV co-treatment abrogated this downregulation.
Figure 3. Effect of tyrosine phosphatase inhibition on expression profiles of cell cycle-related genes.
HLF cells were incubated without or with 1 μM Cr(VI) for 24 h in the absence or presence of 10 μM SOV, and RNA was isolated and hybridized to GeneChip Human Genome U133 Plus 2.0 Array as described in the Materials and Methods section. Data set was filtered by 1-way ANOVA with 5% FDR and transcripts associated with cell cycle and cell proliferation (according to GO Biological Process) were selected and further filtered by expression fold cutoff at 1.5. Seventy-one transcripts are shown here for Groups A-E. Data are the median, 25th and 75th percentile, and range of the log10 expression values from 4 independent RNA samples per treatment group. *: Statistically significant difference from control at p<0.001; +: Statistically significant difference in the presence vs. the absence of SOV at p<0.001. F) Number of genes in each respective group that were identified as positive or negative regulators of cell cycle progression. No/unknown indicates the number of genes for which there was either no known association with cell proliferation or for which the data were ambiguous in terms of positive or negative regulation.
We further classified the 71 transcripts as either positive (n=27) or negative (n=26) regulators of cell cycle progression, based on an extensive literature search and Entrez Gene and PubMed (NCBI, NLM, NIH) classifications (Supplemental Table 2). A designation of either no/unknown function (n=18) was used for the respective following criteria: no apparent cell proliferation function was identified; gene expression reported to correlate with both positive and negative regulation of cell cycle progression; relationship of gene expression to cell cycle progression was cell context-specific, with no known function in lung fibroblasts; no information could be obtained.
As shown in Figure 3F, the majority of the gene expression data was concordant with cell survival/cell growth data as shown in Figures 1 and 2 (i.e., genes that were upregulated by the PTP inhibitor were more likely to have a positive effect on cell cycle progression and vice versa). This is evidenced in Groups C and E (gene expression upregulated by SOV co-treatment), respectively, in which 6/8 and 10/13 transcripts with known function were positive effectors of cell cycle progression. Conversely, in Groups B and D (gene expression down regulated by SOV co-treatment), the respective 8/10 and 13/15 transcripts with known function were negative regulators of cell cycle progression. In Group A (similar upregulation of genes by Cr(VI) alone and with SOV co-treatment), 7/7 transcripts with known function were positive effectors of cell cycle progression, however the Cr-induced expression of these transcripts was not significantly altered by the PTP inhibitor.
3.4 SOV enhances Cr mutagenesis at the HPRT locus
Since the effect of SOV on Cr(VI)-induced clonogenic lethality was not due to decreased Cr-DNA adduct burden, we hypothesized that increased genomic instability would accompany this apparent loss of checkpoint control. Therefore, we measured the effect of SOV on mutation frequency at the HPRT locus after exposure to either Cr(VI) for 24 h or MMS (as a positive control for the mutagenesis assay) for 1 h. HPRT mutation frequency was corrected for cell survival. As shown in Table 1A, in HLF cells, 2 μM Cr(VI) led to a 2.9-fold increase in mutation frequency, which decreased with 3 μM Cr(VI), likely due to high toxicity, consistent with previous reports [16;17]. 10 μM SOV consistently elevated the Cr-induced mutation frequency (p<0.05 for 1 μM Cr(VI); p=0.0550 for 3 μM Cr(VI)) in HLF cells when compared to that in the absence of SOV. MMS induced a respective 4.1- and 6.5-fold increase in mutation frequency, which attained statistical significance with 500 μM MMS, and was not affected by SOV. As shown earlier, SOV enhanced clonogenic survival after Cr(VI) exposure. Interestingly, the PTP inhibitor had no protective effect against MMS-induced clonogenic lethality, which was around 78−83% at 250 μM MMS in the presence or absence of SOV. Moreover, the mutant colonies generated after Cr(VI) + SOV co-treatment contained around 1.5−2.0-fold more cells than those in the presence of Cr(VI) alone (data not shown).
Table 1. Effect of tyrosine phosphatase inhibition on Cr(VI) and MMS mutation frequency at the HPRT locus.
A) HLF cells were incubated without and with 1−3 μM Cr(VI) for 24 h or 250−500 μM MMS for 1 h in the absence and presence of 10 μM SOV for 24h, and were washed and grown exponentially following treatment. For mutant selection, cells were plated in hypoxanthine-free EMEM containing 10% FBS and 30−60 μM 6-TG. In parallel, cells were plated in nonselective medium to determine cloning efficiency, After 21 days, colonies were stained with crystal violet, and colonies with >20 cells were counted and HPRT mutation frequency was calculated per million clonable cells. Data are means ± SE of 4−7 independent experiments. B) CHO cells were incubated without and with either 6−12 μM Cr(VI) for 24 h or 500 μM MMS for 1 h in the absence and presence of 10 μM SOV for 24h, and were washed and grown exponentially following treatment. For mutant selection, cells were plated in hypoxanthine-free EMEM containing 10% FBS and 30−60 μM 6-TG. In parallel, cells were plated in nonselective medium to determine cloning efficiency, After 8 days, colonies were stained and HPRT mutation frequency was calculated as described in A. Data are means ± SE of 2 experiments.
| SOV, 0 μM | SOV, 10 μM | |||||
|---|---|---|---|---|---|---|
| A. HLF cells | Mutation Frequency × 10−6 | Clonogenic Survival, % | Mutation Frequency × 10−6 | Clonogenic Survival, % | ||
| Mean (SE) | Fold control | Mean (SE) | Mean (SE) | Fold control | Mean (SE) | |
| Cr, 0 μM | 47.8 (21.4) | 37.0 (18.4) | ||||
| Cr, 1 μM | 56.1 (25.5) | 1.9 | 45.8 (7.9) | 140.9 (86.5) | 11.2+ | 65.4 (7.7) |
| Cr, 2 μM | 124.2 (76.8) | 2.9 | 10.3 (4.0) | 209.8 (144.9) | 3.1 | 22.7 (9.2) |
| Cr, 3 μM | 15.1 (11.5) | 0.5 | 2.0 (1.2) | 104.4 (53.1) | 7.2# | 9.0 (4.2) |
| MMS, 0 μM | 32.9 (22.1) | |||||
| MMS, 250 μM | 77.8 (35.0) | 4.1 | 78.4 (8.2) | 43.0 (18.2) | 2.3 | 82.9 (9.3) |
| MMS, 500 μM | 63.5 (36.0) | 6.5* | 59.2 (20.4) | |||
| SOV, 0 μM | SOV, 10 μM | |||||
|---|---|---|---|---|---|---|
| B. CHO cells | Mutation Frequency × 10−6 | Clonogenic Survival, % | Mutation Frequency × 10−6 | Clonogenic Survival, % | ||
| Mean (SE) | Fold control | Mean (SE) | Mean (SE) | Fold control | Mean (SE) | |
| Cr, 0 μM | 18.1 (2.3) | 10.1 (1.5) | ||||
| Cr, 6 μM | 28.0 (7.4) | 1.5 | 58.0 (1.0) | 36.2 (14.4) | 3.5 | 64.5 (6.5) |
| Cr, 9 μM | 54.1 (3.1) | 3.0* | 33.0 (19.0) | 52.9 (23.7) | 5.0 | 39.0 (16.0) |
| Cr, 12 μM | 2.2 (0.8) | 0.1* | 3.0 (2.0) | 35.6 (4.4) | 3.5*+ | 22.5 (0.5) |
| MMS, 0 μM | 9.9 (1.8) | 20.2 (3.8) | ||||
| MMS, 500 μM | 51.1 (3.6) | 5.3* | 76.0 (8.0) | 82.4 (5.7) | 4.2* | 74.0 (2.0) |
: Statistically significant difference from SOV 0 μM at p<0.05
: p=0.0550.
: Statistically significant difference from control at p<0.05.
We also examined the effect of SOV on HPRT mutation frequency in Chinese hamster ovary (CHO) cells, a standard model for forward mutagenesis analysis at this locus (Table 1 B). Mutation frequency increased at 6 and 9 μM Cr(VI), reaching statistical significance after exposure to 9 μM Cr(VI). Mutation frequency declined after exposure to 12 μM Cr(VI), presumably due to toxicity, and similar to that observed in HLF cells. Similar to its effect in HLF cells, 500 μM MMS induced a 5-fold induction in mutation frequency in CHO cells. SOV enhanced Cr(VI)-induced mutation frequency, reaching statistical significance in cells exposed to 12 μM Cr(VI), despite the toxicity-driven decrease in mutagenesis by Cr(VI) alone. At 12 μM Cr(VI) SOV increased Cr(VI)-induced mutation frequency by around 16-fold, which was accompanied by a 7-fold increase in clonogenic survival at the same Cr(VI) concentration. Again, the PTP inhibitor was without effect on MMS-induced mutagenesis.
4. Discussion
The results of the present study suggest that maintenance of tyrosine phosphorylation through PTP inhibition by SOV increases clonogenic survival in the presence of Cr(VI)-induced genotoxic insult. Our data strongly indicate that the lack of a protracted growth arrest after Cr(VI) treatment, in the presence of SOV, is the mechanism of enhanced clonogenic survival. The enhanced survival induced by SOV after Cr(VI) exposure was not due to decreased Cr-DNA adduct burden. Importantly, our data show that the SOV enhances Cr(VI)-induced mutation frequency. Taken together, these results underline the potential for genomic instability as a result of loss of checkpoint control and highlight a potential role for PTPs in early neoplastic progression after initial genotoxic stress.
There is considerable evidence that dysregulated protein tyrosine phosphorylation is responsible for the maintenance of proliferative signals and is involved in the early stages of neoplasia [1;18]. While protein tyrosine kinases catalyze the addition of phosphate, PTPs catalyze the removal (for review, see [19]). Consequently, certain PTPs have been proposed to function as tumor suppressors in cancer development, and have been regarded to have therapeutic target potential [19-21]. Notably, the present study is the first report that PTP inhibition by SOV results in a non-lethal bypass of growth arrest in the face of genotoxic insult. Vanadate is widely used as a PTP inhibitor, acting as a reversible transition-state analog of phosphate, thus mimicking the phosphorous group of phosphotyrosine, and binding to the active site of the PTP [22]. SOV has been shown to activate tyrosine phosphorylation in vivo, in association with its PTP inhibitory activity [23], which we have also found in the present study. SOV has been reported to activate both AKT and ERK pathways [24-26], which is presumably through PTP inhibition, highlighting a role for tyrosine phosphorylation in the regulation cell survival. SOV has also been reported to inhibit both Na+/K+-ATPase [27]as well as dynein ATPase activity [28], which would suggest a non-PTP inhibitor function for SOV, however the concentrations of SOV used in these respective studies were around 30-fold higher than those used in the present study.
Previous reports have shown vanadate-induced PTP inhibition to enhance survival by decreasing apoptosis in primary mammary epithelial cells [29] as well as in rat brain and cardiac models of ischemic injury [30-32]. However, in the present in vitro study we observed no effect of SOV on Cr(VI)-induced apoptosis. It has also been reported that SOV was sufficient to enhance S phase progression in murine 3T3 and epithelial cells [33;34], although SOV treatment alone had no apparent effect on cell proliferation at concentrations ≤10 μM in the present studies. Furthermore, at concentrations ≥ 25 μM, SOV decreased clonogenic survival in our studies, consistent with reports of vanadate cytotoxity at higher concentrations [35;36].
As early as 4 days after Cr exposure, SOV was associated with an enhanced rate of proliferation. This is consistent with our finding that SOV induced a bypass of both the Cr-induced G1/S and G2/M checkpoints (data not shown). We analyzed the effect of SOV on changes in gene expression induced by 1μM Cr(VI) after 24h, which is the concentration and time period in which we observed checkpoint bypass in our aforementioned studies. Taken together, the data suggest that potential transcriptional mechanisms by which SOV bypasses Cr(VI)-induced growth arrest may be through both up regulation of genes that positively regulate cell cycle progression and down regulation of cell cycle inhibiting genes. Moreover, at least for a subset of genes, SOV appears to abrogate the Cr(VI)-induced upregulation of cell cycle inhibiting genes, as well as to abrogate the Cr(VI)-induced downregulation of genes that positively regulate cell cycle progression.
Resistance to growth inhibition is considered to be an early event in the multi-step process of cancer progression [37]. Dysregulation of cell cycle checkpoint regulators and/or survival signaling pathways have been reported to override growth arrest after DNA damage, but this is often at the expense of cellular survival. Activation of Cdk1 after etoposide treatment results in a DNA damage checkpoint override and subsequent sensitization, leading to cell death in HeLa cells [38]. Inhibition of checkpoint effector pathways by caffeine has been shown to increase sensitization to DNA damaging agents leading to cell death in several cell lines [39;40]. A recent report indicates that both the overexpression of c-MYC and the activating mutation of RAS were able to override the G2-restriction point in serum-starved mouse embryonic fibroblasts; however c-MYC overexpression resulted in increased apoptosis, whereas RAS activation resulted in continued proliferation in the absence of serum [41]. Studies by Knauf et al., demonstrated that expression of HRASV12 in rat thyroid PCCL3 cells accelerated cell cycle progression after ionizing radiation, corresponding to an increase in chromosomal instability [42;43]. Moreover, studies by Kandel et al. report a G2/M override by AKT was accompanied by the accumulation of mutations after exposure to 6-TG in Rat1a cells [44]. Indeed, our data suggest that PTP inhibition by SOV overrides Cr-induced growth arrest, enhances the rate of Cr-induced forward mutations at the HPRT locus in two different cell types, consistent with enhanced clonogenic survival. Studies by Mailhes et al. found that vanadate induced premature anaphase in oocytes as well as aneuploidy and polyploidy in mouse bone marrow cells [45]. These data are consistent with those of other studies reporting aneuploidy in vivo in bone marrow cells as well as micronuclei in lymphocytes after vanadium exposure [46-48]. However, SOV exposure alone was not found to be mutagenic in our studies. Moreover, SOV co-treatment had no effect on either MMS-induced mutagenesis or clonogenic lethality in the respective HLF and CHO cell lines, which strongly points to a correlation of genotoxin-induced genomic instability with cell survival. This could be related to genotoxin-specific DNA damage response pathways. As described below, Cr(VI) induces a complex array of DNA damage, while MMS exposure results in single base lesions. Indeed, as shown in our present and previous studies [49], exposure to Cr(VI) (1 μM) induced a prolonged growth arrest for over 6 days after end of the treatment. Notably, SOV significantly increased cell proliferation in the presence of Cr(VI), as early as 5 days after exposure. In contrast, MMS-induced single base lesions, which should be rapidly repaired, are associated with a transient S phase arrest in human fibroblasts [50]. Thus, the positive effect of SOV on cell cycle progression occurs during a period in which cells are still repairing Cr(VI)-DNA damage, while the associated enhanced survival indicates that potentially lethal Cr(VI)-induced damage may have already been repaired. Taken together, these studies support a role for SOV-mediated PTP inhibition in cell cycle checkpoint override, leading to genomic instability.
SOV increased clonogenic survival after Cr(VI) exposure but had no effect on clonogenic survival after MMS exposure. As detailed earlier, this may be related to genotoxin-specific cellular damage. Under physiological conditions, Cr exists as an oxyanion that enters the cell through an anion transporter, after which Cr(VI) is metabolically reduced to genotoxic species by cellular reducing agents such as glutathione and ascorbate [51]. The reduction process leads to a complex array of DNA damage that includes Cr-monoadducts to both DNA bases and sugar-phosphate backbone, strand breaks, oxidized bases, DNA-protein crosslinks, abasic sites, Asc-Cr(VI) ternary adducts and DNA-Cr-DNA interstrand crosslinks (ICLs) (for review, see [8]). We and others have highlighted the importance of both the ATM and the nucleotide excision repair pathways in repair of Cr-induced DNA lesions [49;52-57]. In contrast, MMS is a monofunctional DNA methylating agent, and base excision repair has been shown to be critical for MMS-induced lesions [58]. Therefore, it remains possible that the effect of SOV on Cr-induced survival is related to the activation of DNA damage response/repair pathways that are not affected by MMS exposure. Although Cr-DNA adduct levels were unaffected by SOV, as measured immediately after Cr(VI) exposure, this does not exclude the possibility that SOV may alter DNA damage repair pathways or repair kinetics.
Acquired resistance to genotoxin-induced death is a hallmark of neoplastic progression. Therefore, dysregulation of survival signaling may promote cell survival in the face of genotoxin-induced damage, which in turn may contribute to genomic instability and promote early stage carcinogenesis. The delineation of the molecular circuitry that promotes cell survival in the face of genotoxin-induced damage will serve to improve our understanding of carcinogenic mechanisms of action while potentially identifying biomarkers for risk assessment, and may have the added benefit of identifying molecular targets for rational drug design in anti-cancer therapy.
Supplementary Material
Supplemental Figure 1: Phosphotyrosine protein expression after SOV exposure: HLF cells were incubated with 10 or 25 μM SOV for 24h. Following this period, cells were gently scraped into RIPA buffer (1% Igepal, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) containing a cocktail of protease and phosphatase inhibitors, and cellular lysates were processed for immunoblotting. Equal amounts of protein from 3 independent experiments were separated by SDS-PAGE, and electroblotted onto PVDF membranes (Bio-Rad, Hercules, CA). Blots were probed with a monoclonal phosphotyrosine antibody (clone 4G10, Upstate Biotechnology, Lake Placid, NY), followed by a horseradish peroxidase-linked goat anti-mouse secondary antibody. The secondary antibody was detected by enhanced chemiluminescence (NEN, Boston, MA) after exposure to x-ray film.
Supplemental Figure 2: Effect of tyrosine phosphatase inhibition on apoptosis after exposure to Cr(VI): HLF cells were incubated without or with 1−6 μM Cr(VI) for 24 h in the absence or presence of 10 or 25 μM SOV followed by culture in complete medium without Cr(VI) for an additional 24 h. Apoptosis was assessed as the proportion of cells with translocated phosphatidylserine, as determined by binding of fluorescent annexin V. Data are expressed as % of apoptotic cells and are the means ± SE of 3 independent experiments. *, **: Statistically significant difference from control at p<0.05, p<0.01.
Supplemental Table 1: Effect of SOV on Cr-DNA adducts. HLF cells were incubated with 1 or 2 μM Na251CrO4 for 24 h, in the absence or presence of 10 μM SOV, and DNA was isolated. DNA-associated radioactivity (Cr adducts) was determined by scintillation counting. Data are expressed as pmol Cr/μg DNA, and are the means ± SE of 2 independent experiments, performed in triplicate.
Supplemental Table 2: Transcripts associated with cell cycle and cell proliferation. HLF cells were incubated without or with 1 μM Cr(VI) for 24 h in the absence (control, Ctl) or presence of 10 μM SOV, and RNA was isolated using RNAbee followed by a secondary purification on Qiagen RNAeasy Mini columns. RNA quality was assessed by Agilent bioanalyzer and RNA Integrity Numbers (RIN) for all samples were >9.9. Labeled cRNA was hybridized to GeneChip Human Genome U133 Plus 2.0 Array and arrays were scanned with an Agilent laser scanner. The raw expression data were analyzed by GeneSpring (Silicon Genetics) for differentially regulated genes after respective treatment. Data set was filtered by 1-way ANOVA with 5% FDR and 1,880 genes out of total 54,675 transcripts were chosen. Transcripts associated with cell cycle and cell proliferation (according to GO Biological Process) were selected and further filtered by expression fold cutoff at 1.5. Expression data are the average of the normalized log10 values from 4 independent RNA samples per treatment, and are grouped as follows: A) genes upregulated by Cr(VI) treatment both in the absence and presence of SOV, as compared to control; B) genes upregulated by Cr(VI) treatment as compared to both control and Cr(VI) + SOV co-treatment; C) genes that were upregulated by Cr(VI) + SOV co-treatment, as compared to both control and Cr(VI) treatment alone; D) genes downregulated by Cr(VI) + SOV co-treatment, when compared to both control and Cr(VI) treatment alone; E) genes were downregulated by Cr(VI) treatment as compared to both control and Cr(VI) + SOV co-treatment. Genes were further classified as either positive or negative regulators of cell cycle progression. A designation of no/unknown was used for the respective following criteria: no apparent cell proliferation function was identified; gene expression reported to correlate with both positive and negative regulation of cell cycle progression; relationship of gene expression to cell cycle progression was cell context-specific, with no known function in lung fibroblasts; no information could be obtained.
Acknowledgements
The authors thank Dr. Norman Lee and Dr. Bernard Bouscarel for their helpful discussions. This work was supported by NIH grants ES05304 and ES09961 to SRP, and CA107972 to SC as well as a grant from the Catherine Birch McCormick Genomics Center to SC.
Footnotes
Conflict of interest statement
None.
Reference List
- 1.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat.Rev.Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez F, Sellers WR. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochimica et Biophysica Acta. 2000;1470:M21–M35. doi: 10.1016/s0304-419x(99)00032-3. [Review] [125 refs] [DOI] [PubMed] [Google Scholar]
- 3.Chu Y, Solski PA, Khosravi-Far R, Der CJ, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. Journal of Biological Chemistry. 1996;271:6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 4.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat.Rev.Mol.Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 5.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat.Rev.Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 6.Chromium, nickel and welding. IARC Monogr Eval.Carcinog.Risks Hum. 1990;49:1–648. 1−648. [PMC free article] [PubMed] [Google Scholar]
- 7.Langard S. One hundred years of chromium and cancer: a review of epidemiological evidence and selected case reports. Am J Ind Med. 1990;17:189–215. doi: 10.1002/ajim.4700170205. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien TJ, Ceryak S, Patierno SR. Complexities of chromium carcinogenesis: role of cellular response, repair and recovery mechanisms. Mutat.Res. 2003;533:3–36. doi: 10.1016/j.mrfmmm.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Plunkett ER. Handbook of Industrial Toxicology. Chemical Publishing; New York, NY: 1976. [Google Scholar]
- 10.Cassarett and Doull's Toxicology. Maxwell-MacMillan-Pergamon; New York, New York: 1991. [Google Scholar]
- 11.Ceryak S, Zingariello C, O'Brien T, Patierno SR. Induction of pro-apoptotic and cell cycle-inhibiting genes in chromium(VI)-treated human lung fibroblasts: Lack of effect of ERK. Mol Cell Biochem. 2004;255:139–149. doi: 10.1023/b:mcbi.0000007270.82431.3e. [DOI] [PubMed] [Google Scholar]
- 12.Hamdan S, Morse B, Reinhold D. Nickel subsulfide is similar to potassium dichromate in protecting normal human fibroblasts from the mutagenic effects of benzo[a]pyrene diolepoxide. Environmental & Molecular Mutagenesis. 1999;33:211–218. doi: 10.1002/(sici)1098-2280(1999)33:3<211::aid-em5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard DE, Singh J, Carlisle DL, Patierno SR. Cyclosporin A inhibits chromium(VI)-induced apoptosis and mitochondrial cytochrome c release and restores clonogenic survival in CHO cells. Carcinogenesis. 2000;21:2027–2033. doi: 10.1093/carcin/21.11.2027. [DOI] [PubMed] [Google Scholar]
- 14.Pritchard DE, Ceryak S, Ha L, Fornsaglio JL, Hartman SK, O'Brien TJ, Patierno SR. Mechanism of apoptosis and determination of cellular fate in chromium(VI)-exposed populations of telomerase-immortalized human fibroblasts. Cell Growth Differ. 2001;12:487–496. [PubMed] [Google Scholar]
- 15.Vilcheck SK, O'Brien TJ, Pritchard DE, Ha L, Ceryak S, Fornsaglio JL, Patierno SR. Fanconi anemia complementation group A cells are hypersensitive to chromium(VI)-induced toxicity. Environmental Health Perspectives. 2002;110(Suppl 5):773–777. doi: 10.1289/ehp.02110s5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JL, Hsieh YC, Wu CW, Lee TC. Mutational specificity of chromium(VI) compounds in the hprt locus of Chinese hamster ovary-K1 cells. Carcinogenesis. 1992;13:2053–2057. doi: 10.1093/carcin/13.11.2053. [DOI] [PubMed] [Google Scholar]
- 17.Cheng L, Liu S, Dixon K. Analysis of repair and mutagenesis of chromium-induced DNA damage in yeast, mammalian cells, and transgenic mice. Environmental Health Perspectives. 1998;106(Suppl 4):1027–1032. doi: 10.1289/ehp.98106s41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 19.Easty D, Gallagher W, Bennett DC. Protein tyrosine phosphatases, new targets for cancer therapy. Curr.Cancer Drug Targets. 2006;6:519–532. doi: 10.2174/156800906778194603. [DOI] [PubMed] [Google Scholar]
- 20.Yi T, Lindner D. The role and target potential of protein tyrosine phosphatases in cancer. Curr.Oncol.Rep. 2008;10:114–121. doi: 10.1007/s11912-008-0019-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhang ZY. Protein tyrosine phosphatases: prospects for therapeutics. Curr.Opin.Chem Biol. 2001;5:416–423. doi: 10.1016/s1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- 22.Gordon JA. Use of vanadate as protein-phosphotyrosine phosphatase inhibitor. Methods Enzymol. 1991;201:477–482. doi: 10.1016/0076-6879(91)01043-2. 477−82. [DOI] [PubMed] [Google Scholar]
- 23.Scheving LA, Thomas JR, Zhang L. Regulation of intestinal tyrosine phosphorylation and programmed cell death by peroxovanadate. Am.J Physiol. 1999;277:C572–C579. doi: 10.1152/ajpcell.1999.277.3.C572. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto J, Morioka M, Hasegawa Y, Kawano T, Yoshinaga Y, Maeda T, Yano S, Kai Y, Fukunaga K, Kuratsu J. Sodium orthovanadate enhances proliferation of progenitor cells in the adult rat subventricular zone after focal cerebral ischemia. J Pharmacol.Exp.Ther. 2006;318:982–991. doi: 10.1124/jpet.106.104562. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, Bhatt AJ, Fratkin JD, Rhodes PG. Neuroprotective effects of sodium orthovanadate after hypoxic-ischemic brain injury in neonatal rats. Brain Res.Bull. 2008;76:102–108. doi: 10.1016/j.brainresbull.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Wu DN, Pei DS, Wang Q, Zhang GY. Down-regulation of PTEN by sodium orthovanadate inhibits ASK1 activation via PI3-K/Akt during cerebral ischemia in rat hippocampus. Neurosci.Lett. 2006;404:98–102. doi: 10.1016/j.neulet.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Todorov LD, Mihaylova-Todorova ST, Choe SM, Westfall DP. Facilitation of the purinergic contractile response of the guinea pig vas deferens by sodium orthovanadate. J Pharmacol.Exp.Ther. 2005;312:407–416. doi: 10.1124/jpet.104.072991. [DOI] [PubMed] [Google Scholar]
- 28.Wang XM, Huang TH, Xie QD, Zhang QJ, Ruan Y. Effect of dynein inhibitor on mouse oocyte in vitro maturation and its cyclin B1 mRNA level. Biomed.Environ.Sci. 2004;17:341–349. [PubMed] [Google Scholar]
- 29.Furlong F, Finlay D, Martin F. PTPase inhibition restores ERK1/2 phosphorylation and protects mammary epithelial cells from apoptosis. Biochem.Biophys.Res.Commun. 2005;336:1292–1299. doi: 10.1016/j.bbrc.2005.08.260. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Tian LH, Xie HF, Shi W, Feng H, Li J, Chen FW. [Effects of CD147 on the production of matrix metalloproteinase-9 by fibroblasts and the invasion of melanoma cells] Zhong.Nan.Da.Xue.Xue.Bao.Yi.Xue.Ban. 2005;30:249–252. [PubMed] [Google Scholar]
- 31.Yang P, Dankowski A, Hagg T. Protein tyrosine phosphatase inhibition reduces degeneration of dopaminergic substantia nigra neurons and projections in 6-OHDA treated adult rats. Eur.J Neurosci. 2007;25:1332–1340. doi: 10.1111/j.1460-9568.2007.05384.x. [DOI] [PubMed] [Google Scholar]
- 32.Bhuiyan MS, Takada Y, Shioda N, Moriguchi S, Kasahara J, Fukunaga K. Cardioprotective effect of vanadyl sulfate on ischemia/reperfusion-induced injury in rat heart in vivo is mediated by activation of protein kinase B and induction of FLICE-inhibitory protein. Cardiovasc.Ther. 2008;26:10–23. doi: 10.1111/j.1527-3466.2008.00039.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith JB. Vanadium ions stimulate DNA synthesis in Swiss mouse 3T3 and 3T6 cells. Proc.Natl.Acad.Sci.U.S.A. 1983;80:6162–6166. doi: 10.1073/pnas.80.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones TR, Reid TW. Sodium orthovanadate stimulation of DNA synthesis in Nakano mouse lens epithelial cells in serum-free medium. J Cell Physiol. 1984;121:199–205. doi: 10.1002/jcp.1041210125. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Leonard SS, Huang C, Vallyathan V, Castranova V, Shi X. Role of reactive oxygen species and MAPKs in vanadate-induced G(2)/M phase arrest. Free Radical Biology & Medicine. 2003;34:1333–1342. doi: 10.1016/s0891-5849(03)00145-x. [DOI] [PubMed] [Google Scholar]
- 36.Huang C, Zhang Z, Ding M, Li J, Ye J, Leonard SS, Shen HM, Butterworth L, Lu Y, Costa M, Rojanasakul Y, Castranova V, Vallyathan V, Shi X. Vanadate induces p53 transactivation through hydrogen peroxide and causes apoptosis. Journal of Biological Chemistry. 2000;275:32516–32522. doi: 10.1074/jbc.M005366200. [DOI] [PubMed] [Google Scholar]
- 37.Hahn WC, Weinberg RA. Rules for making human tumor cells. N.Engl.J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 38.Jiang H, Luo S, Li H. Cdk5 activator-binding protein C53 regulates apoptosis induced by genotoxic stress via modulating the G2/M DNA damage checkpoint. J Biol Chem. 2005;280:20651–20659. doi: 10.1074/jbc.M413431200. [DOI] [PubMed] [Google Scholar]
- 39.Powell SN, DeFrank JS, Connell P, Eogan M, Preffer F, Dombkowski D, Tang W, Friend S. Differential sensitivity of p53(−) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]
- 40.Bache M, Pigorsch S, Dunst J, Wurl P, Meye A, Bartel F, Schmidt H, Rath FW, Taubert H. Loss of G2/M arrest correlates with radiosensitization in two human sarcoma cell lines with mutant p53. Int.J Cancer. 2001;%20:110–117. doi: 10.1002/ijc.1002. 96. [DOI] [PubMed] [Google Scholar]
- 41.Foijer F, Simonis M, van Vliet M, Wessels L, Kerkhoven R, Sorger PK, Te RH. Oncogenic pathways impinging on the G2-restriction point. Oncogene. 2008;27:1142–1154. doi: 10.1038/sj.onc.1210724. [DOI] [PubMed] [Google Scholar]
- 42.Knauf JA, Ouyang B, Knudsen ES, Fukasawa K, Babcock G, Fagin JA. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J Biol Chem. 2006;281:3800–3809. doi: 10.1074/jbc.M511690200. [DOI] [PubMed] [Google Scholar]
- 43.Saavedra HI, Knauf JA, Shirokawa JM, Wang J, Ouyang B, Elisei R, Stambrook PJ, Fagin JA. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene. 2000;19:3948–3954. doi: 10.1038/sj.onc.1203723. [DOI] [PubMed] [Google Scholar]
- 44.Kandel ES, Skeen J, Majewski N, Di Cristofano A, Pandolfi PP, Feliciano CS, Gartel A, Hay N. Activation of Akt/protein kinase B overcomes a G(2)/m cell cycle checkpoint induced by DNA damage. Molecular & Cellular Biology. 2002;22:7831–7841. doi: 10.1128/MCB.22.22.7831-7841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mailhes JB, Hilliard C, Fuseler JW, London SN. Vanadate, an inhibitor of tyrosine phosphatases, induced premature anaphase in oocytes and aneuploidy and polyploidy in mouse bone marrow cells. Mutat.Res. 2003;538:101–107. doi: 10.1016/s1383-5718(03)00108-6. [DOI] [PubMed] [Google Scholar]
- 46.Ciranni R, Antonetti M, Migliore L. Vanadium salts induce cytogenetic effects in in vivo treated mice. Mutat.Res. 1995;343:53–60. doi: 10.1016/0165-1218(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 47.Migliore L, Zotti-Martelli L, Scarpato R. Detection of chromosome loss and gain induced by griseofulvin, estramustine, and vanadate in binucleated lymphocytes using FISH analysis. Environ.Mol.Mutagen. 1999;34:64–68. [PubMed] [Google Scholar]
- 48.Migliore L, Scarpato R, Falco P. The use of fluorescence in situ hybridization with a beta-satellite DNA probe for the detection of acrocentric chromosomes in vanadium-induced micronuclei. Cytogenet.Cell Genet. 1995;69:215–219. doi: 10.1159/000133967. [DOI] [PubMed] [Google Scholar]
- 49.Ha L, Ceryak S, Patierno SR. Chromium (VI) activates ataxia telangiectasia mutated (ATM) protein: Requirement of ATM for both apoptosis and recovery from terminal growth arrest. Journal of Biological Chemistry. 2003;278:17885–17894. doi: 10.1074/jbc.M210560200. [DOI] [PubMed] [Google Scholar]
- 50.Horton JK, Stefanick DF, Kedar PS, Wilson SH. ATR signaling mediates an S-phase checkpoint after inhibition of poly(ADP-ribose) polymerase activity. DNA Repair (Amst) 2007;6:742–750. doi: 10.1016/j.dnarep.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Flora S. Threshold mechanisms and site specificity in chromium(VI) carcinogenesis. Carcinogenesis. 2000;21:533–541. doi: 10.1093/carcin/21.4.533. [Review] [94 refs] [DOI] [PubMed] [Google Scholar]
- 52.Ha L, Ceryak S, Patierno SR. Generation of S phase-dependent DNA double strand breaks by Cr(VI) exposure: involvement of ATM in Cr(VI) induction of {gamma}-H2AX. Carcinogenesis. 2004 doi: 10.1093/carcin/bgh242. [DOI] [PubMed] [Google Scholar]
- 53.O'Brien TJ, Brooks BR, Patierno SR. Nucleotide excision repair functions in the removal of chromium-induced DNA damage in mammalian cells. Mol.Cell Biochem. 2005;279:85–95. doi: 10.1007/s11010-005-8225-0. [DOI] [PubMed] [Google Scholar]
- 54.Reynolds M, Peterson E, Quievryn G, Zhitkovich A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J Biol Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 55.Xie H, Wise SS, Holmes AL, Xu B, Wakeman TP, Pelsue SC, Singh NP, Wise JP., Sr Carcinogenic lead chromate induces DNA double-strand breaks in human lung cells. Mutat.Res. 2005;586:160–172. doi: 10.1016/j.mrgentox.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wakeman TP, Kim WJ, Callens S, Chiu A, Brown KD, Xu B. The ATM-SMC1 pathway is essential for activation of the chromium[VI]-induced S-phase checkpoint. Mutat.Res. 2004;554:241–251. doi: 10.1016/j.mrfmmm.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Sobol RW, Wilson SH. Mammalian DNA beta-polymerase in base excision repair of alkylation damage. Prog.Nucleic Acid Res.Mol.Biol. 2001;68:57–74. doi: 10.1016/s0079-6603(01)68090-5. 57−74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Phosphotyrosine protein expression after SOV exposure: HLF cells were incubated with 10 or 25 μM SOV for 24h. Following this period, cells were gently scraped into RIPA buffer (1% Igepal, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) containing a cocktail of protease and phosphatase inhibitors, and cellular lysates were processed for immunoblotting. Equal amounts of protein from 3 independent experiments were separated by SDS-PAGE, and electroblotted onto PVDF membranes (Bio-Rad, Hercules, CA). Blots were probed with a monoclonal phosphotyrosine antibody (clone 4G10, Upstate Biotechnology, Lake Placid, NY), followed by a horseradish peroxidase-linked goat anti-mouse secondary antibody. The secondary antibody was detected by enhanced chemiluminescence (NEN, Boston, MA) after exposure to x-ray film.
Supplemental Figure 2: Effect of tyrosine phosphatase inhibition on apoptosis after exposure to Cr(VI): HLF cells were incubated without or with 1−6 μM Cr(VI) for 24 h in the absence or presence of 10 or 25 μM SOV followed by culture in complete medium without Cr(VI) for an additional 24 h. Apoptosis was assessed as the proportion of cells with translocated phosphatidylserine, as determined by binding of fluorescent annexin V. Data are expressed as % of apoptotic cells and are the means ± SE of 3 independent experiments. *, **: Statistically significant difference from control at p<0.05, p<0.01.
Supplemental Table 1: Effect of SOV on Cr-DNA adducts. HLF cells were incubated with 1 or 2 μM Na251CrO4 for 24 h, in the absence or presence of 10 μM SOV, and DNA was isolated. DNA-associated radioactivity (Cr adducts) was determined by scintillation counting. Data are expressed as pmol Cr/μg DNA, and are the means ± SE of 2 independent experiments, performed in triplicate.
Supplemental Table 2: Transcripts associated with cell cycle and cell proliferation. HLF cells were incubated without or with 1 μM Cr(VI) for 24 h in the absence (control, Ctl) or presence of 10 μM SOV, and RNA was isolated using RNAbee followed by a secondary purification on Qiagen RNAeasy Mini columns. RNA quality was assessed by Agilent bioanalyzer and RNA Integrity Numbers (RIN) for all samples were >9.9. Labeled cRNA was hybridized to GeneChip Human Genome U133 Plus 2.0 Array and arrays were scanned with an Agilent laser scanner. The raw expression data were analyzed by GeneSpring (Silicon Genetics) for differentially regulated genes after respective treatment. Data set was filtered by 1-way ANOVA with 5% FDR and 1,880 genes out of total 54,675 transcripts were chosen. Transcripts associated with cell cycle and cell proliferation (according to GO Biological Process) were selected and further filtered by expression fold cutoff at 1.5. Expression data are the average of the normalized log10 values from 4 independent RNA samples per treatment, and are grouped as follows: A) genes upregulated by Cr(VI) treatment both in the absence and presence of SOV, as compared to control; B) genes upregulated by Cr(VI) treatment as compared to both control and Cr(VI) + SOV co-treatment; C) genes that were upregulated by Cr(VI) + SOV co-treatment, as compared to both control and Cr(VI) treatment alone; D) genes downregulated by Cr(VI) + SOV co-treatment, when compared to both control and Cr(VI) treatment alone; E) genes were downregulated by Cr(VI) treatment as compared to both control and Cr(VI) + SOV co-treatment. Genes were further classified as either positive or negative regulators of cell cycle progression. A designation of no/unknown was used for the respective following criteria: no apparent cell proliferation function was identified; gene expression reported to correlate with both positive and negative regulation of cell cycle progression; relationship of gene expression to cell cycle progression was cell context-specific, with no known function in lung fibroblasts; no information could be obtained.