SUMMARY
The transcription factors that regulate endothelial cell development have been a focus of active research for several years, and many players in the endothelial transcriptional program have been identified. This review discusses the function of several major regulators of endothelial transcription, including members of the Sox, Ets, Forkhead, GATA, and Kruppel-like families. This review also highlights recent developments aimed at unraveling the combinatorial mechanisms and transcription factor interactions that regulate endothelial cell specification and differentiation during vasculogenesis and angiogenesis.
The vascular system is essential for embryonic development and adult life, and aberrant vascularization is associated with numerous diseases, including cancer, atherosclerosis, retinopathy, and stroke. Vasculogenesis, the de novo formation of endothelial cells from mesodermal precursors, occurs prior to the onset of blood circulation and results in the formation of the extra-embryonic yolk sac vasculature, the paired aortae, endocardium, and primary vascular plexus of the embryo (Flamme et al., 1997; Patan, 2004). In mammals, vascular progenitors first appear in the yolk sac, where mesodermal precursors of both hematopoietic and endothelial lineages differentiate into solid clumps known as blood islands (Flamme et al., 1997; Patan, 2004). The outer cells of these blood islands become flattened and differentiate into endothelial cells, while the inner cells become hematopoietic cells (Fig. 1). Subsequent fusion of blood islands results in the formation of a vascular plexus (Flamme et al., 1997; Patan, 2004; Fig. 1). Within the embryo, endothelial precursor cells, or angioblasts, aggregate to form the ventral and dorsal aortae, and the vitelline arteries and veins. At the same time, proendocardial cells migrate and line up along the intestinal portal to form a single endocardial tube (Flamme et al., 1997).
Figure 1.
Schematic representation of endothelial development from mesodermal progenitors. Endothelial cells development in the extraembryonic mesoderm in the yolk sac within blood islands, which contain an inner layer of hematotoietic cells and outer layer of angioblasts. From these blood islands, a primitive vascular plexus is formed. Within the embryo itself, angioblasts arise from mesodermal progenitors to form the aortae, cardinal veins and the endocardium (not depicted). The primitive embryonic and extraembryonic vasculature is then extensively remodeled via angiogenic processes. Mature, differentiated arteries, veins and lymphatic vessels are formed from the remodeled embryonic vasculature.
Following the initial formation of the primitive embryonic and extra embryonic vasculature through the process of vasculogenesis, these vascular systems are rapidly expanded and remodeled (Fig. 1). This process, referred to as angiogenesis, involves endothelial cell sprouting, vessel branching, and intussusception from existing blood vessels (Flamme et al., 1997; Patan, 2004). In addition, the blood vasculature becomes further specialized into arteries, veins and capillaries. Arteries consist of a layer of endothelial cells surrounded by multiple layers of smooth muscle cells. This provides vessels with a degree of contractility and provides strong structural support to the vessels due to the presence of elastic fibers in the smooth muscle walls of the vessels. In contrast, veins contain a thinner smooth muscle layer with fewer elastic fibers (Rossant and Hirashima, 2003). Capillaries and post-capillary venules are associated adluminally with pericytes, which are mesodermally-derived cells that provide a degree of contractility and help regulate permeability in the microvasculature (Hirschi and D'Amore, 1996). Although it was long hypothesized that specification of arteries and veins was controlled by mechanical stimuli, such as sheer stress and flow, the discovery of the differential expression of the genes encoding ephrin-B2 and its receptor EphB4 in arteries and veins, respectively, prior to the establishment of embryonic circulation strongly suggests that arterial and venous fates are at least partly genetically determined early in development (Adams et al., 1999; Wang et al., 1998).
A subset of endothelial cells within the developing embryo become further specialized into the cells of the lymphatic vasculature (Oliver and Alitalo, 2005). Lymphatic endothelial cells first appear as a polarized subset of cells lining the anterior cardinal vein, and then in more caudally located embryonic veins. These specialized endothelial cells then migrate from the veins to form primitive lymph sacs (Fig. 1). From these structures, lymphatic endothelial cells divide and sprout to give rise to the entire lymphatic network, although these lymph sacs are not required for the initial formation of the lymph nodes (Oliver and Srinivasan, 2008; Vondenhoff et al., 2009).
Numerous studies have examined the signaling molecules involved in vasculogenesis and angiogenesis, and it is well established that vascular endothelial growth factors (VEGFs) and their receptors are critical cell non-autonomous regulators of endothelial cell and blood vessel formation (Ferrara, 2004; Olsson et al., 2006). However, the transcriptional mechanisms through which the expression of genes downstream of VEGF receptors and the receptor genes themselves is activated and maintained in endothelial cells remain important questions in vascular biology. In addition, how VEGF and other signaling pathways influence the array of transcription factors involved in the endothelial gene expression program remains to be fully elucidated.
Endothelial Transcription Factors
Tal1 and GATA2 are Key Regulators of Hematopoietic and Endothelial Transcription
Many transcription factors are known to play an important role in the activation and maintenance of endothelial gene expression (Table 1). Given the common origin of blood and endothelial cells, it is perhaps not surprising several of the factors important for the early development of endothelial cells are also important for hematopoietic development (Table 2). For example, the bHLH transcription factor Tal1 (SCL) is essential for both blood and endothelial cell development (Bloor et al., 2002). Tal1 is expressed early during embryogenesis in the development of hematopoietic, endothelial, and neuronal precursor cells, and disruption of Tal1 in either mouse or zebrafish results in severe defects in the development of the vascular system (Green et al., 1992; Kallianpur et al., 1994; Patterson et al., 2005; Visvader et al., 1998; Fig. 2). Interestingly, blood vessels do form in the absence of Tal1, suggesting that this transcription factor may not be required for the initial specification of endothelial cells (Patterson et al., 2005; Visvader et al., 1998). On the other hand, over expression of Tal1 induces expression of several endothelial genes in zebrafish, suggesting that this factor may be sufficient for endothelial gene activation in vivo, at least in this context (Gering et al., 1998). Consistent with this dominant role for Tal1, several endothelial-specific gene enhancers are activated by Tal1 through essential E-box binding elements (Table 1).
Table 1.
A partial list of developmentally regulated endothelial enhancers and promoters. The list includes regulatory elements that were validated in transgenic embryos and have one or more identified, validated binding sites for endothelial transcription factors. Relevant references for each promoter/enhancer are listed. GS, gel shift assay (EMSA); Ch, chromatin immunoprecipitation; M(Tg), mutagenesis in transgenic embryos; M(C), mutagenesis in cell culture assays; TA, trans-activation assays in cell culture. The asterisks indicate factors identified by ChIP only.
| Gene | Enhancer Location |
Binding Sites | Binding Site Validation |
References |
|---|---|---|---|---|
| Mef2c (F10) | Intronic | FOX:ETS (Etv2/FoxC2); ETS |
GS; Ch; M(Tg); TA | (De Val et al., 2008) |
| Mef2c (F7) | Intronic | ETS | GS; M(Tg) | (De Val et al., 2004) |
| Flk1 | Upstream | FoxH1 | GS; M(C) | (Choi et al., 2007) |
| Flk1 | Promoter | HIF2α; ETS | M(Tg); TA | (Kappel et al., 2000) |
| Flk1 | Intronic | E box (Tal1); GATA; FOX:ETS (Etv2/FoxC2) |
GS; Ch; M(Tg); TA | (Kappel et al., 2000) (De Val et al., 2008) |
| Tal1 | Downstream | ETS (Fli-1/Elf-1/Etv2); GATA; FOX:ETS |
GS; Ch; M(Tg) | (De Val et al., 2008) (Gottgens et al., 2002) |
| Tal1 | Upstream | ETS (Fli-1/Elf-1) | GS; Ch; M(Tg); M(C) | (Gottgens et al., 2004) |
| Endoglin | Upstream | ETS (Fli-1/Erg/Elf) | Ch; M(Tg); M(C) | (Pimanda et al., 2006) |
| Endoglin | Intronic | ETS (Fli-1) | Ch | (Pimanda et al., 2008) |
| LMO2 | Promoter | ETS (Fli-1/Elf-1/Ets-1) | Ch; M(C) | (Landry et al., 2005) |
| Fli1 | Intronic | ETS; GATA; E box (Tal1)* | Ch; M(Tg); M(C) | (Pimanda et al., 2007) |
| Tie2 | Promoter | ETS (Ets-1/Elf-1); FOX:ETS (Etv2/FoxC2) |
GS; Ch; M(Tg); TA | (Dube et al., 1999) (Schlaeger et al., 1997) (De Val et al., 2008) (Minami et al., 2003) |
| Tie1 | Promoter | ETS (Ets-1/Ets-2/Nerf2) | M(Tg); TA | (Iljin et al., 1999) (Korhonen et al., 1995) |
| Flt1 | Promoter | ETS | M(C); TA | (Minami et al., 2002) (Morishita et al., 1995) (Wakiya et al., 1996) |
|
Ve-cadherin (Cdh5) |
Promoter | ETS (Ets-1); GATA2*; FOX:ETS (Etv2/FoxC2); E box (Tal1) |
GS; Ch; M(Tg); TA | (De Val et al., 2008) (Deleuze et al., 2007) |
| Gata2 | Intronic | E box (Tal1) | GS; M(Tg) | (Khandekar et al., 2007) |
| Gata2 | Upstream | ETS (Fli-1); GATA2; E box (Tal1)* |
GS; Ch; M(Tg) | (Kobayashi-Osaki et al.,2005) (Pimanda et al., 2007) |
|
Prox1 (lymphatic) |
Promoter | Sox18 | GS; Ch; M(C); TA | (Francois et al.,2008) |
| ECE1 | Intronic | FOX:ETS (Etv2/FoxC2) | GS; Ch; M(Tg); TA | (De Val et al., 2008) |
| FLT4 | Intronic | FOX:ETS (Etv2/FoxC2) | GS; Ch; M(Tg); TA | (De Val et al., 2008) |
| PDGFRβ | Intronic | FOX:ETS (Etv2/FoxC2) | GS; Ch | (De Val et al., 2008) |
| FOXP1 | Intronic | FOX:ETS (Etv2/FoxC2) | GS; Ch | (De Val et al., 2008) |
| NRP1 | Intronic | FOX:ETS (Etv2/FoxC2) | GS; Ch; M(Tg); TA | (De Val et al., 2008) |
| NOTCH4 | Promoter/ Intronic |
FOX:ETS (Etv2/FoxC2) | GS; Ch | (De Val et al., 2008) (Wu et al., 2005) |
| LYL1 | Intronic | ETS (Fli-1/Elf-1/Erg)*; GATA2* |
Ch | (Chan et al., 2007) |
| EPCR | Promoter | ETS; GATA; E box (Tal1) | M(C) | (Mollica et al., 2006) |
| ICAM-2 | Upstream | GATA | GS; M(C); TA | (Cowan et al., 1998) |
Table 2.
A partial list of mouse and zebrafish knockouts and knockdowns with endothelial phenotypes. E, embryonic day; MO, morpholino knockdown. Relevant references are indicated.
| Gene | Mutation | Phenotype | References |
|---|---|---|---|
| Tal1 (Scl) | mouse knockout; zebrafish MO |
lethal in the mouse at E9.5; failure of hematopoiesis; endothelial specification occurs; defective vascular remodeling |
(Patterson et al., 2005; Visvader et al., 1998) |
| Gata2 | mouse knockout | lethal by E10.5; normal vascular development; failure of primitive erythropoiesis |
(Tsai et al., 1994) |
| foxc1a | zebrafish MO | decrease in intersomitic vessel sprouting at 24 hpf | (De Val et al., 2008) |
| foxc1b | zebrafish MO | decrease in intersomitic vessel sprouting at 24hpf | (De Val et al., 2008) |
| foxc1a;b | zebrafish MO | absence of intersomitic vessel sprouts, diminished axial vessel formation at 24hpf |
(De Val et al., 2008) |
| Foxc2 | mouse knockout | lymphatic vessel hyperplasia, dysfunction and abnormal lymphatic patterning |
(Fang et al., 2000) |
| Foxc1/c2 | mouse knockout | lethal by E9.5; defective vascular remodeling, arteriovenous malformations, loss of arterial markers; defective heart development; defects in somitogenesis |
(Kume et al., 2001; Seo et al., 2006) |
| Foxo1 | mouse knockout | lethal by E10.5; defective vascular remodeling; cardiac looping defects | (Furuyama et al., 2004; Hosaka et al., 2004) |
| Foxo3 | mouse knockout | enhanced post-natal vessel formation | (Hosaka et al., 2004; Potente et al., 2005) |
| Foxo4 | mouse knockout | no vascular or other defects observed | (Hosaka et al., 2004) |
| Foxf1 | mouse knockout | defective mesodermal differentiation; defects in yolk sac vascular patterning |
(Mahlapuu et al., 2001) |
| Mef2c | mouse knockout | defective cardiac looping and myocardial differentiation; failure of vascular remodeling |
(Lin et al., 1998) |
| Klf2 | mouse knockout | hemorrhage due to incorrect tunica media formation, incorrect vessel stabilization; embryonic heart failure |
(Kuo et al., 1997;Lee et al.,2006) |
| Klf4 | mouse knockout | no obvious defects in endothelial cell development or function | (Katz et al., 2002;Segre etal.,1999) |
| Ets1 | mouse knockout; zebrafish MO |
decrease in intersomitic vessel sprouting, loss of trunk circulation at 24 hpf in zebrafish; no obvious vascular defects in the mouse; loss of NK cells in the spleen |
(Barton et al., 1998;Pham et al., 2007) |
| Fli1 | mouse knockout; zebrafish MO |
lethal in the mouse by E12.5 with hemorrhage and disruption of vessel tissue integrity;defective hematopoiesis but endothelial specification and initial differentiation is normal; slight decrease in intersomitic vessel sprouting in zebrafish morphants |
(Pham et al., 2007; Spyropoulos et al., 2000) |
| TEL | mouse knockout | lethal by E12; failure of vascular remodeling | (Wang et al., 1997) |
| Etv2 (Er71) | mouse knockout | lethal by E9.5; apparent loss of endothelial specification; complete lack of developed vasculature; absence of early vascular markers |
(Ferdous et al., 2009;Lee et al., 2008a) |
| etsrp | zebrafish mutation; zebrafish MO |
absence of intersomitic vessel sprouts, loss of trunk circulation at 24 hpf; defects in hematopoiesis |
(Pham et al., 2007; Sumanas and Lin, 2006) |
|
etsrp; foxc1a |
zebrafish MO | absence of intersomitic vessel sprouts, diminished axial vessel formation |
(De Val et al., 2008) |
| Lmo2 | mouse knockout; chimeric mice; zebrafish MO |
lethal in the mouse by E10.5; disorganization of vascular development after E9, defective vascular remodeling; decrease in intersomitic vessel sprouts and axial vessel formation in zebrafish |
(Patterson et al., 2005; Warren et al., 1994; Yamada et al., 2000) |
| Rbpj | mouse knockout | failure of vascular remodeling, loss of arterial markers; defects in somitogenesis and cardiac development |
(Krebs et al., 2004) |
| Hey1 | mouse knockout; | no obvious defects reported | (Fischer et al., 2004b) |
|
Hey2 (grl) |
mouse knockout; zebrafish mutation |
cardiac defects in the mouse; development of arteriovenous shunts, loss of aorta, increased venous marker expression, decreased expression of arterial markers in the zebrafish |
(Fischer et al., 2004a; Weinstein et al., 1995; Zhong et al., 2001) |
| Hey1/2 | mouse knockout | Failure of vascular remodeling, absence of arterial markers | (Fischer et al., 2004b); (Kokubo et al., 2005) |
| Sox17 | mouse knockout | Limited defects in anterior dorsal aorta, enlarged cardinal vein; defective endoderm development; aberrant cardiac looping |
(Kanai-Azuma et al., 2002; Sakamoto et al., 2007) |
| Sox18 | zebrafish MO | Mild subcutaneous edema, defective lymphatic patterning; blood vasculature unaffected |
(Francois et al., 2008) |
| Sox17;18 | mouse knockout | Limited defects in anterior dorsal aorta and head vasculature | (Sakamoto et al., 2007) |
| Sox7;18 | zebrafish MO | Lack of trunk and tail circulation, fusions between major axial vessels, errors in arteriovenous specification |
(Cermenati et al., 2008; Pendeville et al., 2008) |
|
Nr2f2 (Coup-TFII) |
mouse knockout; conditional ko |
Lethal by E11.5; hemorrhage, thin and dialated vessels, ectopic expression of arterial markers, decrease of venous markers; defective vascular remodeling; cardiac defects |
(Pereira et al., 1999;You et al., 2005) |
| Prox1 | mouse knockout | lethal by E14.5; no lymphatic vessels; massive edema; blood vasculature unaffected |
(Wigle and Oliver, 1999) |
Figure 2.
Schematic depiction of the vascular and hematopoietic phenotypes in Tal1-null mouse embryos at E9.5. A representation of the yolk sac vasculature is shown. Compared to wild-type embryos (A, C), Tal1 mutant embryos show an absence of blood cells (white circles). Remodeling of the vascular endothelium is defective in mutant embryos (B), but endothelial cells (white ovals) are present in the mutant embryos (D), suggesting that initial specification occurs properly. Arrowheads mark endothelium in A, B and endothelial cells in C, D. Based on data published in (Visvader et al., 1998).
The zinc finger transcription factor GATA2 is also an important regulator of both hematopoietic and endothelial genes. GATA2 is the most abundantly expressed GATA factor in endothelial cells (Lee et al., 1991), and numerous endothelial enhancers contain GATA binding sites, which are bound directly by GATA2 (Table 1). In addition, experiments in embryonic stem cells demonstrated the importance of GATA2 in the development of Flk-1+/ Tal1+ hemangioblast-like cells and in the induction of endothelial-specific genes (Lugus et al., 2007). Together, these studies support the notion that GATA2 is an early regulator of hematopoietic and endothelial development and that this transcription factor may be involved in the specification of hemangioblast progenitors from the mesoderm early in embryonic development.
Forkhead Proteins are Important Regulators of Endothelial Transcription
Members of at least five different subfamilies of Forkhead (Fox) transcription factors are expressed in endothelial cells or their precursors. These include members of the FoxC, FoxF, FoxH, and FoxO families (Papanicolaou et al., 2008). Although no Forkhead proteins are specific to endothelial cells or their progenitors, several play essential roles in vascular biology and endothelial transcription. Targeted disruption of Foxo1 in mice causes vascular remodeling defects and mid-gestational lethality (Furuyama et al., 2004; Hosaka et al., 2004; Kume et al., 2001; Table 2). While it is clear that FoxO1 is required for vascular development, the mechanism through which it controls endothelial gene expression remains unresolved. FoxO factors are generally thought to bind to and function through insulin response elements (IREs) (Dejana et al., 2007; Furuyama et al., 2003). However, FoxO regulation can occur independently of these elements, and FoxO1 also binds to other divergent motifs (De Val et al., 2008; Paik et al., 2007; Potente et al., 2005; Ramaswamy et al., 2002). Interestingly, FoxO1 functions as both a positive and negative regulator of transcription, suggesting that this factor may act as a transcriptional switch in the endothelium (Daly et al., 2004; Paik et al., 2007).
FoxF1 and FoxH1 are also involved in endothelial gene regulation. Inactivation of Foxf1 in mice results in a severe vascular phenotype and embryonic lethality (Mahlapuu et al., 2001; Table 2). Interestingly, Foxf1 is not expressed within endothelial cells of the differentiated embryonic vasculature. Rather, Foxf1 is expressed earlier in the splanchnic mesoderm prior to endothelial cell specification and may regulate BMP signaling, which is essential for vascular development (Astorga and Carlsson, 2007). In contrast, FoxH1 may play an inhibitory role on vascular development. In the zebrafish, FoxH1 over expression impairs vascular development and is a negative regulator of flk1 expression through direct enhancer binding (Choi et al., 2007).
The FoxC family of Forkhead proteins is essential for vascular development as Foxc1−/−;Foxc2−/− mice have severe vascular defects (Kume et al., 2001; Fig. 3). Although endothelial cells are specified in embryos lacking FoxC1 and FoxC2, some studies have highlighted the requirement for these factors in early endothelial development (De Val et al., 2008; Kume et al., 2001). Additionally, FoxC1 and FoxC2 also have an important function in arterial and lymphatic endothelial cell specification, and may be important downstream effectors of Notch signaling (Hayashi and Kume, 2008; Seo et al., 2006). More recently, we have demonstrated an important role for FoxC proteins as cofactors for Ets proteins in the combinatorial regulation of endothelial gene expression (De Val et al., 2008; discussed below).
Figure 3.
Defective differentiation of arterial endothelial cells and abnormal artery development in Foxc1; Foxc2 double mutant mice at E9. Foxc1;Foxc2 double mutant mice exhibit arteriovenous malformations (white arrow in panel B) and abnormal expression of venous markers, such as COUP-TFII in arteries (C, D). The white arrow in (C) marks the dorsal aorta (da) in the wild type embryo, which expresses PECAM-1, but not COUP-TFII. The white arrowhead in (C) marks the cardinal vein (cv) in the wild type embryo, which properly expresses both PECAM-1 and COUP-TFII. The red arrow in (D) marks PECAM-1+ arterial endothelial cells, which aberrantly express COUP-TFII and other venous markers in double mutant embryos. nt, neural tube. The images shown were originally published in (Seo et al., 2006).
Krüppel-like Factors Regulate Endothelial Genes in Response to Injury and Stress
Members of the Krüppel-like factor (KLF) transcription factor family appear to function in endothelial cells after initial specification and differentiation have already occurred (Atkins and Jain, 2007)). Klf2 expression is induced by sheer stress and regulates the expression of several genes important for maintaining vascular tone in response to flow (Dekker et al., 2002; Dekker et al., 2005; Lee et al., 2006; SenBanerjee et al., 2004). In addition, Klf2 null mice die by embryonic day (E)14.5 due to hemorrhage caused by incorrect vessel stabilization and defective tunica media formation (Kuo et al., 1997; Lee et al., 2006; Table 2). Although less is known about their involvement in vascular biology, KLF4 and KLF6 are also expressed in endothelial cells and their expression is increased by sheer stress and vascular injury (Atkins and Jain, 2007; Botella et al., 2002; Hamik et al., 2007; Kojima et al., 2000; Yet et al., 1998). Interestingly, Krüppel-like factors are also expressed in endothelial cells during vasculogenesis and early angiogenesis, but it remains to be determined exactly how members of this family function in the early pathways that regulate endothelial cell specification and differentiation.
Ets Transcription Factors are Central Regulators of Endothelial Gene Expression
Although many transcription factors play important roles in vascular development, none appear to be as centrally involved in the transcriptional programs controlling endothelial cell development as Ets proteins. To date, all characterized endothelial enhancers and promoters contain multiple, essential ETS binding sites (Table 1), and ETS motifs are strongly associated with endothelial genes throughout the human genome (Bernat et al., 2006; De Val et al., 2008). At least 19 different Ets factors are expressed in human endothelial cells, and over 12 are present in endothelial cells in zebrafish (Hollenhorst et al., 2004; Liu and Patient, 2008). Within the Ets family, Ets-1, Elf-1, Fli-1, Tel, and Erg each have well characterized roles in endothelial gene expression, and each bind to the enhancers and activate the expression of numerous endothelial genes (Table 1). Knock down of either Ets1 or Erg expression in endothelial cells in culture results in decreased endothelial cell migration and tube formation (Birdsey et al., 2008; Iwasaka et al., 1996). Intriguingly, germline deletion or mutation of the majority of individual Ets genes in either mouse or zebrafish model systems has resulted in little or no vascular phenotype or has caused defects only in later vascular remodeling, while vasculogenesis remained largely intact (Barton et al., 1998; Hart et al., 2000; Pham et al., 2007; Spyropoulos et al., 2000; Wang et al., 1997; Table 2). This is most likely due to redundancy among Ets factors in endothelial development. The exception to this apparent redundancy among Ets factors in endothelial development is observed when the function of the Ets protein Etv2 (ER71, Etsrp71) is removed in mice or when its ortholog Etsrp is knocked down in zebrafish (Ferdous et al., 2009; Lee et al., 2008a; Sumanas et al., 2008). In contrast to other Ets genes, inactivation of Etv2/Etsrp causes profound impairment of vasculogenesis, suggesting a central role for this factor in endothelial specification (discussed below).
The redundancy among the majority of Ets factors in endothelial biology probably reflects the fact that these proteins bind to identical or nearly identical cis-acting elements. The DNA binding domain of Ets transcription factors, also known as the Ets domain, is highly conserved among all members of the family, and all Ets proteins bind to the same invariant GGA(A/T) core sequence (Graves and Petersen, 1998). Although there are flanking sequence requirements in addition to the invariant core sequence, the majority of Ets proteins show preferential binding to similar extended consensus sequences (Graves et al., 1996; Gunther and Graves, 1994; Landry et al., 2005; Pimanda et al., 2006). Furthermore, no Ets factors are exclusively expressed in endothelial cells, and Ets proteins function in many developmental processes other than the endothelial program, including hematopoiesis, neuronal maturation, and bone development (Bartel et al., 2000; Dalla Torre di Sanguinetto et al., 2008; Maroulakou and Bowe, 2000; Sharrocks, 2001). In addition, nearly two-thirds of mammalian Ets factors are almost ubiquitous in their expression in adult tissues, and ETS binding sites are not specific to endothelial-expressed gene loci (Hollenhorst et al., 2004). Indeed, it has been estimated that 5–15% of gene promoters, many for housekeeping genes, are bound by Ets proteins (Hollenhorst et al., 2007). This has raised the question as to how Ets proteins contribute to endothelial-specific gene expression. Accordingly, it has been hypothesized that Ets factors must regulate endothelial-specific transcription by functioning in combination with other transcription factors. While this notion is likely to be correct, recent studies have implicated two individual Ets factors, Fli-1 and Etv2, as early, essential regulators of endothelial cell specification.
The Ets Protein Fli-1 is an Early Regulator of Endothelial Development
The Ets transcription factor Fli-1 is expressed very early in cells of the hematopoietic and endothelial lineages in mice and zebrafish (Brown et al., 2000; Melet et al., 1996). However, unlike other early endothelial markers such as Tal1, Gata2, and Tie2, Fli1 is expressed in cloche mutant zebrafish, which lack differentiated endothelial cells and also have impaired hematopoietic development (Brown et al., 2000; Liao et al., 1997; Stainier et al., 1995). This early expression in cloche mutants, even in the absence of clearly identifiable endothelial cells, suggests that Fli-1 may be one of the earliest transcription factors involved in endothelial and hematopoietic progenitor cell development. Gain-of-function experiments in zebrafish further support a role for Fli-1 in early endothelial and hematopoietic specification. Injection of an mRNA encoding a constitutive activator form of Fli-1 (Fli-1-VP16) induced expression of Tal1, Gata2, and other hemangioblast markers but did not rescue the phenotype in Tal1 morphants, suggesting that Fli-1 acts upstream of Tal1 in the hemangioblast and endothelial programs (Liu et al., 2008).
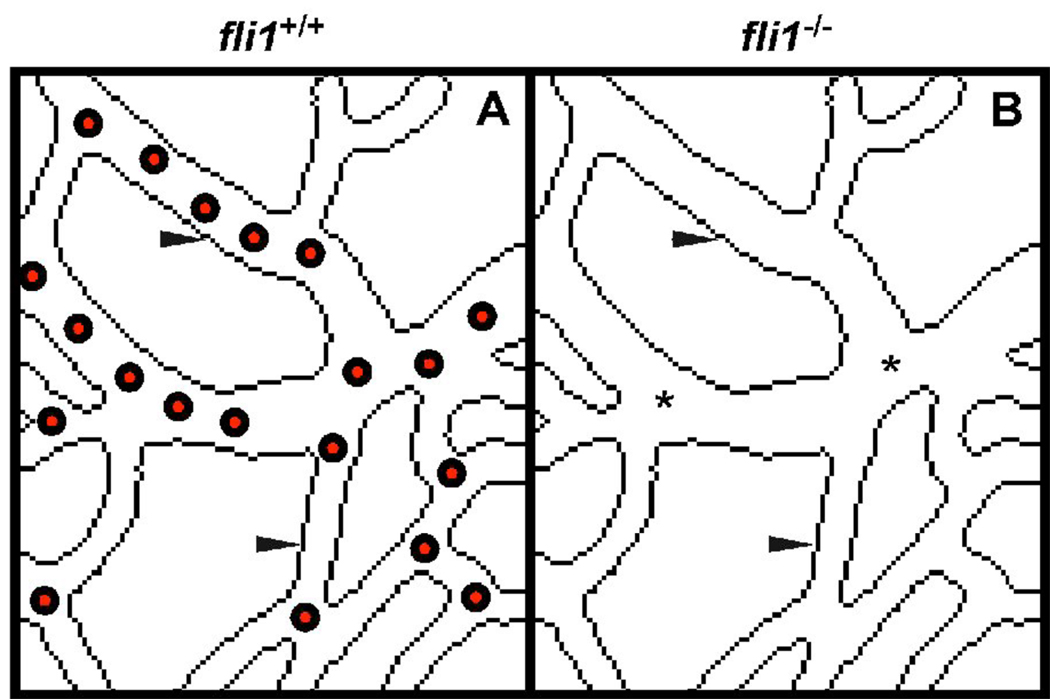
Fli1 also appears to function upstream of Gata2 in the hematopoietic and endothelial programs. Morpholino knock down experiments in Xenopus embryos at sites of primitive and definitive hemangioblast populations showed that inhibition of Gata2 had no effect on Fli1 expression, whereas Fli-1 knock down moderately reduced Gata2 expression (Liu et al., 2008). Knock down of either Gata2 or Fli1 inhibited expression of other endothelial markers, including Tal1, Lmo2, and flk1, suggesting that the two factors both function early in the program, but that Fli1 may act upstream of Gata2 in regulation of hemangioblast gene expression (Liu et al., 2008). Finally, Ets sites within gene enhancers for Gata2, Tal1, and Fli1 itself are essential for expression, and each of these enhancers is bound by Fli-1, further suggesting that Fli-1 functions upstream in the hemangioblast program in a feed-forward or recursive mechanism (Donaldson et al., 2005; Liu et al., 2008; Pimanda et al., 2007). Importantly, disruption of Fli1 in either mouse or zebrafish models does not result in significant vascular defects (Fig. 4), possibly due to the continued expression of closely related factors Erg (in mouse) and Fli1b (in zebrafish) (Hart et al., 2000; Pham et al., 2007; Spyropoulos et al., 2000). Alternatively, this may suggest that Fli-1 plays a predominant role in hemangioblast and subsequent hematopoietic development, whereas other Ets factors, particularly Etv2 (discussed below), may function in hemangioblast development and subsequent endothelial specification.
Figure 4.
Schematic representations of mouse yolk sacs at E11 from wild type (A) and Fli1 mutant (B) embryos. Endothelial cells are properly specified and the vasculature is initially formed normally in mutant embryos. However, mutant embryos have a nearly complete absence of blood cells (circles in A), indicating a profound defect in hematopoiesis. Based on data published in (Spyropoulos et al., 2000).
The Ets Transcription Factor Etv2 is an Essential Regulator of Endothelial Development
Several recent studies have shown that the Ets-related factor Etv2 (ER71, Etsrp71), which was previously thought to be testis-specific, is essential for the development of endothelial and blood lineages in the mouse (Ferdous et al., 2009; Lee et al., 2008a; Sumanas et al., 2008). Etv2 expression is present at the very early stages of vascular development in the mouse, with expression detected in the blood islands of the yolk sac, and the earliest vessels in the embryo. Notably, Etv2 expression begins decreasing within endothelial cell populations by E9.5 and is essentially extinguished in those lineages by E10.5, suggesting an involvement of this transcription factor in early vascular development (Ferdous et al., 2009; Lee et al., 2008a; Lugus et al., 2007). Etv2 null mice have severe defects in vasculogenesis and hematopoiesis. Etv2−/− embryos die at midgestation and lack any detectable embryonic vessels, blood islands in the yolk sac, or endothelial progenitors (Ferdous et al., 2009; Lee et al., 2008a; Fig. 5). Expression of early vascular markers, such as flk1, Pecam, and Tie2-lacZ is almost completely abolished in the absence of Etv2, and endothelial cells are apparently not specified if Etv2 is not present (Ferdous et al., 2009; Lee et al., 2008a).
Figure 5.
Etv2 null mice fail to specify endothelial cells. Immunohistochemical staining for Flk1 (A, B) at E9.5 shows normal endothelial cell specification and differentiation and vessel remodeling in wild type embryos (A), whereas Etv2 (Er71) mutants have no detectable expression of Flk1 (B). Similarly, blood-filled vessels are not present in the yolk sac of Etv2 mutant embryos (D) at a stage when they are obvious in wild type controls (C, arrowheads). This is due to loss of detectable endothelial cells and profound defects in hematopoiesis. (E, F) Schematic representations of wild type (E) and Etv2 (Etsrp71) (F) mutant embryos, indicating that the endocardium is not properly specified in mutant embryos, as evidenced by the loss of expression of Endomucin and other endothelial markers (denoted in blue in E). The images shown in (A–D) were originally published in (Lee et al., 2008a). The schematic images shown in (E, F) are based on data published in (Ferdous et al., 2009).
Consistent with an early role for Etv2 in the endothelial cell transcriptional hierarchy, Etv2 expression is highly enriched in flk1+/Tal1+ hemangioblast-like embryonic stem cells, and Etv2 is also a potent inducer of Flk1+ mesoderm in embryonic stem cells (Lee et al., 2008a). Furthermore, Etv2 is a potent activator of several early endothelial genes, including flk1, Tal1, Mef2c, Pecam, and Tie2 and has been demonstrated to activate these genes through direct promoter or enhancer binding (De Val et al., 2008; Ferdous et al., 2009); (Table 1).
In zebrafish, the Ets transcription factor Etsrp is the likely ortholog of Etv2 (Pham et al., 2007; Sumanas et al., 2008; Sumanas and Lin, 2006). Expression of etsrp is restricted to vascular endothelial cells and their precursors, and knock down of etsrp causes severe defects in vascular development, although with variable penetrance (Pham et al., 2007; Sumanas et al., 2005; Sumanas and Lin, 2006). In addition, y11 mutant zebrafish, which have a defective etsrp gene, lack intersegmental vessels at 24 hours post-fertilization (hpf), and blood vessels in these embryos fail to undergo correct tubular morphogenesis (Pham et al., 2007; Sumanas et al., 2008). Notably, forced expression of Etsrp or mouse Etv2 in zebrafish leads to expansion of both hemangioblast and vascular endothelial lineages (Sumanas et al., 2008). Taken together, the mouse and zebrafish studies highlight a central, early role for Etv2/Etsrp in endothelial specification and suggest that this transcription factor acts at or near the top of the transcriptional network controlling endothelial cell development (Fig. 6).
Figure 6.
Different stages in endothelial development are regulated by distinct sets of transcription factors. This model depicts the steps within endothelial cell development from mesodermal progenitors and hemangioblasts to differentiated arterial, venous, and lymphatic endothelium and the various transcription factors associated with their development. This model depicts the speculation that Fli-1, GATA2, and Tal1 control differentiation of hematopoietic cells from hemangioblasts, while Etv2 and FoxC proteins control the differentiation of endothelial cells from that progenitor population and that Etv2 likely sits at the top of this transcriptional cascade.
Combinatorial Regulation of Endothelial Transcription
Regulation by Multiple Factors through Clustered Binding Sites
As noted above, no transcription factor is known to be expressed exclusively in endothelial cells or their progenitors. This has led to the suggestion that endothelial-specific gene expression is controlled combinatorially by multiple transcription factors that have overlapping expression in the endothelium. Indeed, many endothelial-specific enhancers contain clustered binding sites for multiple transcription factors, suggesting that endothelial specificity may be achieved through the combination of these sites and their cognate binding proteins. For example, several characterized endothelial and hematopoietic enhancers contain conserved binding sites for Ets and GATA transcription factors (Table 1), suggesting that these two factors may function together in the activation of these genes. Indeed, the Ets factors Fli-1 and Elf-1 physically interact with GATA factors, and Ets and GATA factors synergistically activate a megakaryocyte enhancer from the GPIX gene and the endogenous ANG2 gene in HeLa cells (Eisbacher et al., 2003; Gottgens et al., 2002; Pimanda et al., 2006; Simon et al., 2008).
A genome-wide search for ETS and GATA sites in a similar arrangement as that found in the Tal1 endothelial and hematopoietic stem cell enhancer identified two additional enhancers in the human FLI1 and PRH loci, which were each sufficient to direct expression to endothelial and hematopoietic lineages in transgenic mice (Donaldson et al., 2005). However, Ets and GATA factors are coexpressed in many tissues other than the vasculature, and the 67 ETS/GATA clusters identified in the genome-wide scan were not enriched in their association with endothelial or hematopoietically expressed genes (Donaldson et al., 2005), suggesting that other cis-acting elements must also be required to confer endothelial specificity. Indeed, ETS and GATA sites have been found in association with Tal1 E-boxes in enhancers from the fli1, Gata2, flk1, and EPCR genes, and it has been proposed that GATA2, Fli-1, and Tal1 form a self-reinforcing gene regulatory circuit for early hematopoietic and endothelial cell gene expression (Kappel et al., 2000; Mollica et al., 2006; Pimanda et al., 2007).
Although the mechanism through which Tal1, GATA2, and Ets transcription factors function remains unresolved, their role in endothelial gene co-regulation suggests that the two may function as part of a biochemical complex (Pimanda et al., 2007). Additional support for this notion comes from the observation that the LIM domain protein, LMO2, also associates with GATA2 and Tal1 as part of a multifactorial complex (Lahlil et al., 2004; Wadman et al., 1997). LMO2 functions as a transcriptional cofactor without directly binding DNA and is coexpressed with Tal1 and GATA2 in blood and endothelial progenitors in zebrafish (Liu et al., 2008). LMO2, Tal1, and GATA2 function together to activate the VE-cadherin (Cdh5) promoter, and ectopic expression of Tal1 and Lmo2 together in zebrafish embryos results in the induction of endothelial markers (Deleuze et al., 2007; Gering et al., 2003). Interestingly, the VE-cadherin promoter also contains several conserved ETS sites (Prandini et al., 2005), suggesting that all of these factors may function together via their clustered binding sites to confer endothelial specificity.
Combinatorial Regulation by Multiple, Distinct Ets Family Members
Endothelial enhancers usually have multiple, conserved Ets sites, which occur in clusters (Table 1). Often, more than one of these Ets sites is required for enhancer function, and many endothelial enhancers appear to be bound by more than one individual Ets family member (De Val et al., 2004; De Val et al., 2008; Gottgens et al., 2004; Gottgens et al., 2002; Landry et al., 2005; Pimanda et al., 2006; Prandini et al., 2005). These observations have led to the hypothesis that multiple, discrete Ets proteins may function combinatorially to regulate endothelial gene expression. In support of this idea, Pham et al. showed that combined knock down of four distinct Ets genes caused a much more severe vascular phenotype in zebrafish than knock down of any of the individual Ets genes alone (Pham et al., 2007). Although these studies suggest a possible interaction among Ets proteins, these observations may simply reflect that redundancy among Ets family members exists in zebrafish, rather than cooperativity or combinatorial regulation. Additional genetic and biochemical studies are needed to resolve whether Ets proteins function cooperatively with themselves or other members of the Ets family.
Combinatorial Regulation through Composite Binding Sites
As noted earlier in this review, Ets proteins are central regulators of the endothelial transcriptional program. However, the widespread distribution of Ets factors in the embryo, combined with their ability to bind to identical cis-acting elements, presents a challenge to understanding how endothelial specificity by members of this transcription factor family may be achieved. It has also been hypothesized that Ets proteins may bind to more divergent sequences than the canonical ETS motif by interacting with DNA in conjunction with other factors (Hollenhorst et al., 2007). Indeed, recent studies by our group demonstrate that Ets factors function combinatorially with FoxC transcription factors through a composite DNA binding site, the FOX:ETS motif, which consists of an ETS site and a noncanonical Forkhead site (De Val et al., 2008). We found that evolutionarily-conserved FOX:ETS motifs were strongly associated with endothelial genes throughout the human genome, and essential FOX:ETS motifs were identified within numerous previously characterized endothelial enhancers and promoters, including those from Tal1, Tie2, flk1, and VE-cadherin (Cdh5) (De Val et al., 2008). The FOX:ETS motif is bound robustly by FoxC2 and Etv2, and the two proteins bind the element simultaneously, suggesting cooperativity by these factors.
Consistent with a cooperative role for FoxC2 and Etv2 in endothelial gene activation, the two factors synergistically activate all of the enhancers or promoters that contain a bona fide FOX:ETS motif and were examined experimentally, including Mef2c, Notch4, Tal1, Tie2, flk1, and VE-cadherin (De Val et al., 2008). Interestingly, FoxC2 or Etv2 alone only weakly activated the endothelial enhancer elements; strong synergy required both factors together. Importantly, coexpression of FoxC2 and Etv2 together in a normally avascular region of Xenopus embryos resulted in strong induction of endothelial genes and ectopic vessel formation, while neither factor alone caused this effect (De Val et al., 2008). Thus, these recent studies suggest a mechanism for endothelial gene activation and vascular development depending on two factors, neither of which is specific to endothelial cells, converging on a common, composite cis-acting motif. It will be interesting to determine if this is a general mechanism for endothelial gene activation and if Ets proteins function combinatorially with factors other than FoxC proteins by co-binding DNA through composite sites, as has been proposed (De Val et al., 2008; Hollenhorst et al., 2007).
Transcriptional Control of Endothelial Subtype Specification
Arteries and veins exhibit differences in gene expression prior to the onset of blood circulation, indicating an important role for genetic pathways in arteriovenous differentiation, and extensive work in zebrafish and mouse model systems has demonstrated an essential role for Notch signaling in arteriovenous identity (Krebs et al., 2004; Krebs et al., 2000; Lawson et al., 2001; Villa et al., 2001). Notch receptors are specifically expressed in arterial cells, and Notch signaling functions in endothelial cells, at least in part, to suppress venous fate (Lawson et al., 2001; Villa et al., 2001). After binding by their cognate ligands, Notch receptors are cleaved, and the Notch intracellular domain (NICD) translocates to the nucleus where it interacts with its obligate canonical signaling partner RBP-J (CBF1), resulting in activation of Notch target genes, such as Hey1 and Hey2 (Roca and Adams, 2007).
Transcription Factors Involved in Arterial Specification
The Notch target genes Hey1 and Hey2 encode members of the hairy and enhancer of split-related family of bHLH transcription factors and are mammalian orthologs of the zebrafish gridlock (grl) gene (Kokubo et al., 2005). Hey proteins are important moderators of Notch signaling in arteriovenous specification. grl is strongly expressed in the developing dorsal aorta but not the axial vein in zebrafish, and grl mutants have defective development of the dorsal aorta caused by arteriovenous shunts (Weinstein et al., 1995; Zhong et al., 2001). Knock down of grl results in loss of the dorsal aorta, with a concomitant increase in the size of axial vein. In addition, expression of arterial markers is reduced in grl mutants while expression of venous markers is increased (Zhong et al., 2001). Conversely, over expression of grl causes a reduction in the size of the axial vein. Taken together, these observations have led to the suggestion that a gridlock-dependent pathway controls the formation of the dorsal aorta in zebrafish by determining arterial-venous identity (Zhong et al., 2001). Similarly, compound Hey1;Hey2 double knockout mice die at midgestation with severe vascular defects and lack the expression of arterial markers (Fischer et al., 2004b; Kokubo et al., 2005), further supporting a role for Hey transcription factors and upstream Notch signaling in arterial identity.
In addition to their role in vasculogenesis in cooperation with Ets proteins, the Forkhead transcription factors FoxC1 and FoxC2 are essential for arteriovenous specification. Foxc1/Foxc2 compound null mice display vascular fusions between arteries and veins, and the expression of many arterial markers, including Notch1, Notch4, Dll4, Jagged1, and Hey2, are absent in these mice even though the expression of pan-endothelial markers is maintained (Kume et al., 2001; Seo et al., 2006). Furthermore, expression of either FoxC1 or FoxC2 in fibroblast and endothelial cell lines increases expression of arterial-specific genes, and the promoter regions of the arterial marker genes Hey2 and Dll4 are bound and activated by FoxC proteins (Hayashi and Kume, 2008; Seo et al., 2006). Since the expression of Foxc1 and Foxc2 is not restricted to the arterial compartment, it has been proposed that FoxC-specific regulation of arterial genes may be achieved in combination with NICD and RBP-J, which implicates FoxC proteins in the VEGF and Notch signaling pathways (Hayashi and Kume, 2008).
Members of the F-subgroup Sox transcription factor family, Sox7, Sox17, and Sox18, may also be required for correct arteriovenous specification. Knock down of the orthologs of sox7 and sox18 together in zebrafish causes severe vascular defects (Cermenati et al., 2008; Pendeville et al., 2008). sox7; sox18 morphants lack trunk and tail circulation, exhibit multiple fusions between the major axial vessels, and have aberrant expression of arterial markers (Cermenati et al., 2008; Pendeville et al., 2008). A dominant-negative form of Sox18 is the cause of the ragged mouse mutant, which exhibits cardiovascular phenotypes (James et al., 2003; Pennisi et al., 2000a; Pennisi et al., 2000b), and inactivation of Sox18 on an inbred background results in defects in lymphatic vasculature and embryonic lethality (Francois et al., 2008).
COUP-TFII Regulates Differentiation of Venous Endothelium
COUP-TFII (encoded by the Nr2f2 gene), a nuclear receptor bound and activated by retinoic acid, which may be its endogenous ligand, functions as a key regulator of venous identity by suppressing Notch signaling (Kruse et al., 2008; You et al., 2005). COUP-TFII expression in the endothelium is limited to veins and lymphatics, and deletion of the Nr2f2 gene in mice results in embryonic lethality due to vascular defects. Notably, the absence of COUP-TFII causes inappropriate expression of arterial markers, including Nrp1 and Notch1, in veins, supporting a role for this transcription factor in venous identity (Pereira et al., 1999; You et al., 2005). Further evidence for a role for COUP-TFII in venous identity comes from gain-of-function studies in mice. Over expression of COUP-TFII in all endothelial cells caused defects in angiogenesis, large fused vessels, and lack of arterial-venous distinction (You et al., 2005). Based on these and other studies, it has been proposed that COUP-TFII acts by suppressing Nrp1 expression and inhibiting Notch signaling, which results in suppression of arterial-specific genes and activation of venous genes (You et al., 2005). However, this model remains to be tested and the mechanisms through which COUP-TFII suppresses arterial fate remain to be elucidated.
Transcriptional Regulation of Lymphangiogenesis
The SRY-box containing transcription factor Sox18 may act as a molecular switch inducing the differentiation of lymphatic endothelial cells. The human disease hypotrichosis-lymphedema-telangiectasia, characterized by chronic swelling of the extremities due to dysfunction of the lymphatic vessels, is caused by SOX18 mutation (Irrthum et al., 2003). Similarly, Sox18 null mice die by E15 with massive edema, and Sox18 heterozygous mice display defects in their lymphatic vasculature (Francois et al., 2008). Over expression of Sox18 in embryonic stem cells or cultured endothelial cells results in increased expression of lymphatic markers, including Prox1 (Francois et al., 2008). Interestingly, Sox18 expression precedes Prox1 expression in the dorsolateral sector of the anterior cardinal veins, the first site of lymphangiogenesis in mice, suggesting that Sox18 may function upstream of Prox1 and be among the earliest regulators of lymphatic endothelial cell specification. In support of this notion, a 4-kb Prox1 promoter fragment, which recapitulates endogenous Prox1 expression in transgenic mice, is bound and activated by Sox18 (Francois et al., 2008).
The homeodomain transcription factor Prox1 has often been described as the master regulator of lymphangiogenesis (Adams and Alitalo, 2007; Hong and Detmar, 2003; Hong et al., 2002; Oliver and Alitalo, 2005). Prox1 expression is first observed in a subset of venous endothelial cells, where lymphangiogenesis initiates (Adams and Alitalo, 2007; Hong and Detmar, 2003; Oliver and Alitalo, 2005; Wigle and Oliver, 1999). Although Prox1 expression is not restricted to endothelial cells, within the vasculature, it is only detected in lymphatic endothelial cells (Rodriguez-Niedenfuhr et al., 2001; Wigle and Oliver, 1999). Prox1 null mice fail to develop any lymphatic vasculature and die by E15 due to massive edema, even though the blood vasculature is not affected (Wigle and Oliver, 1999). These loss-of-function and expression data point to an essential role for Prox1 in early lymphangiogenesis. Furthermore, Prox1 is sufficient to promote a lymphatic fate. Forced expression of Prox1 in cultured blood endothelial cells increases the expression of genes specific to the lymphatic endothelium, such as Podoplanin (Pdpn) and flt4 (Vegfr3), and the concomitant down regulation of blood endothelial genes (Hong et al., 2002; Petrova et al., 2002).
Despite the obvious importance of Prox1 in the specification of lymphatic endothelial cells, surprisingly few putative direct transcriptional targets of Prox1 have been identified (Mishima et al., 2007; Shin et al., 2006). It has recently been proposed that Prox1 may activate the expression of lymphatic endothelial genes in combination with COUP-TFII, which is a key regulator of venous identity, as discussed earlier in this review. COUP-TFII and Prox1 proteins are coexpressed in sprouting lymphatic endothelial cells and newly formed lymphatic vessels, and the two factors physically interact (Lee et al., 2008b). Prox1 and COUP-TFII synergistically activate the expression of Fgfr3, a known Prox1 target, which is more prominently expressed in lymphatic endothelial cells than in blood endothelial cells (Lee et al., 2008b; Shin et al., 2006). In addition, siRNA knock down of the two factors causes significant reduction in Fgfr3 and flt4 expression (Lee et al., 2008b). Although the precise cis-motifs required for Prox1/COUP-TFII regulation of gene expression have not been identified, the synergy between the two factors provides further evidence that tissue-specific gene activation can be achieved by transcription factors with broader expression than just the endothelium through combinatorial activation.
Concluding Remarks and Future Perspectives
The cell autonomous regulatory networks that control endothelial development are rapidly being decoded, and many transcription factors involved in endothelial specification and differentiation have been identified. Numerous studies have highlighted the roles of individual transcription factors in endothelial development and gene expression through genetic, biochemical, and cell culture-based approaches, but many important questions remain unanswered. In particular, the combinatorial mechanisms through which these transcription factors achieve endothelial specificity are only beginning to be deciphered. Our recent identification of a combinatorial mechanism involving members of the Forkhead and Ets transcription factors (De Val et al., 2008) suggests a paradigm for how endothelial genes may be regulated through composite cis-acting elements, but it is unclear whether other endothelial transcription factors will utilize a similar mechanism. Other recent studies have suggested more conventional mechanisms for how endothelial-specific gene regulation may be achieved through the combined actions of several transcription factors interacting independently with their respective binding sites in endothelial enhancers and promoters (Kappel et al., 2000; Khandekar et al., 2007; Pimanda et al., 2007). Although these mechanisms are certainly not mutually exclusive, it will be important to define how different classes of endothelial transcription factors function biochemically and combinatorially to achieve specificity. Similarly, it will be essential to place endothelial transcription factors into a regulatory hierarchy and to define even earlier regulators of the endothelial program. Recent studies discussed in this review have identified several key endothelial factors as early regulators of endothelial specification, but how these factors themselves are regulated transcriptionally and posttranscriptionally remains to be determined. The Etv2 gene is regulated, at least in part, by the homeodomain transcription factor, Nkx2-5, which activates Etv2 through its proximal promoter in endocardial progenitors (Ferdous et al., 2009). It will be important to determine if Nkx2-5 regulates other early endothelial factors in the endocardium and how the same endothelial factors are transcriptionally activated in other endothelial progenitor populations.
Several signaling pathways are well known for their cell non-autonomous roles in vasculogenesis and angiogenesis, but how these pathways affect transcription factor function during development remains largely undefined. For example, it is likely that VEGF signaling influences the post-translational modifications of numerous endothelial transcription factors, but this notion remains to be established. Interestingly, recent studies show that VEGF-activated PI3K and MAPK signaling pathways influence the transcriptional activity of the FoxC transcription factors (Hayashi and Kume, 2008). Ets transcription factors are also likely to be widely regulated posttranslationally during endothelial development. Indeed, Ets-1 DNA binding affinity is regulated by phosphorylation of multiple independent sites that function additively, suggesting that calcium-dependent signaling controls a graded DNA binding response (Pufall et al., 2005). It will be important to determine how other endothelial Ets factors are regulated by phosphorylation and whether this influences protein-DNA or protein-protein interactions. This observation is consistent with earlier observations that the MAP kinase ERK promotes arterial specification while PI3K signaling promotes venous specification (Hong et al., 2006). Given the role of FoxC1 and FoxC2 in arterial specification, these studies suggest that post-translational modification of the FoxC factors may be central to arteriovenous differentiation. These studies may also reconcile the seemingly disparate roles for FoxC1 and FoxC2 as cofactors for Etv2 in early vasculogenesis and subsequent arteriovenous specification by allowing the FoxC proteins to participate in differential interactions based on phosphorylation state. It is likely that other endothelial transcription factor are also regulated by phosphorylation and other posttranslational modifications during development, and decoding these pathways and how they control endothelial development will be an essential area for future investigation.
The mechanism through which endothelial transcription factors modify chromatin to render endothelial loci accessible and other genes inaccessible is also a topic that remains incompletely understood. As noted in this review, combinatorial control by multiple transcription factors is likely to account for endothelial specificity. However, it is also likely that histone modifications, particularly acetylation, will play a major role in opening chromatin and contributing to endothelial specificity, but this hypothesis remains to be tested fully and the role of chromatin modify enzymes in endothelial-specific gene regulation has not been examined in detail (Matouk and Marsden, 2008). Since many of the transcription factors involved in vasculogenesis, including numerous Ets proteins, are expressed outside endothelial cells and yet do not activate endothelial genes in those other lineages, the influence of chromatin accessibility in endothelial gene activation needs to be resolved.
Finally, aberrant blood vessel growth is a significant cause of age-related macular degeneration, and excessive blood vessel growth underlies proliferative diabetic retinopathy (Andreoli and Miller, 2007; Simo et al., 2006). In addition, vascularization of solid tumors is essential for their growth and eventual metastasis (Carmeliet and Jain, 2000; Stacker et al., 2002). Current therapies for inhibiting angiogenic vessel growth in these and other disorders are primarily aimed at blocking VEGF and other signaling pathways (Ferrara, 2004; Goh et al., 2007). It is attractive to speculate that blocking essential endothelial transcription factors or their interactions may be an alternative approach for inhibiting vascularization. As we unravel the transcriptional networks, upstream signaling pathways, and chromatin modifications involved in endothelial cell development and corresponding vessel growth, new molecular targets for positively and negatively modulating vessel growth should emerge.
Acknowledgements
Work in the Black lab is supported by grants from the NIH and March of Dimes. SDV is supported by a CVRI Postdoctoral Fellowship. We appreciate the many scientists who made helpful comments on this review.
REFERENCES
- Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, Risau W, Klein R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoli CM, Miller JW. Anti-vascular endothelial growth factor therapy for ocular neovascular disease. Curr Opin Ophthalmol. 2007;18:502–508. doi: 10.1097/ICU.0b013e3282f0ca54. [DOI] [PubMed] [Google Scholar]
- Astorga J, Carlsson P. Hedgehog induction of murine vasculogenesis is mediated by Foxf1 and Bmp4. Development. 2007;134:3753–3761. doi: 10.1242/dev.004432. [DOI] [PubMed] [Google Scholar]
- Atkins GB, Jain MK. Role of Kruppel-like transcription factors in endothelial biology. Circ Res. 2007;100:1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Bernat JA, Crawford GE, Ogurtsov AY, Collins FS, Ginsburg D, Kondrashov AS. Distant conserved sequences flanking endothelial-specific promoters contain tissuespecific DNase-hypersensitive sites and over-represented motifs. Hum Mol Genet. 2006;15:2098–2105. doi: 10.1093/hmg/ddl133. [DOI] [PubMed] [Google Scholar]
- Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor AJ, Sanchez MJ, Green AR, Gottgens B. The role of the stem cell leukemia (SCL) gene in hematopoietic and endothelial lineage specification. J Hematother Stem Cell Res. 2002;11:195–206. doi: 10.1089/152581602753658402. [DOI] [PubMed] [Google Scholar]
- Botella LM, Sanchez-Elsner T, Sanz-Rodriguez F, Kojima S, Shimada J, Guerrero-Esteo M, Cooreman MP, Ratziu V, Langa C, Vary CP, et al. Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood. 2002;100:4001–4010. doi: 10.1182/blood.V100.12.4001. [DOI] [PubMed] [Google Scholar]
- Brown LA, Rodaway AR, Schilling TF, Jowett T, Ingham PW, Patient RK, Sharrocks AD. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev. 2000;90:237–252. doi: 10.1016/s0925-4773(99)00256-7. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- Cermenati S, Moleri S, Cimbro S, Corti P, Del Giacco L, Amodeo R, Dejana E, Koopman P, Cotelli F, Beltrame M. Sox18 and Sox7 play redundant roles in vascular development. Blood. 2008;111:2657–2666. doi: 10.1182/blood-2007-07-100412. [DOI] [PubMed] [Google Scholar]
- Chan WY, Follows GA, Lacaud G, Pimanda JE, Landry JR, Kinston S, Knezevic K, Piltz S, Donaldson IJ, Gambardella L, et al. The paralogous hematopoietic regulators Lyl1 and Scl are coregulated by Ets and GATA factors, but Lyl1 cannot rescue the early Scl−/− phenotype. Blood. 2007;109:1908–1916. doi: 10.1182/blood-2006-05-023226. [DOI] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304:735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan PJ, Tsang D, Pedic CM, Abbott LR, Shinkel TA, d'Apice AJ, Pearse MJ. The human ICAM-2 promoter is endothelial cell-specific in vitro and in vivo and contains critical Sp1 and GATA binding sites. J Biol Chem. 1998;273:11737–11744. doi: 10.1074/jbc.273.19.11737. [DOI] [PubMed] [Google Scholar]
- Dalla Torre di Sanguinetto SA, Dasen JS, Arber S. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr Opin Neurobiol. 2008;18:36–43. doi: 10.1016/j.conb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Daly C, Wong V, Burova E, Wei Y, Zabski S, Griffiths J, Lai KM, Lin HC, Ioffe E, Yancopoulos GD, et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1) Genes Dev. 2004;18:1060–1071. doi: 10.1101/gad.1189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, Black BL. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, et al. Combinatorial regulation of endothelial transcription by Ets and Forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejana E, Taddei A, Randi AM. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim Biophys Acta. 2007;1775:298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- Dekker RJ, van Thienen JV, Rohlena J, de Jager SC, Elderkamp YW, Seppen J, de Vriess CJ, Biessen EA, van Berkel TJ, Pannekoek H, et al. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am J Pathol. 2005;167:609–618. doi: 10.1016/S0002-9440(10)63002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleuze V, Chalhoub E, El-Hajj R, Dohet C, Le Clech M, Couraud PO, Huber P, Mathieu D. TAL-1/SCL and its partners E47 and LMO2 up-regulate VE-cadherin expression in endothelial cells. Mol Cell Biol. 2007;27:2687–2697. doi: 10.1128/MCB.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson IJ, Chapman M, Kinston S, Landry JR, Knezevic K, Piltz S, Buckley N, Green AR, Gottgens B. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum Mol Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- Dube A, Akbarali Y, Sato TN, Libermann TA, Oettgen P. Role of the Ets transcription factors in the regulation of the vascular-specific Tie2 gene. Circ Res. 1999;84:1177–1185. doi: 10.1161/01.res.84.10.1177. [DOI] [PubMed] [Google Scholar]
- Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M, Chong BH. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol Cell Biol. 2003;23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, Gorski JL, Seaver LH, Glover TW. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet. 2000;67:1382–1388. doi: 10.1086/316915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci U S A. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- Fischer A, Klamt B, Schumacher N, Glaeser C, Hansmann I, Fenge H, Gessler M. Phenotypic variability in Hey2 −/− mice and absence of HEY2 mutations in patients with congenital heart defects or Alagille syndrome. Mamm Genome. 2004a;15:711–716. doi: 10.1007/s00335-004-2389-x. [DOI] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004b;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamme I, Frolich T, Risau W. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J Cell Physiol. 1997;173:206–210. doi: 10.1002/(SICI)1097-4652(199711)173:2<206::AID-JCP22>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Francois M, Caprini A, Hosking B, Orsenigo F, Wilhelm D, Browne C, Paavonen K, Karnezis T, Shayan R, Downes M, et al. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456:643–647. doi: 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Nakayama K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. Embo J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M, Yamada Y, Rabbitts TH, Patient RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. doi: 10.1242/dev.00875. [DOI] [PubMed] [Google Scholar]
- Goh PP, Sze DM, Roufogalis BD. Molecular and cellular regulators of cancer angiogenesis. Curr Cancer Drug Targets. 2007;7:743–758. doi: 10.2174/156800907783220462. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Broccardo C, Sanchez MJ, Deveaux S, Murphy G, Gothert JR, Kotsopoulou E, Kinston S, Delaney L, Piltz S, et al. The scl +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5' bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol Cell Biol. 2004;24:1870–1883. doi: 10.1128/MCB.24.5.1870-1883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R, Green AR. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. Embo J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves BJ, Gillespie ME, McIntosh LP. DNA binding by the ETS domain. Nature. 1996;384:322. doi: 10.1038/384322a0. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- Green AR, Lints T, Visvader J, Harvey R, Begley CG. SCL is coexpressed with GATA-1 in hemopoietic cells but is also expressed in developing brain. Oncogene. 1992;7:653–660. [PubMed] [Google Scholar]
- Gunther CV, Graves BJ. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994;14:7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, Favier R, Bernstein A. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Kume T. Foxc transcription factors directly regulate Dll4 and Hey2 expression by interacting with the VEGF-Notch signaling pathways in endothelial cells. PLoS ONE. 2008;3:e2401. doi: 10.1371/journal.pone.0002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KK, D'Amore PA. Pericytes in the microvasculature. Cardiovasc Res. 1996;32:687–698. [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CC, Peterson QP, Hong JY, Peterson RT. Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr Biol. 2006;16:1366–1372. doi: 10.1016/j.cub.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, Detmar M. Prox1, master regulator of the lymphatic vasculature phenotype. Cell Tissue Res. 2003;314:85–92. doi: 10.1007/s00441-003-0747-8. [DOI] [PubMed] [Google Scholar]
- Hong YK, Harvey N, Noh YH, Schacht V, Hirakawa S, Detmar M, Oliver G. Prox1 is a master control gene in the program specifying lymphatic endothelial cell fate. Dev Dyn. 2002;225:351–357. doi: 10.1002/dvdy.10163. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljin K, Dube A, Kontusaari S, Korhonen J, Lahtinen I, Oettgen P, Alitalo K. Role of ets factors in the activity and endothelial cell specificity of the mouse Tie gene promoter. Faseb J. 1999;13:377–386. doi: 10.1096/fasebj.13.2.377. [DOI] [PubMed] [Google Scholar]
- Irrthum A, Devriendt K, Chitayat D, Matthijs G, Glade C, Steijlen PM, Fryns JP, Van Steensel MA, Vikkula M. Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am J Hum Genet. 2003;72:1470–1478. doi: 10.1086/375614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaka C, Tanaka K, Abe M, Sato Y. Ets-1 regulates angiogenesis by inducing the expression of urokinase-type plasminogen activator and matrix metalloproteinase-1 and the migration of vascular endothelial cells. J Cell Physiol. 1996;169:522–531. doi: 10.1002/(SICI)1097-4652(199612)169:3<522::AID-JCP12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- James K, Hosking B, Gardner J, Muscat GE, Koopman P. Sox18 mutations in the ragged mouse alleles ragged-like and opossum. Genesis. 2003;36:1–6. doi: 10.1002/gene.10190. [DOI] [PubMed] [Google Scholar]
- Kallianpur AR, Jordan JE, Brandt SJ. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood. 1994;83:1200–1208. [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- Kappel A, Ronicke V, Damert A, Flamme I, Risau W, Breier G. Identification of vascular endothelial growth factor (VEGF) receptor-2 (Flk-1) promoter/enhancer sequences sufficient for angioblast and endothelial cell-specific transcription in transgenic mice. Blood. 1999;93:4284–4292. [PubMed] [Google Scholar]
- Kappel A, Schlaeger TM, Flamme I, Orkin SH, Risau W, Breier G. Role of SCL/Tal-1, GATA, and ets transcription factor binding sites for the regulation of flk-1 expression during murine vascular development. Blood. 2000;96:3078–3085. [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar M, Brandt W, Zhou Y, Dagenais S, Glover TW, Suzuki N, Shimizu R, Yamamoto M, Lim KC, Engel JD. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development. 2007;134:1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, Lim KC, Engel JD, Yamamoto M. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Hayashi S, Shimokado K, Suzuki Y, Shimada J, Crippa MP, Friedman SL. Transcriptional activation of urokinase by the Kruppel-like factor Zf9/COPEB activates latent TGF-beta1 in vascular endothelial cells. Blood. 2000;95:1309–1316. [PubMed] [Google Scholar]
- Kokubo H, Miyagawa-Tomita S, Nakazawa M, Saga Y, Johnson RL. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev Biol. 2005;278:301–309. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Korhonen J, Lahtinen I, Halmekyto M, Alhonen L, Janne J, Dumont D, Alitalo K. Endothelial-specific gene expression directed by the tie gene promoter in vivo. Blood. 1995;86:1828–1835. [PubMed] [Google Scholar]
- Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, et al. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Kinston S, Knezevic K, Donaldson IJ, Green AR, Gottgens B. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106:2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, Weinstein BM. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008a;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, Mericko P, Stadtfeld M, Zhou D, Cheng L, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Lee ME, Temizer DH, Clifford JA, Quertermous T. Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J Biol Chem. 1991;266:16188–16192. [PubMed] [Google Scholar]
- Lee S, Kang J, Yoo J, Ganesan SK, Cook SC, Aguilar B, Ramu S, Lee J, Hong YK. Prox1 physically and functionally interacts with COUP-TFII to specify lymphatic endothelial cell fate. Blood. 2008b doi: 10.1182/blood-2008-03-145789. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Bisgrove BW, Sawyer H, Hug B, Bell B, Peters K, Grunwald DJ, Stainier DY. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124:381–389. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- Liu F, Patient R. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ Res. 2008;103:1147–1154. doi: 10.1161/CIRCRESAHA.108.179713. [DOI] [PubMed] [Google Scholar]
- Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Lugus JJ, Chung YS, Mills JC, Kim SI, Grass J, Kyba M, Doherty JM, Bresnick EH, Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- Melet F, Motro B, Rossi DJ, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol Cell Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami T, Donovan DJ, Tsai JC, Rosenberg RD, Aird WC. Differential regulation of the von Willebrand factor and Flt-1 promoters in the endothelium of hypoxanthine phosphoribosyltransferase-targeted mice. Blood. 2002;100:4019–4025. doi: 10.1182/blood-2002-03-0955. [DOI] [PubMed] [Google Scholar]
- Minami T, Kuivenhoven JA, Evans V, Kodama T, Rosenberg RD, Aird WC. Ets motifs are necessary for endothelial cell-specific expression of a 723-bp Tie-2 promoter/enhancer in Hprt targeted transgenic mice. Arterioscler Thromb Vasc Biol. 2003;23:2041–2047. doi: 10.1161/01.ATV.0000089326.63053.9A. [DOI] [PubMed] [Google Scholar]
- Mishima K, Watabe T, Saito A, Yoshimatsu Y, Imaizumi N, Masui S, Hirashima M, Morisada T, Oike Y, Araie M, et al. Prox1 induces lymphatic endothelial differentiation via integrin alpha9 and other signaling cascades. Mol Biol Cell. 2007;18:1421–1429. doi: 10.1091/mbc.E06-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollica LR, Crawley JT, Liu K, Rance JB, Cockerill PN, Follows GA, Landry JR, Wells DJ, Lane DA. Role of a 5'-enhancer in the transcriptional regulation of the human endothelial cell protein C receptor gene. Blood. 2006;108:1251–1259. doi: 10.1182/blood-2006-02-001461. [DOI] [PubMed] [Google Scholar]
- Morishita K, Johnson DE, Williams LT. A novel promoter for vascular endothelial growth factor receptor (flt-1) that confers endothelial-specific gene expression. J Biol Chem. 1995;270:27948–27953. doi: 10.1074/jbc.270.46.27948. [DOI] [PubMed] [Google Scholar]
- Oliver G, Alitalo K. The lymphatic vasculature: recent progress and paradigms. Annu Rev Cell Dev Biol. 2005;21:457–483. doi: 10.1146/annurev.cellbio.21.012704.132338. [DOI] [PubMed] [Google Scholar]
- Oliver G, Srinivasan RS. Lymphatic vasculature development: current concepts. Ann N Y Acad Sci. 2008;1131:75–81. doi: 10.1196/annals.1413.006. [DOI] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou KN, Izumiya Y, Walsh K. Forkhead transcription factors and cardiovascular biology. Circ Res. 2008;102:16–31. doi: 10.1161/CIRCRESAHA.107.164186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patan S. Vasculogenesis and angiogenesis. Cancer Treat Res. 2004;117:3–32. doi: 10.1007/978-1-4419-8871-3_1. [DOI] [PubMed] [Google Scholar]
- Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–3511. doi: 10.1182/blood-2004-09-3547. [DOI] [PubMed] [Google Scholar]
- Pendeville H, Winandy M, Manfroid I, Nivelles O, Motte P, Pasque V, Peers B, Struman I, Martial JA, Voz ML. Zebrafish Sox7 and Sox18 function together to control arterial-venous identity. Dev Biol. 2008;317:405–416. doi: 10.1016/j.ydbio.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Pennisi D, Bowles J, Nagy A, Muscat G, Koopman P. Mice null for sox18 are viable and display a mild coat defect. Mol Cell Biol. 2000a;20:9331–9336. doi: 10.1128/mcb.20.24.9331-9336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, Muscat G, Abbott C, Koopman P. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet. 2000b;24:434–437. doi: 10.1038/74301. [DOI] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova TV, Makinen T, Makela TP, Saarela J, Virtanen I, Ferrell RE, Finegold DN, Kerjaschki D, Yla-Herttuala S, Alitalo K. Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. Embo J. 2002;21:4593–4599. doi: 10.1093/emboj/cdf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda JE, Chan WY, Donaldson IJ, Bowen M, Green AR, Gottgens B. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a -8-kb enhancer. Blood. 2006;107:4737–4745. doi: 10.1182/blood-2005-12-4929. [DOI] [PubMed] [Google Scholar]
- Pimanda JE, Chan WY, Wilson NK, Smith AM, Kinston S, Knezevic K, Janes ME, Landry JR, Kolb-Kokocinski A, Frampton J, et al. Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code. Blood. 2008;112:4512–4522. doi: 10.1182/blood-2008-05-157560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, Landry JR, Wood AD, Kolb-Kokocinski A, Green AR, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A, Kollipara R, DePinho RA, Zeiher AM, Dimmeler S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandini MH, Dreher I, Bouillot S, Benkerri S, Moll T, Huber P. The human VE-cadherin promoter is subjected to organ-specific regulation and is activated in tumour angiogenesis. Oncogene. 2005;24:2992–3001. doi: 10.1038/sj.onc.1208483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21:2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Niedenfuhr M, Papoutsi M, Christ B, Nicolaides KH, von Kaisenberg CS, Tomarev SI, Wilting J. Prox1 is a marker of ectodermal placodes, endodermal compartments, lymphatic endothelium and lymphangioblasts. Anat Embryol (Berl) 2001;204:399–406. doi: 10.1007/s00429-001-0214-9. [DOI] [PubMed] [Google Scholar]
- Rossant J, Hirashima M. Vascular development and patterning: making the right choices. Curr Opin Genet Dev. 2003;13:408–412. doi: 10.1016/s0959-437x(03)00080-7. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Hara K, Kanai-Azuma M, Matsui T, Miura Y, Tsunekawa N, Kurohmaru M, Saijoh Y, Koopman P, Kanai Y. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun. 2007;360:539–544. doi: 10.1016/j.bbrc.2007.06.093. [DOI] [PubMed] [Google Scholar]
- Schlaeger TM, Bartunkova S, Lawitts JA, Teichmann G, Risau W, Deutsch U, Sato TN. Uniform vascular-endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc Natl Acad Sci U S A. 1997;94:3058–3063. doi: 10.1073/pnas.94.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Fujitas H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Shin JW, Min M, Larrieu-Lahargue F, Canron X, Kunstfeld R, Nguyen L, Henderson JE, Bikfalvi A, Detmar M, Hong YK. Prox1 promotes lineage-specific expression of fibroblast growth factor (FGF) receptor-3 in lymphatic endothelium: a role for FGF signaling in lymphangiogenesis. Mol Biol Cell. 2006;17:576–584. doi: 10.1091/mbc.E05-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo R, Carrasco E, Garcia-Ramirez M, Hernandez C. Angiogenic and antiangiogenic factors in proliferative diabetic retinopathy. Curr Diabetes Rev. 2006;2:71–98. doi: 10.2174/157339906775473671. [DOI] [PubMed] [Google Scholar]
- Simon MP, Tournaire R, Pouyssegur J. The angiopoietin-2 gene of endothelial cells is up-regulated in hypoxia by a HIF binding site located in its first intron and by the central factors GATA-2 and Ets-1. J Cell Physiol. 2008;217:809–818. doi: 10.1002/jcp.21558. [DOI] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacker SA, Baldwin ME, Achen MG. The role of tumor lymphangiogenesis in metastatic spread. Faseb J. 2002;16:922–934. doi: 10.1096/fj.01-0945rev. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Sumanass S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Villa N, Walker L, Lindsell CE, Gasson J, Iruela-Arispe ML, Weinmaster G. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech Dev. 2001;108:161–164. doi: 10.1016/s0925-4773(01)00469-5. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vondenhoff MF, van de Pavert SA, Dillard ME, Greuter M, Goverse G, Oliver G, Mebius RE. Lymph sacs are not required for the initiation of lymph node formation. Development. 2009;136:29–34. doi: 10.1242/dev.028456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. Embo J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiya K, Begue A, Stehelin D, Shibuya M. A cAMP response element and an Ets motif are involved in the transcriptional regulation of flt-1 tyrosine kinase (vascular endothelial growth factor receptor 1) gene. J Biol Chem. 1996;271:30823–30828. doi: 10.1074/jbc.271.48.30823. [DOI] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. Embo J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- Weinstein BM, Stemple DL, Driever W, Fishman MC. Gridlock, a localized heritable vascular patterning defect in the zebrafish. Nat Med. 1995;1:1143–1147. doi: 10.1038/nm1195-1143. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- Wu J, Iwata F, Grass JA, Osborne CS, Elnitski L, Fraser P, Ohneda O, Yamamoto M, Bresnick EH. Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol Cell Biol. 2005;25:1458–1474. doi: 10.1128/MCB.25.4.1458-1474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Pannell R, Forster A, Rabbitts TH. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc Natl Acad Sci U S A. 2000;97:320–324. doi: 10.1073/pnas.97.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yet SF, McA'Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, Layne MD, Chin MT, Wang H, Perrella MA, et al. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Zhong TP, Childs S, Leu JP, Fishman MC. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414:216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]