Abstract
Satellite cells are tissue-specific stem cells responsible for skeletal muscle growth and regeneration. Although satellite cells were identified almost 50 years ago, the identity of progenitor populations from which they derive remains controversial. We developed MyoDiCre knockin mice, and used Cre/lox lineage analysis to determine whether satellite cell progenitors express MyoD, a marker of myogenic commitment. Recombination status of satellite cells was determined by confocal microscopy of isolated muscle fibers and by electron microscopic observation of muscle tissue fixed immediately following isolation, using R26R-EYFP and R26R (β-gal) reporter mice, respectively. We show that essentially all adult satellite cells associated with limb and body wall musculature, as well as the diaphragm and extraocular muscles, originate from MyoD+ progenitors. Neonatal satellite cells were Cre-recombined, but only a small minority exhibited ongoing Cre expression, indicating that most satellite cells had expressed MyoD prenatally. We also show that satellite cell development in MyoD-null mice is not due to functional compensation by MyoD non-expressing lineages. The results suggest that satellite cells are derived from committed myogenic progenitors, irrespective of the anatomical location, embryological origin, or physiological properties of associated musculature.
Introduction
Satellite cells are tissue-specific stem cells that are responsible for skeletal muscle growth and repair. In response to injury, satellite cells enter the cell cycle, activate expression of muscle regulatory factors (MRFs) such as MyoD and Myf-5, and execute the myogenic program, eventually restoring muscle structure and function (reviewed by Zammit, 2008). This postnatal myogenesis bares many similarities to embryonic muscle development, including striking molecular and functional parallels between embryonic myogenic precursors (myoblasts), and satellite cells and their daughters (Parker et al., 2003). Indeed, until recently, satellite cells were widely believed to derive from the fetal myoblast pool, becoming anatomically and functionally partitioned during late fetal development in the chick and mouse (Cossu et al., 1983; Hartley et al., 1992; Bischoff, 1994). Recent data, however, suggest that satellite cells derive from a more primitive progenitor population that expresses the paired box transcription factors, Pax3 and Pax7, but not the MRFs (Kassar-Duchossoy et al., 2005; Relaix et al., 2005), regulators of myogenic commitment and differentiation that function downstream of Pax3 and Pax7 in the trunk and limbs (Kassar-Duchossoy et al., 2005; Tajbakhsh et al., 1997; Relaix et al., 2005). The existence of Pax3/7+;MRF− progenitors was traced back to the somite dermomyotome (Kassar-Duchossoy et al., 2005; Relaix et al., 2005), the embryological structure from which satellite cells of trunk and limb musculature derive (Armand et al., 1983; Gros et al., 2005; Schienda et al., 2006), suggesting a possible lineage relationship with nascent, sublaminar satellite cells at fetal stages.
MyoD and Myf-5 function in partially redundant pathways to control embryonic myogenesis (Rudnicki et al., 1993). The key role of Myf-5 and MyoD in myoblast specification has been shown by their dominant ability to activate the myogenic program in non-muscle cells (Weintraub et al., 1991), and the demonstration that presumptive myogenic cells failing to activate Myf-5 and MyoD due to germline mutations can adopt non-myogenic cell fates (Tajbakhsh et al., 1996; Kablar et al., 1999). Compelling evidence has been presented that embryonic and fetal myogenesis, except for the initial formation of the myotome, also is dependent on Pax3 and Pax7 (Relaix et al., 2005). In the absence of these Pax genes, presumptive myogenic cells do not express the MRFs, they cannot enter the myogenic program, and some adopt non-myogenic fates (Relaix et al., 2005). However, both Pax3 and Pax7 are expressed in the early neural tube, and Pax3 exhibits widespread expression in the paraxial mesoderm and epithelial somite prior to establishment of definitive myogenic lineages (reviewed by Buckingham and Relaix, 2007), indicating that these genes are not sufficient for myogenic programming. Collectively, these data imply that Pax3/7+;MRF− cells that occupy the satellite cell position in fetal muscle and likely represent satellite cell progenitors (Kassar-Duchossoy et al., 2005; Relaix et al., 2005), are not strictly committed to the myogenic program. Consistent with this view, several groups have reported that satellite cells from adult muscles can spontaneously adopt adipogenic (Asakura et al., 2001; Csete et al., 2001; Shefer et al., 2004), osteogenic (Asakura et al., 2001) and fibroblastic (Brack et al., 2007) fates in culture.
Evaluating whether satellite cells express MyoD or other markers of lineage commitment in their developmental history requires methods that provide a permanent record of expression over time. This is particularly true because of the lack of positive markers that distinguish satellite cells progenitors from committed myoblasts, and the consequent inability to exclude the possibility that satellite cells derive from Pax3/7+;MRF+ myoblast-like cells that downregulate MRFs prior to assuming the sublaminar satellite cell position in the fetus. To this end, we developed a MyoDiCre knockin allele and used Cre/lox lineage analysis to address whether the MyoD locus is active in satellite cell progenitors. We show that satellite cells, irrespective of their anatomical location and embryological origin, transit through a developmental stage in which the MyoD locus is active. Analysis of the timing of Cre-dependent satellite cell labeling strongly indicates that the MyoD locus is active in progenitor populations prenatally, suggesting a close lineage relationship between satellite cell precursors and lineage-committed myoblastic populations. Further, we show that satellite cells derive from MyoD+ lineages even in MyoD mutant mice, demonstrating that MyoD is dispensable for satellite cell development within the MyoD+ lineage, and that functional compensation by MyoD non-expressing lineages does not occur to an appreciable extent.
Materials and Methods
Generation of MyoDiCre knockin mice
The targeting vector was designed to replace 3 bp of 5′ untranslated sequence, exon 1 sequences encoding amino acids 1–209, and 47 bp of intron 1, with the improved Cre gene (iCre; Shimshek et al., 2002), in which mammalian codon usage was optimized and CpG content was reduced to lessen the likelihood of epigenetic silencing. iCre was followed by two tandem SV40 polyadenylation sequences (2pA), and a PGKNeo cassette flanked by FRT sequences for excision with Flipase recombinase. Standard subcloning methods and recombineering (Liu et al., 2003) were used for vector construction (Yamamoto et al., 2009). The targeting strategy is outlined in Fig. 1.
Figure 1.
MyoDiCre targeting strategy. (A) MyoD locus showing MyoD exons (black boxes), position of homology arms in the targeting vector (dotted lines), and position of 5′ and 3′ probes (open boxes) used for Southern blot screening for targeted alleles. H, HpaI sites used for Southern screening. (B) Targeting vector design. The iCre gene (Shimshek et al., 2002) followed by two SV40 polyadenylation sequences replaced all coding sequences of exon 1 (which includes the essential basic and helix-loop-helix domains), as well as the first 47 nucleotides of intron 1, resulting in a predicted null allele (Rudnicki et al., 1992). The PGKNeo selection cassette is in reverse orientation relative to MyoD transcription and is flanked by FRT sites to allow FLP recombinase-mediated excision. (C–D) Structure of the targeted MyoD locus before (C; MyoDiCreNeo) and after (D; MyoDiCre) FLP recombination. FLP-mediated excision of PGKNeo was accomplished by crossing MyoDiCreNeo/+ mice to Rosa26FLP/FLP mice (Farley et al., 2000). (E) Southern blot analysis of ES cell clones using the 5′ probe. Targeted clones (lanes 3, 5 and 6) show a 9.3-kb band, specific to the mutant MyoDiCreNeo allele, and an 18.8-kb wild-type band. Fidelity of the targeted allele was confirmed with the 3′ probe (not shown). (F) RT-PCR analysis of MyoD expression. Total RNA was isolated from E12.5 embryos after removal of the head, reverse transcribed, and transcripts were amplified using primers anchored in exons 2 and 3, or anchored in exon 1 and the junction between exons 1 and 2. A minor product was detected in MyoDiCre/iCre embryos with primers specific for exons 2 and 3, which represents read-through transcription from the iCre cassette (not shown). As expected, exon 1 coding sequences were not detected with primers specific to exons 1 and 2.
ES cell electroporation and production of chimeras was performed by the University of Connecticut Gene Targeting and Transgenic Facility (GTTF) using 129S6/C57BL6 hybrid ES cells (D1: established by GTTF). Screening of ES cell clones was performed by Southern blot hybridization with the PCR-generated probes corresponding to sequences outside of the 5′ and 3′ homology arms. The following primers were used for probe amplification: 5′ probe, 5′-TGGTT CAGTTAACTCAGTGGGTTTG-3′ and 5′-ATGCT ATAAACCTCCCATGCCATGC-3′; 3′ probe, 5′-TTCTGGCAGAATGAGTCTGTCTA GG-3′ and 5′-GAGTGTGTAAGGAACCCTACAGAGC-3′. Chimeric mice were crossed to Rosa26FLP/FLP mice (Farley et al., 2000; Jackson Laboratories) to remove the PGKNeo cassette. Germ line transmission of the targeted allele and removal of the PGKNeo cassette were assessed by PCR (see below).
Mouse breeding and genotyping
All lines were maintained by breeding to FVB mice. Experimental mice were generated by crossing Cre heterozygotes with heterozygous (R26R) or homozygous (R26R-EYFP) reporter mice. Leaky expression of Cre in the female germline (Chen et al., 2005) was not observed with the MyoDiCre allele. The MyoDiCre allele was detected by PCR using a forward primer (5′-GCGGATCCGAATTCGAAGTTCC-3′) that lies at the 3′ end of the iCre2pA cassette and a reverse primer in intron 1 of MyoD (5′-TGGGTCTCCAAAGCGACTCC-3′), generating a product of 149 bp. The Hprt1Cre allele and the HSA-Cre transgene were detected using forward (5′-CATTTGGGCCAGCTAAA CAT-3′) and reverse 5′-CGGATCATCAGCTACACCAG-3′) primers to Cre, generating a product of 432 bp. The R26R allele was detected using the lacZ primers, 5′-CCGAAATCCCGAATCTCTATC-3′, and 5′-TTGGCTTCATCCACCACATAC-3′, generating a product of 333 bp. All PCR reactions were conducted using standard conditions for 30 cycles with annealing temperatures of 55°C (lacZ), 56°C (iCre) or 59°C (Cre).
MyoD expression analysis
Total RNA was extracted from E12.5 mouse embryos using the RNeasy Mini Kit (QIAGEN), and treated with RQ1 DNase I (Promega). cDNA was generated using the ProtoScript First Strand cDNA Synthesis Kit using a (dN)9 random oligomer for first strand synthesis according to the manufacturer’s protocol (NEB). We assayed for MyoD transcripts downstream of the targeted insertion using primers anchored in exon 2 (5′-GGCAGAATGGCTA CGACACC -3′) and 3 (5′-CTGGGTTCCCTGTTCTGTGT -3′), which generates a 224-bp product. The absence of exon 1 coding sequence in MyoDiCre/iCre embryos was verified using a primer anchored in exon 1 (5′-CTTCTATG ATGATCCGTGTTTCGAC -3′) and a primer that spans the junction between exons 1 and 2 (5′-CTGTAATCCATCATGCCATCAGA -3′). PCR for β-actin (internal control) was performed with primers 5′-TAGGCACCAGGGTGTGATGG-3′ (for) and 5′-GTAC ATGGCTGGGGTGTTGAA-3′ (rev), generating a 280-bp product.
In situ hybridization and X-gal staining of embryos
Embryo collection, whole mount situ hybridization and X-gal staining were described previously (Yamamoto et al., 2007). No background β-gal activity was observed in embryos having only the MyoDiCre or R26R allele. The iCre probe corresponded to the entire coding sequence of iCre (Shimshek et al., 2002). Wild-type and MyoDiCre/+ embryos were used for MyoD and Cre in situ hybridizations, respectively. Images were captured with a Leica MZFLIII stereomicroscope and a Nikon Coolpix 950 digital camera.
Single muscle fiber isolation
Muscle fibers were isolated from hind limb muscles of juvenile (P14; EDL) and adult (6–10 weeks old) mice as previously described (Shefer et al., 2004). At P0, hind limb fibers were isolated by incubation of the whole hind limb in 0.2% collagenase (Type I, Sigma-Aldrich, Cat# C-0130) for 1 hr after careful removal of the skin. Following collagenase treatment, muscles were transferred to DMEM (Gibco-Invitrogen Life Technologies; Cat. #11995065) containing 10% horse serum and triturated to release individual fibers. Fibers were fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature, washed three times for 20 min each in PBS, pH 7.4, and processed for immunohistochemistry. Absence of endothelial contamination was confirmed by staining with rat anti-mouse CD31 antibody (BD Biosciences; Clone MEC 13.3).
Immunohistochemistry
Muscle fibers
The following antibodies and dilutions were used: Mouse anti-chicken Pax7 monoclonal antibody, 1:3 to 1:10 dilution of hybridoma supernatant (Developmental Studies Hybridoma Bank); rat anti-mouse CD34 monoclonal antibody (eBioscience, RAM34 Cat# 14-0341-82) diluted 1:400; chicken anti-mouse Syndecan 4 (Cornelison et al., 2004) diluted 1:1,000; rabbit anti-Cre (Novagen, Cat# 69050-3) diluted 1:500; mouse anti-mouse MyoD monoclonal antibody (BD Pharmingen; clone: mAb 5.8A) diluted 1:500. Secondary antibodies used include Alexa Fluor-conjugated goat anti-mouse, rabbit or chicken antibodies (Invitrogen) at a dilution of 1:500 or 1:1,000, and 6 μg/ml biotinylated goat anti-rat IgG (BD Pharmingen, Cat# 559286) and 5 μg/ml Alexa Fluor-conjugated streptavidin for CD34.
For Pax7, Cre, and MyoD IHC, muscle fibers were blocked in 2% BSA, 5% goat serum and 0.2% Triton X-100 at room temperature for 45 min, followed by incubation in primary antibodies for 2 hr in fresh blocking solution at room temperature. Following incubation with primary antibodies, fibers were washed three times for 5 min each in PBS (pH 7.4) and incubated in Alexa Fluor-conjugated secondary antibody diluted in blocking buffer for 45 min at room temperature. For simultaneous detection of two proteins, Alexa Fluors were used in the combinations 555 and 633, or 568 and 633. After washing three times for 5 min each, fibers were stained with DAPI and mounted on slides using aqueous mounting medium (Biomeda). For CD34 and Syndecan 4 IHC, fibers were blocked in 5% goat serum and 0.1% Triton X-100 in PBS overnight at 4°C, followed by incubation in primary antibodies for 3 hr in fresh blocking solution at room temperature. After washing as above, fibers were incubated in secondary antibody diluted in blocking buffer for 1 hr. After washing as above, fibers for CD34 detection were incubated with Alexa Fluor-conjugated streptavidin for 1 hr in the blocking solution. Fibers were washed, stained with DAPI and coverslipped as above.
Stained fibers were observed with a Leica TCS SP2 laser scanning spectral confocal microscope equipped with an Argon laser (488 nm excitation) for EYFP, a Helium-Neon green laser (543 nm excitation) for Alexa Fluors 555 and 568, and a Helium-Neon red laser (633 nm excitation) for Alexa Fluor 633 detection. Samples were visualized with 40X oil or 100X oil objectives and captured and analyzed with Leica Confocal Software version 2.61. Each satellite cell was first identified using a standard UV episource to detect Pax7, CD34 or Syndecan 4 staining, and DAPI staining was verified before scanning. A Z-stack was collected for each satellite cell to assess EYFP fluorescence throughout the cell, which aided in scoring both positive and negative cells.
Tissue sections
The following antibodies and dilutions were used: mAb 5.8A to MyoD diluted 1:50; rabbit anti-Cre diluted 1:500; 5 μg/ml biotinylated goat anti-mouse IgG, 1:400 and 1:500 dilution of Alexa Fluor 555-Streptavidin and Alexa Fluor 488-goat anti-rabbit IgG, respectively.
E16.5 MyoDiCre/+;R26R-EYFP embryos were fixed in 4% PFA/PBS for 2 hr at 4°C and washed in PBS several times. After embedding in OCT, 14 μm cryosections were collected on ProbeOne Plus glass slide (Fisher Scientific) and immediately dried under hot air for 1 hr. The slides were rinsed in PBS and incubated in 0.1 μg/ml 4′6′-diamidino-2-phenylindole dihydrochloride (DAPI)/PBS. For antigen retrieval, slides were incubated in methanol for 6 min at −20°, rinsed in PBS three times, autoclaved in 10 mM Sodium Citrate buffer (pH 6.0) for 15 min and rinsed in PBS three times. The sections were then blocked in 1.5% Non-fat dry milk/1.5% BSA/0.1% Triton X-100/PBS (PMSMT) for 2 hr and simultaneously incubated with anti-MyoD and anti-Cre antibodies in PBSMT for 1 hr. After washed in PBS three times, the sections were incubated with 5 μg/ml biotinylated goat anti-mouse IgG in PBSMT for 1 hr and washed in PBS three times. The sections were then incubated in Streptavidin-conjugated Alexa Fluor 555-Streptavidin and Alexa Fluor 488-goat anti-rabbit IgG in PBSMT for 1 hr, incubated in 0.1 μg/ml DAPI/PBS, washed in PBS three times and mounted in Gel/Mount. Sections were imaged with a Nikon E600 microscope and images were captured with a SPOT RT3 camera and software.
Clonal cultures
Cultures were rinsed in PBS, fixed in 4% PFA for 5 min, blocked (5% normal goat serum, 2% BSA, 0.2% Triton X-100 in TBS) for 45 min and incubated in a 1:2 dilution of mAb MF20 (Developmental Studies Hybridoma Bank) hybridoma supernatant for 2 hr. Subsequently, cells were washed three times in PBS and incubated for 45 min in a 1:500 dilution of Alexa fluor 546 goat anti-mouse IgG. After washing in PBS, cells were counterstained with DAPI and imaged under PBS with a Nikon TE2000 inverted microscope. Images were captured with a Retiga EXi camera (QImaging) and Openlab (Improvisions) software.
Transmission electron microscopy
After dissection, the TA muscle from MyoDiCre/+;R26R, Hprt1Cre/+;R26R and HSA-Cre;R26R mice was pinned and fixed in EM-grade 2% paraformaldehyde, 0.25% glutaraldehyde in 0.1M sodium phosphate buffer (pH 7.4) for 3 hr on ice. The muscle was rinsed in buffer, cut into pieces of approximately 1 × 2mm, and incubated overnight at 37°C, with gentle rotation in the dark, in the β-gal substrates, Bluo-gal (Sigma-Aldrich; 1mg/ml in PBS, pH 7.4, containing 10mM each of potassium ferricyanide and potassium ferrocyanide and 2mM MgCl2), or X-gal (Gold Biotechnology; 1mg/ml in PBS, pH 7.4, containing 5mM each of potassium ferricyanide and potassium ferrocyanide, 2mM MgCl2, and 0.1% Tween-20). The tissue pieces were rinsed in 0.1M sodium cacodylate (pH 7.4) followed by a 1 hr incubation in 2% OsO4 at 4°C. After incubation in 8% uranyl acetate in water for 2 hr at 4°C, the tissues were dehydrated in a standard ethanol series and embedded in a resin mixture containing Embed 812, DDSA, NMA and BDMA (Electron Microscopy Sciences). Propylene oxide was not used as a transitioning agent since it results in leaching of the chromogen deposits even with brief incubations (Masahira et al., 2005; unpublished observations). Tissue sections of 70–90nm were cut with a diamond knife and post-stained with 4% uranyl acetate for 8 min to increase contrast. The sections were observed using a Tecnai G2 Spirit Biotwin transmission electron microscope at an accelerating voltage of 80kV. Images were captured using G2 Spirit imaging software.
Cell sorting and clonal cultures
Single cell suspensions were prepared from muscle tissue of MyoDiCre;R26NG mice (Yamamoto et al., 2009) as previously described (van Beijnum et al., 2008), with minor modifications. Briefly, hind limb muscles were pooled, minced under sterile conditions, and incubated for 1 hr at 37°C in a 9:1 mixture of 0.1% collagenase type II (Gibco Invitrogen, #17101-015) and 2.5U/ml dispase-PBS (Gibco Invitrogen, #17105-041) with gentle trituration every 15 min. Digestion was terminated by addition of cold medium [10% FBS, 10% horse serum, 0.5% chick embryo extract (Sera Labs International LTD), Pen/Strep, in DMEM], and the cell suspension was filtered through a 100μm filter and placed on ice. After centrifugation at 2000 rpm for 5 min, cells were re-suspended in PBS, and filtered through a 30μm filter immediately before sorting. Cells were subjected to cytometric analysis and live cells sorted into EGFP+ and EGFP− fractions with a BD FACSAria 2 flow cytometer and FACSDiva 6.1 software. Gating parameters were established with cell suspensions from muscle of wild-type mice and cells were sorted twice to improve fidelity. Cells were plated at clonal density (50 cells/cm2 –200 cells/cm2) in multi-well plates coated with Growth Factor Reduced Matrigel (BD Biosciences). Cultures were fed every 2 days with proliferation medium [10% FBS, 10% HS, 0.5% chick embryo extract (Gibco Invitrogen), Pen/Strep, in DMEM] for 8 days and then switched to differentiation medium (10% HS, Pen/Strep, in DMEM) for 5 days. Colonies were scored for EGFP fluorescence and immunohistochemically stained for MHC expression using mAb MF20.
Results
Development of the MyoDiCre knockin allele
We produced a Cre knockin allele of MyoD (Yamamoto et al., 2009) to investigate the relationship between the satellite cell lineage and committed myogenic progenitors. MyoD was chosen for this analysis for two reasons. First, MyoD is an extensively validated marker for determined myogenic progenitors (Berkes and Tapscott, 2005; Zammit, 2008); both within the major muscle forming regions of the embryo (Sassoon et al., 1989), as well as in rare myogenic cells found in other tissues and organs (Mayer and Leinwand, 1997; Gerhart et al., 2001; Gerhart et al., 2007), expression of MyoD mRNA and protein is restricted to committed myogenic cells. Second, in contrast with the related myogenic factor, Myf-5, MyoD is not expressed by quiescent satellite cells in the adult (Yablonka-Reuveni and Rivera, 1994; Cornelison and Wold, 1997), allowing possible lineage relationships with MyoD-expressing ancestors to be ascertained. In addition, Myf5Cre knockin mice (Tallquist et al., 2000) are of limited value for assessing myogenic programming of satellite cell precursors, since this line exhibits extensive Cre-dependent labeling of non-muscle lineages in the early embryo (Gensch et al., 2008).
To produce MyoDiCre, we replaced coding sequences of exon 1 with iCre (Shimshek et al., 2002), a Cre recombinase gene that had been modified to improve translational efficiency and reduce the likelihood of epigenetic silencing (Fig. 1). Removal of exon 1 produced a MyoD null allele (Rudnicki et al., 1992), as sequences encoding the DNA binding domain and helix-loop-helix protein interaction domain were deleted (Weintraub et al., 1991). Most experiments were conducted with heterozygous mice, which are phenotypically wild type (Rudnicki et al., 1992). To evaluate the specificity of Cre expression, MyoDiCre mice were crossed with the Cre-dependent lacZ reporter line, R26R (Soriano, 1999), and embryos were X-gal stained to reveal the distribution of β-gal. MyoDiCre/+;R26R embryos at embryonic day 10.5 (E10.5) and E11.5 showed robust expression of lacZ that closely matched the distribution of MyoD mRNA (Fig. 2A, B, D, E). No X-gal staining was observed in embryos lacking the MyoDiCre allele (data not shown). The minor delay in appearance of X-gal staining in limb buds and the posterior (younger) somites at E10.5 (Fig. 2A, B) reflected the time required for accumulation of β-gal subsequent to Cre expression, and not a delay in transcription of the MyoDiCre allele, as iCre mRNA was detected in these regions coincident with MyoD mRNA (Fig. 2B, C). At E13.5, staining of the limbs was restricted to the developing muscle beds (Fig. 2F, G). We also assessed the muscle specificity of Cre expression within muscle forming regions of the limb at E16.5 by comparing the distribution of Cre and MyoD proteins. There was a close correspondence between nuclear-localized MyoD and Cre expression, and Cre expression was restricted to the MyoD+ population (Fig. 2H–K). Collectively, these data indicate that Cre expression in the embryo is muscle-specific and a faithful reflection of MyoD transcriptional activity. Cre-dependent recombination also was muscle-specific in adult limbs, except for labeling of an occasional vasculature-associated cell representing approximately 1–5% of cells of the arteriole wall (unpublished observations).
Figure 2.
Cre expression driven by the MyoDiCre knockin allele closely matches endogenous MyoD expression. (A) lacZ expression in MyoDiCre/+;R26R embryos at E10.5 is restricted to skeletal muscle lineages, as revealed by X-gal staining. lacZ expression closely resembled the in situ hybridization pattern for endogenous MyoD mRNA (B), except for a delay in limb buds (black arrowheads in A and B) and the posterior (youngest) somites (red arrowheads), which likely reflects the time required to accumulate detectable levels of β-gal driven by the Cre-recombined R26R locus. Fidelity of iCre expression from the MyoD locus was confirmed by in situ hybridization for iCre mRNA (C), which was essentially identical to the distribution of MyoD mRNA at this stage. Staining of the branchial arches in (A–C) is designated by orange arrowhead. The signal in the otic vesicle in (C) is non-specific background staining. At E11.5, the pattern of X-gal staining (D) closely matched endogenous MyoD expression (E) at all areas of skeletal myogenesis in the head, trunk and limb buds (black arrowheads). At E13.5, X-gal staining in definitive muscle anlagen was robust, as revealed in whole mount (F) and sectioned (G) hind limb preparations. No staining was observed in developing skeletal elements or soft tissues of the hind limb. The section level in (G) is designated by a dashed line in (F). The tibialis anterior (TA), extensor digitorum longus (EDL) and soleus (SOL) muscle rudiments are designated. T, tibia; F, fibula. (H–K) Immunohistochemical detection of MyoD and Cre proteins in a cryosection through the autopod of an E16.5 embryo. All Cre+ cells are also positive for MyoD (I–K). Cells negative for both Cre and MyoD are present in the section (H–K; a region between muscle beds is circumscribed by a dashed line. Five double negative cells within the muscle beds are designated with arrowheads). Scale bars in (G) and (H–K) represent 100 μm and 25 μm, respectively.
Recombination status of quiescent satellite cells derived from adult hind limb muscles
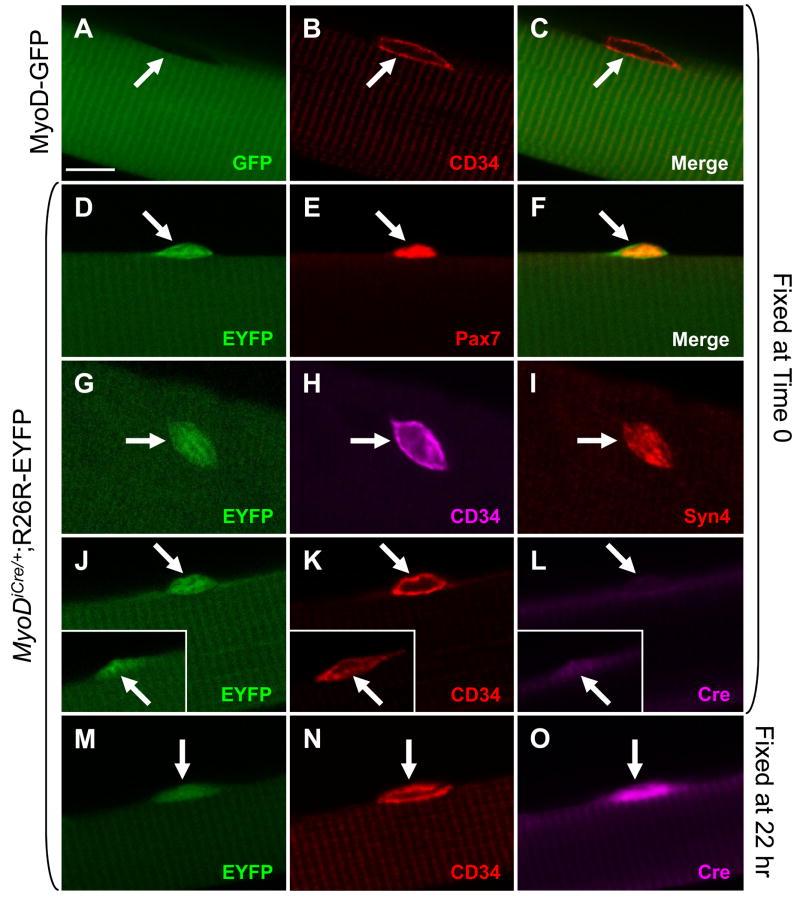
MyoDiCre mice were crossed with the Cre-dependent reporter, R26R-EYFP (Srinivas et al., 2001), and the recombination status of satellite cells associated with individually isolated extensor digitorum longus (EDL) muscle fibers was evaluated by scoring for EYFP fluorescence by confocal microscopy. Because the vast majority of quiescent adult satellite cells do not express MyoD (Yablonka-Reuveni and Rivera, 1994; Cornelison and Wold, 1997), finding EYFP labeling would reflect Cre expression in a precursor population at an earlier developmental period rather than ongoing transcriptional activity of the MyoD locus. EDL muscle fibers were isolated from 6–10 week old mice by gentle collagenase treatment (Shefer and Yablonka-Reuveni, 2005) and fixed immediately following isolation (approximately 1.5 hours after muscle dissection) to minimize satellite cell activation and induction of MyoD transcription (Yablonka-Reuveni and Rivera, 1994). Satellite cells were identified primarily by immunohistochemistry for Pax7, a widely used marker for quiescent satellite cells (Zammit, 2008), and results confirmed using the markers CD34 (Beauchamp et al., 2000) and Syndecan 4 (Cornelison et al., 2004), both of which identify most satellite cells in single fiber preparations. To ensure that intense fiber fluorescence would not interfere with accurate scoring of unrecombined satellite cells, we first conducted control experiments with MyoD-GFP transgenic mice (Yamamoto et al., 2007), which provide a readout of ongoing MyoD transcription. As anticipated, confocal microscopy provided clear discrimination of intensely fluorescent muscle fibers and closely associated, but unlabeled, satellite cells (Fig. 3A–C).
Figure 3.
Analysis of satellite cell recombination status and Cre expression using confocal microscopy. EDL fibers from adult (6–10 weeks) mice are shown. Fibers were either fixed immediately following isolation (Time 0; A–L), or after 22 hr in culture (M–O). (A–C) EGFP (A), CD34 (B) and merged (C) images of an isolated fiber from a MyoD-GFP transgenic mouse (Chen et al., 2001; Yamamoto et al., 2007). The fiber cytoplasm is strongly EGFP+, whereas the CD34+ satellite cell is EGFP−. This control demonstrates that strongly positive fiber cytoplasm does not interfere with detection of unlabeled satellite cells. (D–F) EYFP (D), Pax7 (E) and merged (F) images of a recombined, EYFP+, satellite cell associated with an EDL fiber from a MyoDiCre/+;R26R-EYFP mouse. (G–I) A second example of an EYFP+ satellite cell (G) identified by both CD34 (H) and Syndecan 4 (I) staining. (J–L) 90% of EYFP+/CD34+ satellite cells (J, K; N=120) associated with fibers fixed immediately following isolation were negative for Cre (I). Most of the remaining 10% were weakly positive (J–L, inset). After 22 hr of culture, 99% of EYFP+/CD34+ satellite cells (M, N; N=130), were Cre+, and most were strongly positive (O). Arrows, satellite cells. Scale bar in panel A represents 10 μm. All images were photographed at the same magnification.
Surprisingly, 98% of satellite cells associated with adult EDL fibers were unambiguously positive for EYFP fluorescence (Fig. 3D–I; Table 1; Supplemental Data, Fig. 1), and the signal was easily distinguished from EYFP+ muscle fibers. No differences were observed when Pax7, CD34 or Syndecan 4 were used for cell identification, and the numbers for EDL fibers are pooled in Table 1. Similar results were observed with EDL fibers from 10-month old mice (data not shown). Weaker fluorescence of muscle fibers probably reflects their lower nuclear to cytoplasmic ratio or differences in the strength of the promoter driving EYFP expression. Because recombination of the reporter allele in Cre-expressing cells may not be 100% efficient, the actual percentage of satellite cells that had expressed the MyoDiCre allele may be higher than 98%.
Table 1.
Quantification of satellite cell recombination status (EYFP+) in adult skeletal muscle1
| Muscle Type | # animals | Total # observations2 | Mean # SC per fiber (±S.E.M.)3 | EYFP+4 | EYFP−4 | %EYFP+ |
|---|---|---|---|---|---|---|
| EDL | 14 | 419 | 5.9±0.3 | 411 | 8 | 98% |
| Gastrocnemius | 3 | 256 | N.D.6 | 254 | 2 | 99% |
| Soleus | 5 | 313 | 12.8±1.1 | 311 | 2 | 99% |
| Tibialis anterior | 3 | 266 | 14.2±1.7 | 261 | 5 | 98% |
| Diaphragm | 3 | 187 | N.D.6 | 187 | 0 | 100% |
| Intercostal muscles | 3 | 79 | N.D.6 | 79 | 0 | 100% |
| EOM | 3 | 73 | N.D.6 | 73 | 0 | 100% |
| EDL (MyoDiCre/−)5 | 3 | 446 | 20.3±0.7 | 446 | 0 | 100% |
Mice were 6–10 weeks old
Satellite cells were identified by immunohistochemistry for Pax7. For EDL fibers only, Syndecan 4 and CD34 were also used for satellite cell identification.
Determined by Pax7 immunohistochemistry of intact fibers. S.E.M., standard error of the mean
Satellite cells were scored by confocal microscopy
Homozygous null for MyoD
Not determined due to frequent fiber fragmentation or tangling during processing (EOM)
Satellite cell activation and initiation of MyoD transcription occurs during the first day following muscle fiber isolation (Yablonka-Reuveni and Rivera, 1994; Cornelison and Wold, 1997). To rule out the possibility that EYFP labeling resulted from transcriptional activation of MyoDiCre during fiber isolation, Cre immunohistochemistry was performed on fibers that had been fixed immediately following isolation and compared to fibers that had been cultured for approximately 1 day, as a positive control. As expected, satellite cells on fibers cultured for 22 hr were uniformly Cre+ (Fig. 3M–O; 99% Cre+, N = 130). In contrast, only 10% of satellite cells (N = 120) associated with fibers fixed immediately after isolation were Cre+, and most of these were weakly positive (Fig. 3J–L), suggesting that MyoDiCre was activated in a small percentage of satellite cells during fiber isolation. Satellite cells associated with fibers isolated by gentle teasing of muscles that were fixed immediately after muscle dissection were only rarely Cre+, consistent with this interpretation (data not shown).
Recombination status of satellite cells was also assessed by electron microscopy (EM) of freshly isolated muscle tissue, providing a distinct labeling method and independent criteria for satellite cell identification. In addition, this approach eliminates the possibility of MyoD transcriptional activation and satellite cell labeling during fiber isolation. The tibialis anterior (TA) muscle from adult MyoDiCre/+;R26R mice was fixed immediately after isolation, stained with the β-gal substrate, Bluo-gal, processed for EM, and satellite cells scored for the presence of βgal-catalyzed, electron-dense deposits. As a positive control, β-gal labeling was assessed in Hprt1Cre/+;R26R mice, in which Cre expression is ubiquitous (Tang et al., 2002). In both control and MyoDiCre/+;R26R skeletal muscle, electron dense deposits encircled the satellite cells, which were identified by their surrounding plasmalemma and sublaminar position juxtaposed to the muscle fiber (Fig. 4A–D). These results were confirmed using X-gal as a substrate, which generated a discrete, punctate reaction product clearly contained within the satellite cell cytoplasm (Fig. 4E). Of 44 satellite cells scored by EM, 89% were labeled; unlabeled cells probably resulted from incomplete penetration of the β-gal substrate into the tissue block, as a similar percentage of unlabeled cells was observed in control, Hprt1Cre/+;R26R mice. As a negative control we also addressed whether cells that had initiated the expression of α-skeletal actin could contribute to the satellite cell pool. We reasoned that such cells would be committed to myogenic differentiation and would not represent satellite cell progenitors. HSA-Cre79 (HSA-Cre) transgenic mice (Miniou et al., 1999) were crossed with R26R and R26R-EYFP mice, and satellite cell labeling of muscle from adult offspring was assessed by electron and confocal microscopy, respectively. As anticipated, no satellite cell labeling was observed by either EM (Fig. 4F) or confocal microscopy (N=32; Supplemental Data, Fig. 2), although the muscle fiber cytoplasm was clearly labeled. The absence of Bluo-gal deposits associated with satellite cells of HSA-Cre;R26R muscle processed for EM also provided a technical control, showing that diffusion of β-gal or the Bluo-gal product in muscle fiber cytoplasm did not account for electron dense deposits encircling satellite cells of MyoDiCre/+;R26R muscle (Fig. 4C, D).
Figure 4.
Evaluation of satellite cell recombination status by EM. (A) Section of a Bluo-gal-stained TA muscle from a wild-type mouse. No electron dense precipitate is observed in the single satellite cell (arrow) or fiber cytoplasm. (B) Section from Hprt1Cre/+;R26R TA muscle (positive control) showing electron-dense Bluo-gal precipitate encircling the satellite cell. Precipitate is also observed in the muscle fiber cytoplasm. (C–E) Three examples of satellite cell labeling in sections of MyoDiCre/+;R26R TA muscles. Labeling of both satellite cells and fiber cytoplasm is observed. In (C) and (D) Bluo-gal was used as the substrate, which produces a dense deposit associated with the plasma membrane. In (E), X-gal was used as the substrate, which shows clear punctate deposits within the satellite cell cytoplasm. (F) Section from HSA-Cre;R26R muscle showing Bluo-gal staining in fiber cytoplasm but not in the satellite cell (arrow). Staining of satellite cells and muscle fiber cytoplasm is denoted with white and black arrowheads, respectively. Scale bars represent 0.5 μm.
Satellite cells associated with anatomically and developmentally distinct muscles derive from MyoD+ progenitors
We next addressed whether the MyoD locus was active in satellite cell precursors associated with muscles of different fiber type composition, anatomical location and developmental origin. We evaluated the recombination status of satellite cells associated with additional hind limb muscles, including predominantly fast twitch (gastrocnemius) and slow twitch (soleus) muscles, as well as the diaphragm and intercostal muscles of the body wall. Collectively, between 98% and 100% of satellite cells on single dissociated muscle fibers were EYFP+ (Table 1; data not shown). We also evaluated labeling of satellite cells associated with the extraocular muscles (EOM), which exhibit a number of distinct physiologic and developmental differences compared to trunk and limb musculature. For example, EOM derive from preoptic head mesenchyme rather than somites, they express a number of immature MHC isoforms, and their development is under distinct genetic control (reviewed by McLoon et al., 2007). Nevertheless, 100% of Pax7+ satellite cells associated with individual adult EOM muscle fibers were EYFP+ (Table 1).
Timing of Cre-dependent labeling of satellite cells
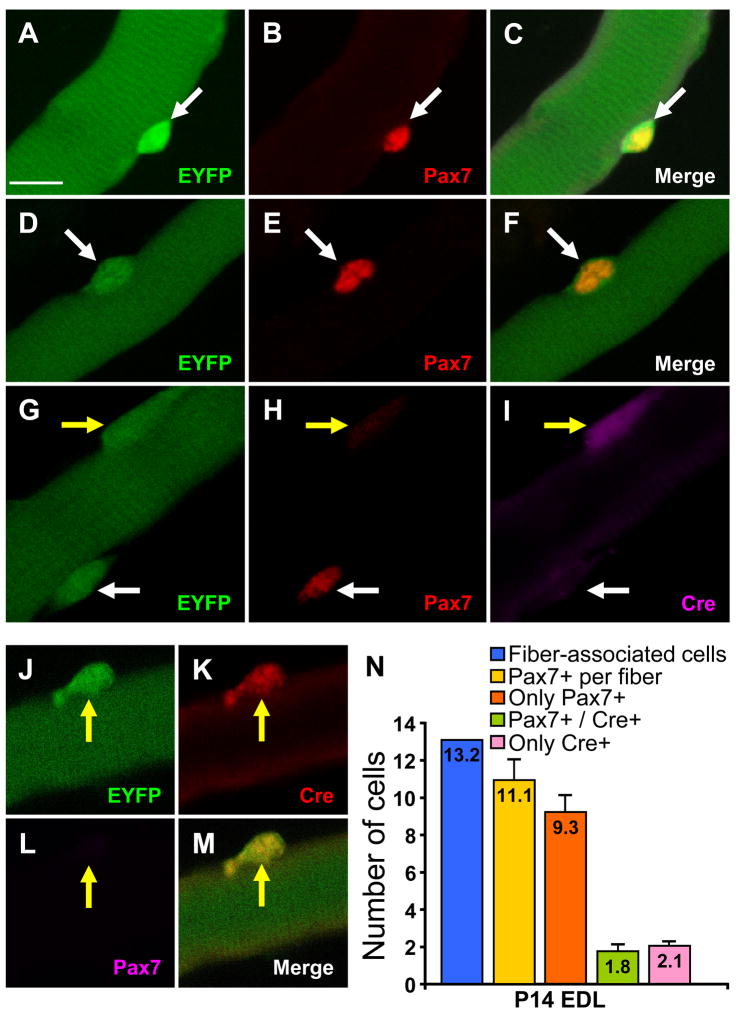
The experiments presented thus far show that all, or nearly all, quiescent satellite cells of adult skeletal muscle are derived from one or more progenitor populations that transit through a MyoD+ stage. However, these data do not distinguish whether MyoD expression occurs prenatally, or during postnatal muscle growth and expansion of the satellite cell pool. To define the developmental window during which satellite cell precursors activate the MyoD locus, we determined the recombination status of satellite cells associated with muscle fibers isolated from neonatal (P0) and juvenile (P14) MyoDiCre/+;R26R-EYFP mice. At P14, 100% of Pax7+ satellite cells associated with EDL muscle fibers were EYFP+, whereas 95% of satellite cells associated with fibers from pooled hind limb muscles isolated on the day of birth were recombined (Fig. 5A–F; Table 2). Of note, 95% of Pax7+ satellite cells at P0 were negative for Cre protein (Fig. 5G–I, N; Table 2), suggesting that MyoD transcription and cell labeling occurred in satellite cell precursors at prenatal stages.
Figure 5.
Recombination status and Cre expression in satellite cells associated with juvenile (P14) and neonatal (P0) skeletal muscle from MyoDiCre/+;R26R-EYFP mice. (A–M) EYFP (A, D, G, J), Pax7 (B, E, H, L), Cre (I, K) and merged (C, F, M) images of a P14 EDL fiber (A–C, J–M) or a P0 fiber isolated from pooled hind limb muscles (D–I). EYFP labeling of satellite cells was 95% and 100% at P0 and P14, respectively. While the majority of EYFP+ fiber-associated cells were Pax7+/Cre− (white arrows in G–I), Pax7−/Cre+ cells were also observed (yellow arrows in g–m). (N) Enumeration of Pax7 and Cre-expressing cells associated with P14 EDL fibers. An average of 11.1 Pax7+ satellite cells per fiber was observed (N=20 fibers), of which 9.3 were Pax7+/Cre− and 1.8 were positive for both markers. In a separate experiment, we determined that 2.1 cells per P14 EDL fiber (N=22 fibers) were Cre+/Pax7−. The average number of fiber-associated cells (Pax7+ and Pax7−) at P14 was 13.2. Bars in (N) represent standard error of the mean. Scale bar in panel A represents 10 μm. All images were photographed at the same magnification.
Table 2.
Quantification of satellite cell recombination status (EYFP+) in juvenile (P14) and neonatal (P0) skeletal muscle fibers
| Muscle Type | # animals | Total # observations | Mean # SC per fiber (±S.E.M.)1 | EYFP+2 | EYFP−2 | %EYFP+ | Cre+ | Cre− | %Cre+ |
|---|---|---|---|---|---|---|---|---|---|
| P14 EDL | 5 | 365 | 11.1±1.1 | 365 | 0 | 100% | 54 | 311 | 15% |
| P0 Hind Limb | 6 | 356 | N.D. | 338 | 18 | 95% | 18 | 338 | 5% |
Determined by Pax7 immunohistochemistry. Additionally, there were 2.1 Cre+/Pax7− fiber-associated cells per fiber at P14. S.E.M., standard error of the mean.
Satellite cells were scored by confocal microscopy
The low percentage of Cre+ cells among the Pax7+ satellite cell population at newborn and juvenile stages was interesting in light of the massive muscle growth and nuclear addition to myofibers that occurs at these stages, and the fact that MyoD is expressed in myogenic precursor cells that are actively engaged in myogenesis. We therefore examined whether a population of cells that express Cre but not Pax7 were associated with single fibers isolated from neonatal and juvenile stages. In fact, fiber-associated cells that were Cre+ and Pax7− or Pax7weak were observed at both P0 and P14 (Fig. 5G–M). Enumeration of the Cre+/Pax7− population revealed an average of 2.1 cells per EDL fiber at P14 (Fig. 5N). A similar number, 1.8 cells per fiber, expressed both Cre and Pax7 (Fig. 5N). As expected, Cre+ cells were positive for MyoD protein (data not shown). Thus, approximately 30% of the fiber-associated cells on P14 EDL fibers were Cre+/MyoD+ (Fig. 5N). Pax7+/Cre− cells and Cre+/MyoD+ cells probably represent quiescent satellite cells and myogenic precursor cells actively engaged in myogenesis, respectively, consistent with the dynamics of Pax7 and MyoD expression in cell culture models of regeneration (Olguin and Olwin, 2004; Zammit et al., 2004).
MyoD is not required for satellite cell specification
Although satellite cell development is not impaired in mice lacking MyoD (Megeney et al., 1996), it is unclear whether MyoD is uninvolved in satellite cell specification or whether compensatory mechanisms function in the absence of MyoD. Therefore, we tested whether MyoD is required cell autonomously for satellite cell development specifically by MyoD-expressing progenitors. EDL fibers were isolated from MyoDiCre/−;R26R-EYFP mice, and Pax7+ satellite cells were scored for EYFP fluorescence using confocal microscopy. Consistent with previous findings (Megeney et al., 1996; Yablonka-Reuveni et al., 1999; Cornelison et al., 2000; Gayraud-Morel et al., 2007), satellite cell numbers per fiber were increased compared to wild type fibers (Table 1). Importantly, all satellite cells associated with MyoD-null fibers were EYFP+ (Table 1), demonstrating that they derived from one or more MyoD-expressing lineages. Thus, compensation by a MyoD-independent cell population cannot account for satellite cell development in MyoD mutant mice.
Prospective isolation of myogenic cells from adult muscle tissue
We next used fluorescence activated cell sorting to test the utility of MyoDiCre mice for prospective isolation of satellite cells and other potential myogenic populations resident in muscle tissue. For this analysis, MyoDiCre mice were crossed with R26NG reporter mice, which is a more robust reporter of Cre activity than R26R-EYFP in adult tissues (Yamamoto et al., 2009), and EGFP+ cells were tested for myogenic activity by clonal analysis in culture. Mononuclear cells were isolated from total hind limb musculature and the EGFP+ gate was set to collect the entire EGFP+ cell fraction (Supplemental Data, Fig. 3) in order to reduce potential selection bias. Cells were grown at clonal density on Matrigel-coated plates, and myogenic differentiation was assessed under low mitogen culture conditions. After 5 days in differentiation medium, 100% of the 214 EGFP+ colonies analyzed exhibited robust myogenic differentiation, as assayed by myotube formation and staining with an anti-sarcomeric myosin antibody (Supplemental Data, Fig. 3). In contrast, analysis of 94 colonies derived from the EGFP-fraction revealed only three colonies with myogenic activity and only these colonies were EGFP+. Whether these myogenic colonies resulted from rare EGFP− cells of the limb that possess myogenic capacity (possibly due to incomplete Cre-mediated recombination in vivo), or reflect the presence of occasional EGFP+ cells in the EGFP− fraction under our sorting conditions, is unclear. These data show that MyoDiCre mice provide a simple, efficient and specific method to prospectively isolate myogenic cell populations from muscle tissue.
Discussion
Lineage analyses in the chick and mouse have revealed that satellite cells of limb and body wall musculature, like embryonic and fetal myoblasts, have a somitic origin (Armand et al., 1983; Gros et al., 2005; Schienda et al., 2006). The lineage relationship between fetal myoblasts and satellite cells, however, remains controversial. The traditional view that satellite cells derive from fetal myoblasts that become “trapped” beneath the basal lamina of growing muscle fibers in the fetus (reviewed by Zammit, 2008) has been challenged in recent years primarily by two sets of observations. First, several groups have reported that adult satellite cells can adopt non-myogenic fates in culture (Asakura et al., 2001; Csete et al., 2001; Shefer et al., 2004; Brack et al., 2007), consistent with the view that at least some satellite cells are not committed to myogenesis. Second, satellite cell progenitors were reported to express Pax3 and Pax7, but not MRFs (Kassar-Duchossoy et al., 2005; Relaix et al., 2005), which regulate myogenic cell fate (Weintraub et al., 1991). Pax3 and Pax7 function upstream of the MRFs in the trunk and limbs (Tajbakhsh et al., 1997; Kassar-Duchossoy et al., 2005; Relaix et al., 2005) and their expression is not restricted to the myogenic lineage (Buckingham and Relaix, 2007), suggesting that satellite cell progenitors are developmentally more primitive than myoblasts and not strictly committed to the myogenic fate.
We developed a MyoDiCre knockin allele (Yamamoto et al., 2009) and used Cre/lox-based lineage analysis to investigate whether satellite cells derive from MyoD+ progenitors. MyoD expression is restricted to cells exhibiting myogenic capacity and is a marker of myogenic commitment (Weintraub et al., 1991; Gerhart et al., 2001; Berkes and Tapscott, 2005; Gerhart et al., 2007), but is not actively expressed by quiescent satellite cells (Yablonka-Reuveni and Rivera, 1994; Cornelison and Wold, 1997). This combination of attributes allowed us to explore myogenic programming of satellite cell precursors by addressing whether satellite cells expressed the MyoDiCre allele in their developmental history. Surprisingly, we found that satellite cells associated with anatomically diverse muscle groups derive from MyoD-expressing lineages. In the limb, where the timing of Cre-dependent recombination was investigated, EYFP labeling was observed at P0, suggesting that the MyoD locus was activated at prenatal stages. These data support the hypothesis that satellite cells, regardless of embryological origin or molecular and functional heterogeneity (Zammit, 2008), are derived from committed myogenic progenitors, although direct proof will require development of methods to purify these progenitors from the fetal muscle beds. The clonal analysis of EGFP+ cells isolated from adult hind limb musculature of MyoDiCre;R26NG mice presented here, and the exclusively myogenic activity of labeled satellite cells in single fiber culture (unpublished observations), are consistent with this view.
Pax3/7+;MRF− progenitors, which can first be identified in the early somite dermomyotome, were proposed to represent progenitor cells that give rise both to MRF+ myoblasts and to MRF-satellite cells (Kassar-Duchossoy et al., 2005; Relaix et al., 2005). Some of these Pax3/7+;MRF− progenitors adopted a satellite cell position beneath the basal lamina of nascent fibers in the fetus, consistent with this view. How can these data be reconciled with the current lineage analysis that demonstrates MyoD transcription in satellite cell progenitors? Satellite cell precursors may activate MyoD transcription but not accumulate detectable levels of MyoD protein. This would be analogous to the populations of cells of the chick epiblast and fetal organs that express MyoD mRNA but not detectable protein, and are stably committed to the skeletal muscle lineage (Gerhart et al., 2001; Gerhart et al., 2007). Alternatively, MyoD may be expressed in satellite cell progenitors prior to formation of the basal lamina toward the end of the fetal period, when satellite cells can first be distinguished from fetal myoblasts by anatomical position. Importantly, MyoD protein has a half-life of only 30–60 minutes (Thayer et al., 1989), and its expression is regulated in the cell cycle (Kitzmann et al., 1998). In this regard, it is unclear to what extent the Pax3/7+;MRF− population represents a stable reservoir of MRF-progenitors that is distinct from MRF+ cells that co-occupy the muscle beds. Currently, there are no molecular markers unique to satellite cells progenitors, and our data is consistent with the possibility that these embryonic progenitors are molecularly and functionally equivalent to myoblasts.
Having established that satellite cells derive from a MyoD+ progenitor pool, we addressed MyoD’s functional role in satellite cell development. In embryonic and fetal myogenesis, MyoD, Myf-5 and Mrf4 collaborate in partially redundant networks to control myoblast cell fate (Rudnicki et al., 1993; Kablar et al., 1997; Kaul et al., 2000; Kassar-Duchossoy et al., 2004). As with embryonic myogenesis, each gene is non-essential for satellite cell specification (Megeney et al., 1996; Gayraud-Morel et al., 2007), although determining whether these MRFs function redundantly has been complicated by the lack of skeletal muscle and perinatal lethality of mice that are deficient for MyoD and Myf-5 or all three genes (Rudnicki et al., 1993; Kaul et al., 2000; Kassar-Duchossoy et al., 2004). Interestingly, however, the Pax7+ progenitor pool was absent at birth in mice lacking skeletal muscle due to mutations in MyoD and Myf-5 (Kassar-Duchossoy et al., 2005), raising the possibility that MyoD and Myf-5 function redundantly to specify or maintain the satellite cell lineage. A priori, redundancy could result from overlapping molecular functions in MyoD+/Myf-5+ progenitor cells, or could reflect regulative behavior of MyoD-dependent and Myf-5-dependent lineages. In this latter regard, recent diphtheria toxin cell ablation studies have identified Myf-5-expressing and non-expressing myogenic populations in the embryo, and have demonstrated remarkable regulative ability of the MyoD+ lineage (presumably Myf-5−) to rescue embryonic and fetal myogenesis in the absence of Myf5-expressing cells (Gensch et al., 2008; Haldar et al., 2008). Based on these considerations, we addressed whether the MyoD gene was essential for satellite cell specification within MyoD-expressing populations, and whether functional compensation by MyoD non-expressing lineages accounted for satellite cell development in MyoD-deficient mice. Our results showed that even in the absence of MyoD, satellite cells derive from one or more MyoD-expressing lineages. These data demonstrate that MyoD function is dispensable for satellite cell specification of the MyoD+ lineage, and that functional compensation by MyoD−/Myf-5+ cells, or other MyoD non-expressing lineages does not occur to an appreciable extent. Given the increase in the numbers of satellite cells in MyoD mutant muscle (Megeney et al., 1996; Yablonka-Reuveni et al., 1999; Gayraud-Morel et al., 2007), these data also suggest that MyoD plays a cell autonomous and non-redundant role in regulating the dynamic balance between proliferation, differentiation and renewal that normally establishes an appropriate satellite cell pool size.
Recent lineage analyses demonstrated satellite cell heterogeneity based on Myf5 expression. Rudnicki and colleagues (Kuang et al., 2007) proposed that the sublaminar satellite cell population is composed of 10% self-renewing stem cells that had never expressed Myf-5, and 90% myogenic precursor cells, defined by ongoing or prior Myf-5 expression, which are primarily responsible for execution of the myogenic program and production of myonuclei. Only the Myf5− satellite stem cells were able to undergo asymmetric cell division, generating both Myf5− and Myf5+ daughters and, correspondingly, were found to be more effective in repopulating the Pax7+ satellite cell compartment following cell transplantation into Pax7 mutant mice (Kuang et al., 2007). Despite this apparent functional compartmentalization, we showed that activation of MyoD transcription represents a common developmental feature of all types of satellite cells, implying that MyoD-dependent programming is compatible with acquisition of the satellite stem cell fate. In addition, immunohistochemistry for Cre protein showed that essentially all (99%) satellite cells in single fiber culture activated the MyoDiCre allele within 1 day of plating, prior to their first division (data not shown). These data strongly suggest that the population of MyoD-expressing satellite cells includes both the Myf5− stem cells and the Myf-5+ precursor cells. Although our experiments did not enumerate MyoD protein-expressing cells, work by others has shown that the vast majority of activated satellite cell and their daughters express MyoD protein (Yablonka-Reuveni and Rivera, 1994; Zammit et al., 2004). Collectively these data strongly suggest that MyoD-dependent programming is compatible with self-renewal, a notion supported by the observation that wild type myoblasts can generate satellite cells after transplantation into injured muscle, albeit inefficiently (Zammit et al., 2006). Using MyoD-dependent cell labeling strategies, it will be interesting to determine in vivo whether self-renewing, Myf5− satellite stem cells give rise to daughters that transiently activate MyoD and return to the MyoD- satellite cell compartment, as suggested by cell culture models of satellite cell renewal (Zammit et al., 2004).
MyoDiCre mice, in addition to their utility for investigating satellite cell precursor biology, provide a valuable new model to re-evaluate satellite cell developmental potential in cell culture, as well as to interrogate the cellular origins of increased fibrosis and adipose infiltration in mouse models of muscular dystrophy. Further, the ability to efficiently and permanently label satellite cells from diverse muscle groups based on prior MyoD transcription provides a simple and efficient means of prospectively isolating satellite cells for translational studies of muscle repair.
Supplementary Material
Acknowledgments
We thank the University of Connecticut Confocal Facility and Electron Microscopy Facility for training, advice and use of instrumentation, members of the Goldhamer lab for comments on the manuscript, Dr. B. Olwin for the Syndecan 4 antibody and Dr. R. Sprengel for the iCre construct. This project was supported by grants from the NIAMS and Muscular Dystrophy Association to D. J. G. J. J. M. was supported, in part by an NIH Supplement to Promote Diversity in Health-Related Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armand O, Boutineau AM, Mauger A, Pautou MP, Kieny M. Origin of satellite cells in avian skeletal muscles. Arch Anat Microsc Morphol Exp. 1983;72:163–81. [PubMed] [Google Scholar]
- Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–53. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–34. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bischoff R. The satellite cell and muscle regeneration. In: Franzini-Armstrong AGEaC., editor. Myology. Vol. 1. McGraw-Hill, Inc.; New York: 1994. pp. 97–118. [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–10. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The Role of Pax Genes in the Development of Tissues and Organs: Pax3 and Pax7 Regulate Muscle Progenitor Cell Functions. Annu Rev Cell Dev Biol. 2007 doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Chen JC, Love CM, Goldhamer DJ. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn. 2001;221:274–88. doi: 10.1002/dvdy.1138. [DOI] [PubMed] [Google Scholar]
- Chen JC, Mortimer J, Marley J, Goldhamer DJ. MyoD-cre transgenic mice: a model for conditional mutagenesis and lineage tracing of skeletal muscle. Genesis. 2005;41:116–21. doi: 10.1002/gene.20104. [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol. 2000;224:122–37. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Wilcox-Adelman SA, Goetinck PF, Rauvala H, Rapraeger AC, Olwin BB. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004;18:2231–6. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191:270–83. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- Cossu G, Molinaro M, Pacifici M. Differential response of satellite cells and embryonic myoblasts to a tumor promoter. Dev Biol. 1983;98:520–4. doi: 10.1016/0012-1606(83)90382-2. [DOI] [PubMed] [Google Scholar]
- Csete M, Walikonis J, Slawny N, Wei Y, Korsnes S, Doyle JC, Wold B. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. J Cell Physiol. 2001;189:189–96. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–10. [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Bast B, Neely C, Iem S, Amegbe P, Niewenhuis R, Miklasz S, Cheng PF, George-Weinstein M. MyoD-positive myoblasts are present in mature fetal organs lacking skeletal muscle. J Cell Biol. 2001;155:381–92. doi: 10.1083/jcb.200105139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Neely C, Elder J, Pfautz J, Perlman J, Narciso L, Linask KK, Knudsen K, George-Weinstein M. Cells that express MyoD mRNA in the epiblast are stably committed to the skeletal muscle lineage. J Cell Biol. 2007;178:649–60. doi: 10.1083/jcb.200703060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–8. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–45. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley RS, Bandman E, Yablonka-Reuveni Z. Skeletal muscle satellite cells appear during late chicken embryogenesis. Dev Biol. 1992;153:206–16. doi: 10.1016/0012-1606(92)90106-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–38. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA. Myogenic Determination Occurs Independently in Somites and Limb Buds. Dev Biol. 1999;206:219–231. doi: 10.1006/dbio.1998.9126. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–71. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–31. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul A, Koster M, Neuhaus H, Braun T. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–9. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447–59. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahira N, Ding L, Takebayashi H, Shimizu K, Ikenaka K, Ono K. Improved preservation of X-gal reaction product for electron microscopy using hydroxypropyl methacrylate. Neurosci Lett. 2005;374:17–20. doi: 10.1016/j.neulet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Mayer DC, Leinwand LA. Sarcomeric gene expression and contractility in myofibroblasts. J Cell Biol. 1997;139:1477–84. doi: 10.1083/jcb.139.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoon LK, Thorstenson KM, Solomon A, Lewis MP. Myogenic precursor cells in craniofacial muscles. Oral Dis. 2007;13:134–40. doi: 10.1111/j.1601-0825.2006.01353.x. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes & Development. 1996;10:1173–83. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27:e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol. 2004;275:375–88. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–53. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–90. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–9. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341:303–7. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, Kunkel LM, Kardon G. Somitic origin of limb muscle satellite and side population cells. Proc Natl Acad Sci U S A. 2006;103:945–50. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Wleklinski-Lee M, Yablonka-Reuveni Z. Skeletal muscle satellite cells can spontaneously enter an alternative mesenchymal pathway. J Cell Sci. 2004;117:5393–404. doi: 10.1242/jcs.01419. [DOI] [PubMed] [Google Scholar]
- Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol Biol. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain [letter] Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–38. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–70. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- van Beijnum JR, Rousch M, Castermans K, van der Linden E, Griffioen AW. Isolation of endothelial cells from fresh tissues. Nat Protoc. 2008;3:1085–91. doi: 10.1038/nprot.2008.71. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rivera AJ. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Developmental Biology. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol. 1999;210:440–55. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009 doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Watt CD, Schmidt RJ, Kuscuoglu U, Miesfeld RL, Goldhamer DJ. Cloning and characterization of a novel MyoD enhancer-binding factor. Mech Dev. 2007;124:715–28. doi: 10.1016/j.mod.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS. All muscle satellite cells are equal, but are some more equal than others? J Cell Sci. 2008;121:2975–82. doi: 10.1242/jcs.019661. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–57. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006;54:1177–91. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.