Abstract
Holocarboxylase synthetase (HCS) catalyzes the binding of the vitamin biotin to carboxylases and histones. Carboxylases mediate essential steps in macronutrient metabolism. For example, propionyl-CoA carboxylase (PCC) catalyzes the carboxylation of propionyl-CoA in the metabolism of odd-chain fatty acids. HCS comprises four putative domains, i.e., the N-terminus, the biotin transfer/ATP binding domain, a putative linker domain, and the C-terminus. Both N- and C-termini are essential for biotinylation of carboxylases by HCS, but the exact functions of these two domains in enzyme catalysis are unknown. Here we tested the hypothesis that N- and C-termini play roles in substrate recognition by HCS. Yeast-two-hybrid (Y2H) assays were used to study interactions between the four domains of human HCS with p67, a PCC-based polypeptide and HCS substrate. Both N- and C-termini interacted with p67 in Y2H assays, whereas the biotin transfer/ATP-binding and the linker domains did not interact with p67. The essentiality of N- and C-termini for interactions with carboxylases was confirmed in rescue experiments with mutant Saccharomyces cerevisiae, using constructs of truncated human HCS. Finally, a computational biology approach was used to model the 3D structure of human HCS and identify amino acid residues that interact with p67. In silico predictions were consistent with observations from Y2H assays and yeast rescue experiments, and suggested docking of p67 near Arg508 and Ser515 within the central domain of HCS.
Keywords: BirA, holocarboxylase synthetase, domains, p67, propionyl-CoA carboxylase
Introduction
Biotin is a water-soluble vitamin that serves as a coenzyme for acetyl-CoA carboxylases 1 and 2 (ACC1 and ACC2), 3-methylcrotonyl-CoA carboxylase (MCC), pyruvate carboxylase (PC), and propionyl-CoA carboxylase (PCC). These carboxylases play pivotal roles in human metabolism [1]. Both ACC1 and ACC2 catalyze the incorporation of bicarbonate into malonyl-CoA, a key regulatory step in fatty acid synthesis (ACC1) and mitochondrial fatty acid transport (ACC2). 3-Methylcrotonyl-CoA carboxylase is involved in the catabolism of the branched-chain amino acid leucine. Pyruvate carboxylase catalyzes the carboxylation of pyruvate to produce oxaloacetate in gluconeogenesis. Finally, PCC is involved in the conversion of propionyl-CoA to methylmalonyl-CoA in the metabolism of odd-chain fatty acids.
Biotin is also attached covalently to at least 11 distinct lysine residues in histones H2A, H3, and H4 [2-5]. Biotinylation of histones participates in the cross-talk among various histone modifications and in gene regulation [6-9]. For example, K12-biotinylated histone H4 co-localizes with K9-dimethylated histone H3 in pericentrometric heterochromatin and repressed genes [9, 10]. In addition to its role in heterochromatin structures and gene repression, biotinylation of histone H4 appears to participate in the cellular response to DNA damage [11], and in the repression of retrotransposons [12].
The attachment of biotin to carboxylases and histones is mediated by holocarboxylase synthetase (HCS, EC 6.3.4.10). The open reading frame of full-length HCS encodes 726 amino acids [13] but the existence of at least three splicing variants (76, 82, and 86 kDa) has been confirmed [14]. HCS has been detected in cytoplasm, mitochondria, cell nuclei, and the nuclear lamina [14, 15].
BirA is the microbial ortholog of HCS and its investigation has provided valuable insights into HCS structure and catalysis, and the amino acid sequences in both proteins are 21% identical. The three-dimensional structure of E. coli BirA has been solved by x-ray crystallography [16] and consists of three domains: an N-terminal domain, a central domain, and a C-terminal domain [17]. The central domains in BirA and HCS contain binding sites for both ATP and biotin and their functions in catalysis are well understood [18-20]. In contrast, the amino acid sequence of the N-termini in BirA and human HCS are fairly distinct, thereby preventing predictions of HCS structure solely based on analogies to the BirA N-terminus. The N-terminus of BirA contains 60 amino acid residues, whereas that of human HCS is up to 446 amino acids long, depending on splicing. It has been speculated that the N-terminal domain in human HCS might play roles in gene regulation, protein-protein interactions (PPI), and substrate recognition [14, 20]. The C-terminal domain in BirA is essential for biotinyl ligase activity [17, 20] and might also participate in substrate recognition [17]. It remains to be determined whether the C-terminus in HCS plays a similar role, given that BirA and HCS have distinct biotinylations targets: biotin carboxyl carrier protein for BirA, and five distinct carboxylases and three major classes of histones for HCS.
The three-dimensional structure of HCS has not yet been determined. Here we provide novel insights into the structure of human HCS and the functions of its domains by using molecular modeling approaches, yeast-two-hybrid (Y2H) assays, and rescue experiments in yeast mutants.
Materials & Methods
Cloning of p67 and fusion to a Gal4 activation domain
The polypeptide p67 comprises the 67 C-terminal amino acids in human PCC (GenBank accession #AAA60035), including the biotin-binding site K669 [18]. p67 is a well-established substrate for studies of HCS [18, 20]. Here we used p67 to investigate domain-domain interactions between substrate and HCS. p67 was cloned from human liver cDNA into the cloning vector pSTBlue (Novagen; San Diego, CA) by using forward primer 5′-gaattcctgcgttccccgatg-3′ and reverse primer 5′-ctgctcgtggagctggaatgaggatcc-3′. The resulting plasmid was named “p67-pSTBlue.” p67-pSTBlue was sequenced in the DNA core facility of the University of Nebraska-Lincoln, and digested using EcoRI and BamHI. The insert encoding p67 was fused to a Gal4 activation domain, using vector pGADT7 (Clontech; Mountain View, CA); this plasmid was named “pGADT7-p67” and its identity was confirmed by sequencing.
Cloning of HCS and its different domains into a Gal4 binding domain vector
Plasmid pUC-CAGG encodes full-length human HCS (GenBank accession #D23672) and was kindly provided by Y. Suzuki (Department of Biochemical Genetics, Tohoku University School of Medicine, Sendai, Japan) [13]. Using pUC-CAGG as a template, full-length human HCS was PCR amplified using the following primers: 5′-actgaattcctcatggaagatagactccac-3′ (forward) and 5′-actgtcgacttaccgccgtttggggaggatg-3′ (reverse). The PCR product was purified, digested with EcoRI and SalI, and sub-cloned directly into a linearized pGBKT7 vector (Clontech, cat.# K1612-1) to produce plasmid “pGBKT7-HCS.” Plasmid identity was confirmed by sequencing.
In silico analysis of human HCS by using the Pfam database [21] predicted four major HCS domains (Fig. 1). The following HCS domains were PCR amplified for studies of interactions between p67 in Y2H assays: the N-terminal domain (M1-F446) was amplified using primers 5′-catggaggccgaattaatggaagatagactccac-3′ (forward) and 5′-gccgctgcaggtcgaccgaaggcctccatgttggtc-3′ (reverse); the biotin transfer/ATP binding (central) domain (F471-S575) was amplified using primers 5′-ccatggaggccgaattatttgccgaagtgacccc-3′ (forward) and 5′-gccgctgcaggtcgagcgtcactgtaataaatatc-3′ (reverse); the putative linker domain (T610-V668) was amplified using primers 5′-catggaggccgaattaaccatctgcatcaacg-3′ (forward) and 5′-gccgctgcaggtcgaccgtggacccagtatcgg-3′ (reverse); and the C-terminal domain (H669-R718) was amplified using primers 5′-catggaggccgaattccacagtggtcagcaagtc-3′ (forward) and 5′-gccgctgcaggtcgacctctcagcatgtcgaaggag-3′ (reverse) primers. PCR products were fused to a Gal4 BD, using pGBKT7 vector that was linearized with EcoRI and SalI and the infusion cloning system (Clontech, cat.# 631774); the four plasmids are denoted pGBKT7-HCS1-446, pGBKT7-HCS471-575, pGBKT7-HCS610-668, and pGBKT7-HCS669-718. Identities of clones were verified by sequencing.
Figure 1.
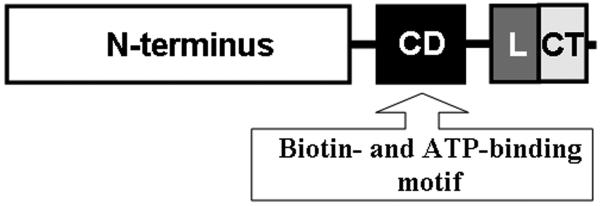
Predictions of four domains in human HCS: N-terminal domain (M1-F446); central domain “CD” (F471-S575) containing biotin transfer and ATP-binding sites; linker domain “L” (T610-V668); and C-terminal domain “CT” (H669-R718). The domains are drawn to scale.
Functional complementation of pGBKT7-HCS and Saccharomyces cerevisiae biotin protein ligase1 (BPL1)
We used a functional complementation approach to verify that HCS was correctly folded and retained its biotin ligase activity when fused to Gal4 BD. The BPL1 gene codes for the HCS ortholog BPL1 in Saccharomyces cerevisiae. The open reading frame of BPL1 plus 500 bp 5′ and 500 bp 3′ flanking sequences was PCR-amplified as a SalI-NotI fragment. This SalI-BPL1-NotI fragment was inserted into pRS416 (CEN-ARS-URA3), resulting in plasmid pLO70. The sequence of the BPL1 gene in pLO70 was confirmed by DNA sequencing. All yeast strains used in this work were derived from W303-1B (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1). W303-1B was transformed to Ura+ with plasmid pLO70, making strain YLO48 (W303-1B + pLO70).
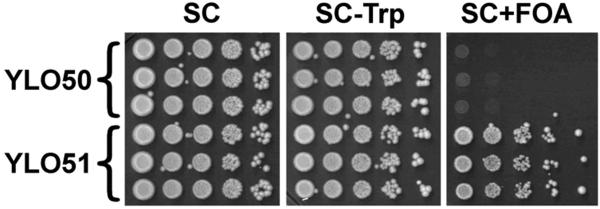
Strain YLO49 (W303-1B, bpl1 Δ::kanMX + pLO70) was made geneticin-resistant by transforming YLO48 with a PCR-generated fragment composed of kanMX flanked by 5′ and 3′ flanking sequences of the BLP1 coding sequence. YLO49 was transformed to Trp+ with pGBKT7 (Gal4 BD), pGBKT7-HCS (Gal4 BD fused with full-length HCS), pGBKT7-HCS1-446 (Gal4 BD fused with the HCS N-terminal domain), pGBKT7-HCS471-575 (Gal4 BD fused with the HCS central domain), pGBKT7-HCS610-668 (Gal4 BD fused with the HCS linker domain), and pGBKT7-HCS669-718 (Gal4 BD fused with the HCS C-terminal domain) to produce strains YLO50 (W303-1B, bpl1 Δ::kanMX + pLO70 + pGBKT7), YLO51 (W303-1B, bpl1 Δ::kanMX + pLO70 + pGBKT7-HCS), YLO52 (W303-1B, bpl1 Δ::kanMX + pLO70 + pGBKT7-HCS1-446), YLO53 (W303-1B, bpl1 Δ::kanMX + pLO70 + pGBKT7-HCS471-575), YLO54 (W303-1B, bpl1 Δ::kanMX + pLO70 + pGBKT7-HCS610-668), and YLO55 (W303-1B, bpl1 Δ::kanMX pLO70 + pGBKT7-HCS669-718) respectively. Ten-fold serial dilutions of three independent clones of each strain were plated on SC (synthetic complete), SC minus tryptophan (SC-Trp) and SC+FOA plates and grown for three days.
In additional rescue studies, the following two truncated forms of HCS were used. HCS lacking the N-terminal domain (M1-F446) was PCR amplified using the pUC-CAGG plasmid as a template and the following primers: 5′-tggaggaattcatgtcatcagaacatttcaacttag-3′ (forward) and 5′-tatatgtcgaccttaccgccgtttggg-3′ (reverse). The PCR products were cloned into pCR-Blunt II-TOPO vector (Invitrogen; Carlsbad, CA) before digestion with EcoRI and SalI and sub-cloning into the pGBKT7 vector for fusion to a Gal4 BD; the resulting vector is denoted pGBKT7-HCS_Ndel. A HCS fusion construct lacking the C-terminal domain (H669-R718) was generated as described for pGBKT7-HCS_Ndel, using the following primers: 5′-tggaggaattcatggaagatagactccacatggataatggactggtaccccaaaagattg-3′ (forward) and 5′-tatatgtcgaccttaccgccgtttggggaggatgaggttgacccagtatcggtaataaag-3′ (reverse). This plasmid is denoted pGBKT7-HCS_Cdel. Plasmids pGBKT7-HCS_Ndel and pGBKT7-HCS_Cdel were used to make strains YLO56 (W303-1B, bpl1 Δ::kanMX + pLO70 + pGBKT7-HCS_Ndel) and YLO57 (W303-1B, bpl1 Δ::kanMX + pL070 + pGBKT7-HCS_Cdel) to determine whether BPL1 mutants could be rescued by N- and C-truncated HCS.
Characterization of interactions between p67 and HCS
Interactions between p67 and HCS were investigated using Y2H assays by co-transforming Saccharomyces cerevisiae strain AH109 (Clontech, cat.# 630444) with various HCS-Gal4 BD fusion plasmids and pGADT7-p67. After transformation, yeast was plated on SD/-Leu, -Trp, +Kan and incubated until colonies became visible. Colonies were then re-suspended in 10 μL YPDA broth and spotted on SD/-Leu, -Trp, -His, -Ade, + Kan and incubated at 30°C in inverted position for 21 days. Plates were monitored daily for appearance of colonies; strong interactors are visible earlier than weak interactors. After 21 days of monitoring, the recovered clones were streaked out on SD/-Leu, -Trp, -His, -Ade, + Kan supplemented with X-α-gal (200 μl of 2 mg/ml in DMF) to screen for LacZ gene activation by X-alpha-Gal assay (Clontech, cat.# 630407).
Modeling of HCS and p67 interactions by computational biology
The 3D structure of full-length human HCS was modeled using PHYRE [22]. The 3D model of human HCS spanning amino acid sequences 393 through 715 was based on the crystal structure of E. coli BirA [23]. The sequences of BirA and human HCS have a FASTA E-value of 4.2e-31 which is considered significant [24]. Similar results were obtained by using the 3D-JIGSAW server [25].
A 3D-model of amino acids 1 to 66 in p67 was obtained by 3D-JIGSAW using biotin carboxyl carrier protein from Pyrococcus horikoshi as a template [26]. The 3D structure of p67 was independently confirmed by PHYRE for amino acids 2 to 65.
In order to assess the interactions between human HCS and p67, the docking of both proteins was analyzed by PATCHDOCK and further refined by FireDock [27]. PATCHDOCK calculates the spatial shape complementation of two structures and provides a list of potential complexes. In our docking calculation, protein surfaces were treated as if they were facing an aqueous phase. The likelihood of unrealistic predictions was further decreased by conducting analogous docking calculations for BirA and its substrate biotin carboxyl carrier protein [23]. Only docking interactions that were similar for HCS and p67 versus BirA and biotin carboxyl carrier protein were considered for further analyses. Data from docking models were refined using data from site-directed mutagenesis experiments with BirA [17]. 3D modeling of HCS/p67 interactions was completed by visual inspection of representations generated by PyMol [28] and Deep View [29].
Results
Functional characterization of the N and C termini of HCS
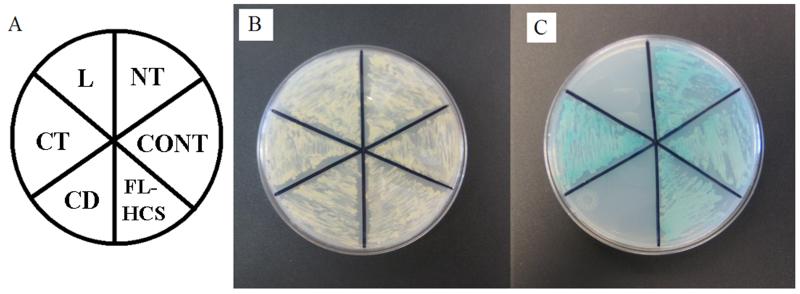
Full-length HCS interacted with p67 in Y2H (Fig. 2). Colonies became visible on plates after 14-15 days of culture, consistent with transient interactions typically observed for enzymes and their substrates [30, 31]. Of the four individual HCS domains tested, only M1-F446 (N-terminus) and H669-R718 (C-terminus) produced detectable interactions with p67. The C-terminal domain interacted with p67 more strongly than the N-terminal domain. In response to transformation with pGBKT7-HCS669-718, yeast colonies became visible on plates after only 4-5 days of culture. In response to transformation with pGBKT7-HCS1-466, yeast colonies became visible on plates after 8-9 days of culture. The observation that individual domains interacted stronger than the full length HCS might be explained by the fact that amino acids involved in establishing the HCS-p67 interaction were exposed and readily accessible in constructs coding for individual domains.
Figure 2.
The N- and C-termini in human HCS interact with p67 in Y2H. N-terminal domain (NT), central domain (CD), linker domain (L), C-terminal domain (CT), and full-length HCS (FL-HCS) were investigated. The interaction between p53 and T antigen was used as positive control (CONT). Panel A: plate layout. Panel B: Growth of the yeast host strain AH109 was transformed with Gal4BD-HCS domains and Gal4AD-p67 on SD/-Leu,-Trp, +Kan medium to confirm co-transformation with both plasmids. Panel C: Activation of reporter genes in AH109 yeast caused by HCS-p67 interactions.
Biological activity of hHCS and HCS domain deletions in yeast
Human HCS is biologically active in yeast. Deletions in YDL141W (the gene coding for BPL1) are known to be lethal for Saccharomyces cerevisiae. In our studies we transformed BPL1 mutants with the plasmid pGBKT7-HCS in which human HCS is fused to a Gal4 BD. Transformation with pGBKT7-HCS rescued BPL1 mutants, whereas non-transformed cells died (Fig. 3). This is consistent with the notion that hHCS functionally complements its yeast ortholog BPL1, and that results from Y2H assays are valid. Transformation of BPL1 mutant yeast with individual HCS domains (pGBKT7-HCS1-446, pGBKT7-HCS471-575, pGBKT7-HCS610-668, and pGBKT7-HCS669-718) or truncated forms of HCS (pGBKT7-HCS_Ndel and pGBKT7-HCS_Cdel) was not sufficient to rescue the mutants. This is consistent with the theory that both N-terminal and C-terminal domains in HCS are crucial for substrate recognition.
Figure 3.
Human HCS functionally complements biotin protein ligase (BPL1) in yeast. The YLO49 strain of S. cerevisiae was created by deleting the BPL1 gene; YLO49 yeast was transformed using an empty Gal4 BD vector (YLO50) or a plasmid coding for human HCS fused to Gal4 BD (YLO51).
3D modeling of HCS
We modeled the 3D structure of human HCS by using the published structure of BirA from E. coli as a template [32]. The proposed structure of HCS spans amino acids 393 to 715, corresponding to the N-terminal, central, and C-terminal domains in BirA. Only a small fragment of the N-terminal domain in human HCS was included in our model because of the absence of a conserved sequence in the BirA template. The overall root mean square deviation (RSMD) between the predicted HCS structure and the BirA template was 3.2 Å, but it improved to 1.2 Å for the biotin-binding site and the HCS region spanning Thr489 to Asn606. The comparably large RSMD for the overall model was caused by an insertion in human HCS, predicting an α- helix in the region from Asn606 to Asn621 compared to a loop in the region from Asn134 to Tyr141 in BirA.
Human HCS and BirA were superimposed to identify conserved amino acid residues that are likely to be important for HCS catalysis. In the central domain of human HCS, the Arg508-Ser515 loop corresponded to the biotin binding site (Arg118-Ser125) in BirA. Our model predicted the existence of a short helix in HCS (Asn606-Asn621), in which valine substituted for tryptophan in the corresponding helix in BirA (Met211-Trp223). Valine668 in the linker region of HCS corresponded to Ile272 in BirA. Histidine-675 and Lys683 in the C-terminal domain of HCS corresponded to Lys277 and Lys283, respectively, in BirA. Although Arg718 in HCS was not included in structure predictions, sequence comparisons suggest that this residue might correspond to Arg317 in BirA.
Next, we predicted the structure of the HCS substrate p67 for subsequent docking calculations. For structure predictions, we used the BirA substrate biotin carboxyl carrier protein as a template. The FASTA E-value for p67 and the biotin carboxyl carrier protein was 4e-19 with an excellent RSMD permitting reliable prediction of the p67 structure at the 0.3 Å level of resolution. In biotin carboxyl carrier protein, Glu119 and Glu147 are important for biotinylation by BirA [17]. These residues are conserved as Glu29 and Glu58 in p67 (Glu691 and Glu720 in PCC).
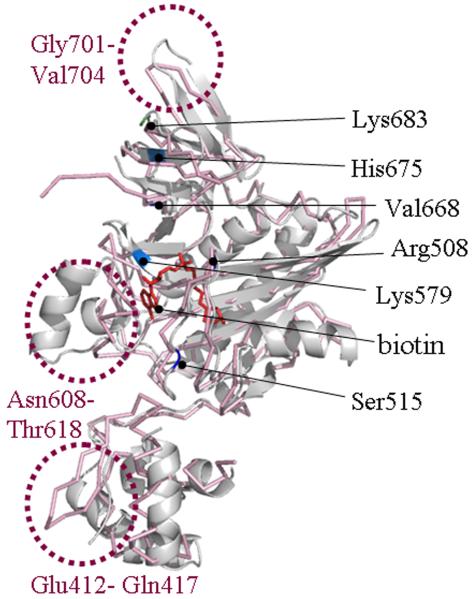
The above observations are consistent with the conservation of individual amino acids and entire domains in biotin protein ligases and their substrates. Our docking calculations suggested that the p67 molecule spatially complemented the cleft along Arg588, Lys579, Ser515 including a potential biotin binding site in human HCS (Fig. 4). Arg508 and Ser515 in human HCS appear to be equally important for binding of p67.
Figure 4.
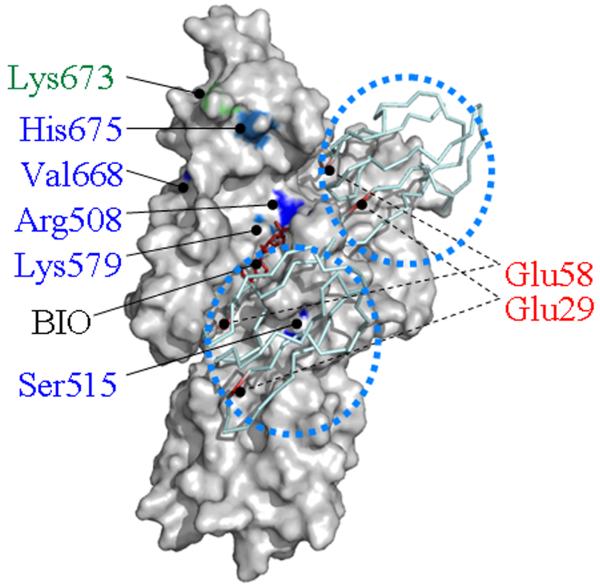
Human HCS contains two potential docking sites for p67 near the biotin (BIO) and ATP binding sites. Two molecules of p67 are depicted as ribbon models and identified by using dotted circles. Functionally important amino acid residues in the central domain and C-terminal domain are highlighted in blue and green, respectively. Functionally important residues in p67 are depicted in red.
Discussion
HCS has a broad substrate specificity and interacts with at least with five different carboxylases and up to five classes of histones [15, 33-35]. Carboxylases and histones do not share any apparent sequence similarities, raising the question of how HCS recognizes its biotinylation targets. While the preparation of pure and bioactive recombinant HCS and its characterization by X-ray crystallography are imminent, no such data are publicly available yet. In the absence of such data, is a reasonable approach to make predictions of HCS structure and substrate interaction based on analogies with BirA. This paper offers novel insights into the structure of HCS and its potential interactions with the p67 substrate by using a unique combination of in silico modeling, Y2H assays, and yeast rescue studies. A similar computational approach was recently used to predict the structure of biotinidase, another enzyme involved in biotin metabolism [36].
This study makes a number of valuable contributions towards the elucidation of HCS structure and catalysis. First, this study provides experimental evidence that the N-terminus domain of HCS participates in p67 binding, based on data from Y2H assays. This is in agreement with previous studies [13, 14, 20], which suggest that amino acids 128 to 446 in the HCS N-terminus are important for biotin ligase activity. The specific roles of amino acids 128 to 446 were not elucidated in these previous studies of HCS catalysis.
Second, this study confirms previous theories by Chapman-Smith et al. that the C-terminus of HCS is important for substrate recognition and for the initiation of the biotin transfer process [17]. Those investigators showed that the C-terminal domain of BirA is essential for biotin carboxyl carrier protein. Likewise, truncation or removal of the C-terminus in human HCS abolishes biotinyl ligase activity of HCS towards biotin carboxyl carrier protein and p67 [20].
Third, our studies demonstrate that both the central and linker domains in HCS do not participate in substrate recognition. This conclusion is based on the absence of detectable interactions between these two domains and p67 in Y2H assays.
Fourth, our laboratory and perhaps others are heavily invested in the identification of proteins that interact with HCS in order to target the enzyme to distinct regions in chromatin [37-39]. Initial screens for HCS-interacting proteins are based on Y2H assays. Our observation that human HCS can rescue BPL1 mutant yeast suggests that human HCS in Y2H is fully functional. Thus, the risk of encountering false positive clones in Y2H assays is reasonably small.
Fifth, this paper reveals potential docking sites for p67 in human HCS, i.e., the cleft along Arg588, Lys579, and Ser515, which act in concert with Arg508 and Ser515. Our prediction for putative docking sites is based on X-ray crystallography data from studies of BirA and biotin carboxyl carrier protein. It is noteworthy that the R508W mutation is one of the most common HCS mutations found in human patients of various ethnicities [40]. Patients carrying this mutations respond exceptionally well to biotin supplementation, consistent with a role of this residue in the binding of substrate and biotin [41].
Sixth, our model also reveals the existence of flexible loops in human HCS, e.g., loops Glu412-Gln417, Asn608-Thr618, Gly701-Val704, and the Phe487-Pro490 turn (Fig. 5). These loops (in particular Gly701-Val704) confer a comparably great degree of structural flexibility and, thus, are of particular interest in the light of the fairly broad substrate specificity of HCS (various carboxylases and histones).
Figure 5.
Human HCS contains flexible loops. The predicted 3D structure of human HCS (gray) was imposed on the structure of BirA (pink) to reveal flexible loops (dotted circles). Functionally important amino acid residues in HCS are highlighted.
Seventh, our in silico model highlighted one arginine residue (R718) in the C-terminal domain of HCS that might have a major role in enzyme-substrate recognition. Our model predicts that R718 likely corresponds to the highly conserved R317 in BirA, which is known to be critical for substrate recognition [17]. In previous studies it has been suggested that R317 in BirA corresponds to R725 rather than R718 in HCS [17]. We do not believe this to be the case, given that that a construct lacking R725 physically interacted with p67 in our Y2H assays. Our modeling studies identified another conserved positively charged residue (His675) that might be important for substrate alignment. Previous studies aligned His675 with a lysine residue (K277) in BirA, and showed that K277 is critical for the correct alignment of biotin carboxyl carrier protein within the active site of BirA [17].
We recognize moderate uncertainties associated with this study. For example, we acknowledge that a simple interaction between single amino acid residues and charged surfaces does not provide sufficient parameters to build a model for enzyme-substrate orientation. However, if coupled with the exploration of the tertiary fold orientation, one can speculate as to which amino acid residues in HCS might be critical for substrate binding in analogy to BirA studies [42, 43].
In conclusion, this paper provides important insights into the structure of human HCS. Our laboratory is currently pursuing the characterization of HCS by X-ray crystallography to address some of the uncertainties associated with the model outlined here.
Acknowledgment
A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945, DK077816, GM62484 and ES015206, USDA grant 2006-35200-17138, and by NSF grants EPS-0701892 and MCB6552870.
Abbreviations
- ACC
acetyl-CoA carboxylase
- Ade
adenine
- Arg
arginine
- Asn
asparagine
- BPL1
biotin protein ligase1
- FOA
5-fluorouracil-6-carboxylic acid monohydrate
- Gal4 BD
Gal4 DNA binding domain
- Glu
glutamic acid
- HCS
holocarboxylase synthetase
- His
histidine
- Kan
kanamycin
- Leu
leucine
- Lys
lysine
- MCC
3-methylcrotonyl-CoA carboxylase
- PC
pyruvate carboxylase
- PCC
propionyl-CoA carboxylase
- PPI
protein-protein interactions
- SD
synthetic defined
- Ser
serine
- Thr
threonine
- Trp
tryptophan
- Tyr
tyrosine
- Y2H
yeast two hybrid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. 9th ed. International Life Sciences Institute; Washington, D.C.: 2006. pp. 314–326. [Google Scholar]
- 2.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur. J. Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 3.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–4259. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Reports. 2008;41:310–315. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J. Nutr. Biochem. 2006;17:225–233. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew YC, Raza AS, Sarath G, Zempleni J. Biotinylation of K8 and K12 co-occurs with acetylation and mono-methylation in human histone H4. FASEB J. 2006;20:A610. abstract. [Google Scholar]
- 7.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J. Nutr. 2006;136:2735–2742. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camporeale G, Zempleni J, Eissenberg JC. Susceptibility to heat stress and aberrant gene expression patterns in holocarboxylase synthetase-deficient Drosophila melanogaster are caused by decreased biotinylation of histones, not of carboxylases. J. Nutr. 2007;137:885–889. doi: 10.1093/jn/137.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J. Nutr. Biochem. 2008;19:400–408. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J. Nutr. Biochem. 2007;18:760–768. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double strand breaks in human JAr choriocarcinoma cells. J. Nutr. 2005;135:2337–2342. doi: 10.1093/jn/135.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, Klinkebiel D, Christman JK, Zempleni J. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J. Nutr. 2008;138:2316–2322. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, Niikawa N, Matsubara Y, Narisawa K. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat. Genet. 1994;8:122–128. doi: 10.1038/ng1094-122. [DOI] [PubMed] [Google Scholar]
- 14.Hiratsuka M, Sakamoto O, Li X, Suzuki Y, Aoki Y, Narisawa K. Identification of holocarboxylase synthetase (HCS) proteins in human placenta. Biochim. Biophys. Acta. 1998;1385:165–171. doi: 10.1016/s0167-4838(98)00032-6. [DOI] [PubMed] [Google Scholar]
- 15.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum. Mol. Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 16.Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman-Smith A, Mulhern TD, Whelan F, Cronan JE, Jr., Wallace JC. The C-terminal domain of biotin protein ligase from E. coli is required for catalytic activity. Protein Sci. 2001;10:2608–2617. doi: 10.1110/ps.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leon-Del-Rio A, Gravel RA. Sequence requirements for the biotinylation of carboxyl-terminal fragments of human propionyl-CoA carboxylase alpha subunit expressed in Escherichia coli. J. Biol. Chem. 1994;269:22964–22968. [PubMed] [Google Scholar]
- 19.Chapman-Smith A, Cronan JEJ. Molecular Biology of Biotin Attachment to Proteins. J. Nutr. 1999;129:477S–484S. doi: 10.1093/jn/129.2.477S. [DOI] [PubMed] [Google Scholar]
- 20.Campeau E, Gravel RA. Expression in Escherichia coli of N- and C-terminally deleted human holocarboxylase synthetase. Influence of the N-terminus on biotinylation and identification of a minimum functional protein. J. Biol. Chem. 2001;276:12310–12316. doi: 10.1074/jbc.M009717200. [DOI] [PubMed] [Google Scholar]
- 21.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–25. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 23.Weaver LH, Kwon K, Beckett D, Matthews BW. Competing protein:protein interactions are proposed to control the biological switch of the E. coli biotin repressor. Protein Sci. 2001;10:2618–2622. doi: 10.1110/ps.32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bates PA, Kelley LA, MacCallum RM, Sternberg MJ. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;5(Suppl):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 26.Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate. J. Biol. Chem. 2008;283:14739–50. doi: 10.1074/jbc.M709116200. [DOI] [PubMed] [Google Scholar]
- 27.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLano WL. The Pymol Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 29.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 31.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc. Natl. Acad. Sci. USA. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hymes J, Fleischhauer K, Wolf B. Biotinylation of biotinidase following incubation with biocytin. Clin. Chim. Acta. 1995;233:39–45. doi: 10.1016/0009-8981(94)05965-u. [DOI] [PubMed] [Google Scholar]
- 34.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem. Mol. Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 35.Hymes J, Wolf B. Human biotinidase isn’t just for recycling biotin. J. Nutr. 1999;129:485S–489S. doi: 10.1093/jn/129.2.485S. [DOI] [PubMed] [Google Scholar]
- 36.Pindolia K, Jensen K, Wolf B. Three dimensional structure of human biotinidase: computer modeling and functional correlations. Mol. Genet. Metab. 2007;92:13–22. doi: 10.1016/j.ymgme.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Hassan YI, Zempleni J. Epigenetic regulation of chromatin structure and gene function by biotin. J. Nutr. 2006;136:1763–1765. doi: 10.1093/jn/136.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan YI, Zempleni J. A tyrosine kinase is involved in the nuclear translocation of human holocarboxylase synthetase; Experimental Biology Meeting; San Diego, CA. 2008; Abstract C119, number 691.14. [Google Scholar]
- 39.Zempleni J, Chew YC, Hassan YI, Wijeratne SSK. Epigenetic regulation of chromatin structure and gene function by biotin: are biotin requirements being met? Nutr. Rev. 2008;66:S46–S48. doi: 10.1111/j.1753-4887.2008.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki Y, Yang X, Aoki Y, Kure S, Matsubara Y. Mutations in the holocarboxylase synthetase gene HLCS. Hum. Mutat. 2005;26:285–290. doi: 10.1002/humu.20204. [DOI] [PubMed] [Google Scholar]
- 41.Dupuis L, Campeau E, Leclerc D, Gravel RA. Mechanism of biotin responsiveness in biotin-responsive multiple carboxylase deficiency. Molec. Genet. Metabol. 1999;66:80–90. doi: 10.1006/mgme.1998.2785. [DOI] [PubMed] [Google Scholar]
- 42.Reche PA. Lipoylating and biotinylating enzymes contain a homologous catalytic module. Protein Sci. 2000;9:1922–1929. doi: 10.1110/ps.9.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reche PA, Howard MJ, Broadhurst RW, Perham RN. Heteronuclear NMR studies of the specificity of the post-translational modification of biotinyl domains by biotinyl protein ligase. FEBS Lett. 2000;479:93–98. doi: 10.1016/s0014-5793(00)01829-9. [DOI] [PubMed] [Google Scholar]