Abstract
With the recent breakthroughs in G protein-coupled receptor structure, one can now compare experimentally determined structures with the most recent modeling and docking methods. A community-wide blind prediction experiment (GPCR Dock 2008) was conducted in coordination with the publication of the human adenosine A2A receptor bound to the ligand ZM241385 crystal structure (Science 322, 1211 (2008)). Twenty-nine participating groups submitted 206 models that were evaluated for the accuracy of the ligand binding mode and the overall receptor model. Several new insights emerged including the critical importance of disulfide bonds in the extracellular loops, helix residue registry, and domain knowledge.
Introduction
Molecular modeling has a pertinent role in rational drug discovery and design1, 2. Reliable three-dimensional models can provide valuable insights into basic principles of molecular recognition and aid in the structure-based drug design approach to lead finding and optimization3. G protein-coupled receptors (GPCRs) are membrane proteins involved in signal transduction pathways and are important therapeutic targets for numerous diseases4, 5. As such, significant structure prediction efforts using methods ranging from de novo to homology-based approaches have been applied to members of the GPCR family6, 7. Until recently, much of GPCR homology modeling efforts have been based on the templates of bovine rhodopsin and bacteriorhodopsin, with refinement of the models by molecular dynamics simulations, ligand docking, and incorporation of additional biochemical and biophysical data8–12. The refinement step is necessary in building accurate models, especially around the ligand-binding site, due to the expected structural differences both among members of the family resulting from the generally low sequence identity and the large diversity of ligands accommodated within the family7, 13–15, and among various conformational states associated with the different ligand efficacy16–18.
The most recently solved GPCR structure is the 2.6 Å crystal structure of the human adenosine A2A receptor bound to an antagonist19. Adenosine receptors belong to the class A rhodopsin-like GPCR family, and are implicated as promising therapeutic targets in a wide range of conditions, including cerebral and cardiac ischaemic diseases, sleep disorders, immune and inflammatory disorders, and cancer20. The A2A structure shows an overall seven transmembrane helix structural similarity to the rhodopsin and adrenergic receptor structures, with shifts in the positions and orientations of the helices and a markedly different structure of the extracellular loops19.
To evaluate the current progress in GPCR structure prediction and docking, we carried out a community-wide blind prediction experiment (GPCR Dock 2008) in coordination with the release of the human adenosine A2A receptor structure in October 200819. GPCR Dock 2008 was organized in a similar manner as the CASP and CAPRI experiments21, 22, and was aimed to assess the current status of GPCR structure modeling and docking, as well as to highlight areas for future efforts in method development. In this paper, we report the results of the assessment together with our analysis of the current status of GPCR structure and ligand docking predictions.
GPCR Dock 2008
In August 2008, prior to the publication of the human adenosine A2A structure in October 200819, participating predictors were asked to blindly predict and submit up to ten ranked models of the human A2A receptor in complex with the ligand ZM241385, starting from the amino acid sequence of the receptor and a 2D structure of the ligand. A total of 63 different individuals initially registered, with 206 models submitted by 29 different individuals in the final data set. Note that 37 of the 206 submitted models were either missing the ligand or had incorrect bond connectivity for the ligand. We assessed the remaining 169 models for the prediction accuracy of the ligand binding mode, and all 206 models for the prediction accuracy of the receptor alone.
Assessment criteria
RMSD (root mean square deviation) is used as a quantitative measure of the similarity between two superimposed atomic coordinates, and is calculated by
where δ is the distance between N pairs of equivalent atoms from the two coordinates. RMSD values (units of Å) can be calculated for any type and subset of atoms, e.g. Cα atoms of proteins (Cα RMSD) for all residues, for residues in the transmembrane helices or the loops; heavy atoms of small-molecule ligands (ligand RMSD).
Z-score is a standard dimensionless score that normalizes a value with respect to the sample mean and standard deviation, and is defined by
where x is the raw value, μ is the mean and σ is the standard deviation.
Assessment criteria are dependent on the purpose of the generated models. Given the primary value of the GPCR structural models in expanding our knowledge in basic molecular recognition and their potential use in design and development of new small molecules, the quality of the models was primarily assessed by the accuracy of the ligand binding mode. Particular attention and care was made towards the fact that the crystal structure is a static structure with positional errors, and the value of modeling is ultimately to guide drug discovery and biological insight. Our numerical measure of accuracy for the ligand binding mode was based on two metrics, ligand RMSD and the number of correct receptor-ligand contacts. Neither metric alone was sufficient to capture the accuracy of prediction around the ligand-binding site; hence, both were used and combined into a z-score scoring scheme to rank the models.
The ligand RMSD between model and crystal structure is calculated as the coordinate root mean-square deviation (RMSD) for the 25 non-hydrogen atoms of ZM241385 after superimposing the protein Cα atoms of the model and the crystal structure. In addition, the ligand RMSD is also calculated excluding the phenoxy group of ZM241385 that has high B-factor values. The number of correct contacts is counted as the number of correctly predicted native contacts observed between protein atoms and the ligand. A native contact is defined as any inter-atomic distance within 4 Å of the ligand in the crystal structure. There are 75 such receptor-ligand contacts, and an additional 15 contacts formed with water.
The models were ranked by assigning a combined mixed z-score to each model. The combined z-score was calculated as the average of z-scores for ligand RMSD and the number of correct contacts: Zcombined=(−ZLigandRMSD+ ZN _ CorrectContacts)/2. The z-scores for ligand RMSD and the number of correct contacts were computed in two passes as follows: i) assign a z-score to each model using the average and standard deviation values from all models, ii) re-compute the average and standard deviation excluding models with z-scores more than two standard deviations above (for ligand RMSD) and below (for the number of correct contacts) the average, iii) re-assign a z-score to each model using the revised average and standard deviation values obtained in step ii. The best model, i.e. the model with the highest combined z-score, from each group was analyzed.
Overall results
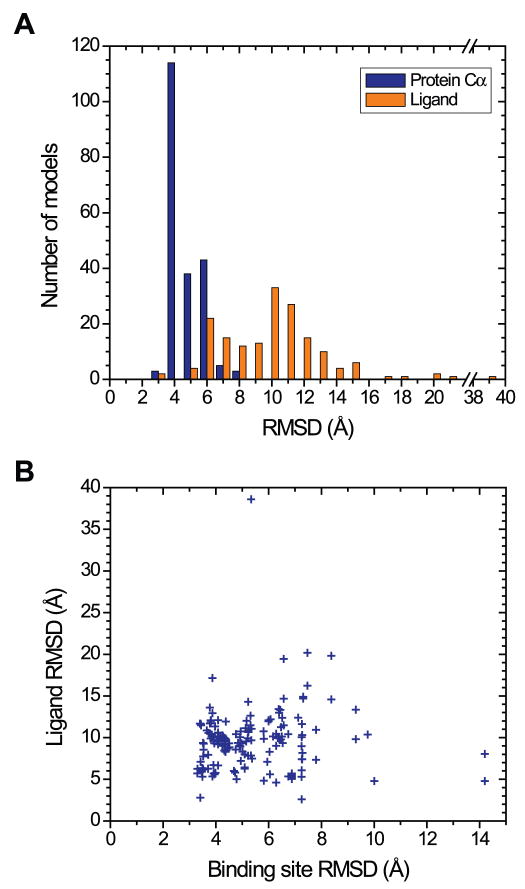
The submitted models show a wide distribution in prediction accuracy of the ligand binding mode, with average values of 9.5 Å (s.d. 3.8 Å) for ligand RMSD and 4 (s.d. 7) for the number of correct contacts (Figure 1A).. These statistics indicate that the majority of the submitted models do not predict the ligand position and the binding interactions very accurately. Furthermore, the lack of strong correlation between ligand RMSD and binding site RMSD (Figure 1B), e.g. models with <4.0 Å binding site RMSD have a range of 2.8 to 17.2 Å ligand RMSD, suggests that the performance of some ligand docking methods is poor and can be improved.
Figure 1.
RMSD of submitted models. (A) Distribution of ligand RMSD (gold bars) and protein Cα RMSD (blue bars) for all models. (B) A scatterplot of ligand RMSD (y axis) versus binding site RMSD (x axis) for all models. The binding site RMSD values are calculated for heavy atoms of the binding site residues (F1685.29, E1695.30, M1775.38, W2466.48, L2496.51, H2506.52, N2536.55, H2646.66, M2707.35) after the models are superimposed to the crystal structure using the protein Cα atoms.
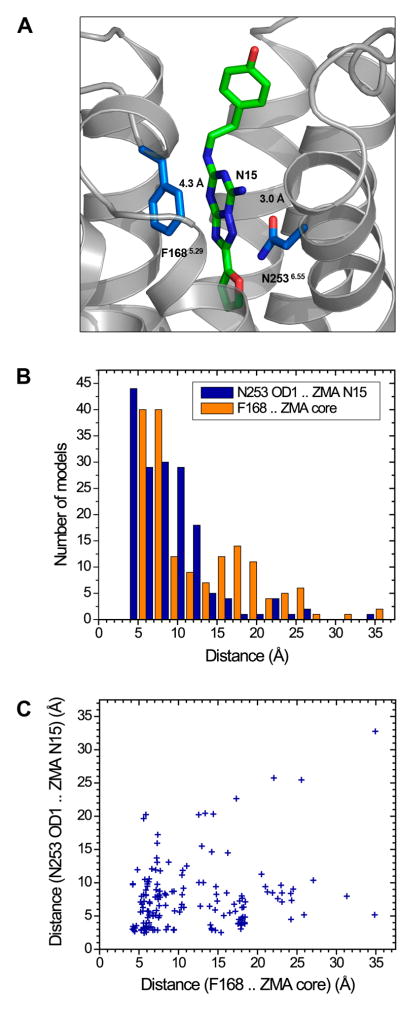
Very few models score well in both ligand RMSD and the number of correct contacts (only 13 out of the 169 total receptor-ligand models have >1 combined z-score, compared to 40 models that score well solely in ligand RMSD, ZligandRMSD <−1.0). For models with relatively low ligand RMSD values but small number of correct contacts, the inaccuracy in binding interactions may be attributed to errors in the sidechain placement of the ligand-binding residues. Although nearly a third of the models capture the hydrogen bonding interaction between the N2536.55 sidechain and the exocyclic N15 atom of the ligand (44 out of 169 models have <4 A N253 OD1 – ZM241385 N15 interaction distance), other key receptor-ligand interactions such as the aromatic stacking interaction between the F1685.29 sidechain and the bicyclic ring of the ligand are not captured well in most models (Figure 2).
Figure 2.
Statistics of the two key receptor-ligand interactions in all models. (A) The hydrogen bonding interaction with N2536.55 and the aromatic stacking interaction with F1685.29 are shown by dashed lines with the distance measurements from the crystal structure. (B) Distribution of the distance for the interaction between the sidechain carbonyl oxygen OD1 atom in N2536.55 and the exocyclic N15 atom of the ligand, and the average interatomic distance for the aromatic stacking interaction between the heavy atoms in the F1685.29 sidechain and the bicyclic ring (atoms C11, N12, N13, C14, N15, N16, N17, C18, N19, C20) of ZM241385. (C) A scatterplot of the distances for the hydrogen bonding interaction (y axis) versus the aromatic stacking interaction (x axis).
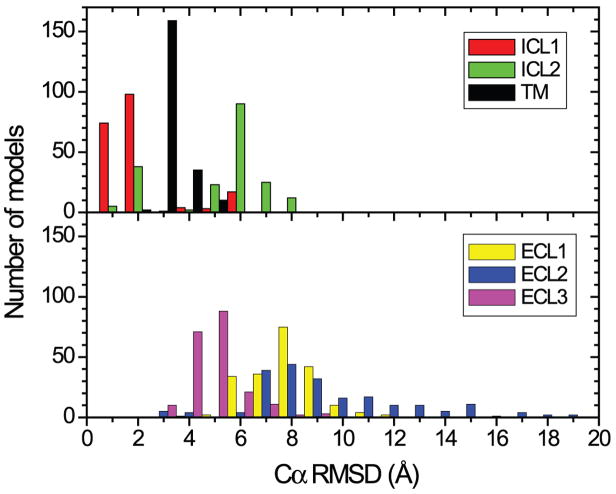
While the overall outcome clearly shows that there are remaining challenges in accurately predicting the ligand binding mode, the quality of the predictions for the receptor alone appear to be much higher: 4.2 ± 0.9 Å for the receptor Cα RMSD, and 2.8 ± 0.5 Å for the transmembrane (TM) helices Cα RMSD; loop regions, with the exception of ICL1 (intracellular loop 1), are not modeled accurately in the majority of the models (Figure 3A,B and Figure 4). It is notable that some groups that accurately predict the TM region of the receptor do not predict the ligand binding mode very well (TM Cα RMSD: 2.0 Å for Pogozheva/Lomize, and 2.1 Å for Horst/Roy), indicating that the methods for modeling the receptor and docking the ligand can be generally considered as distinct steps in the generation of models for the receptor-ligand complex.
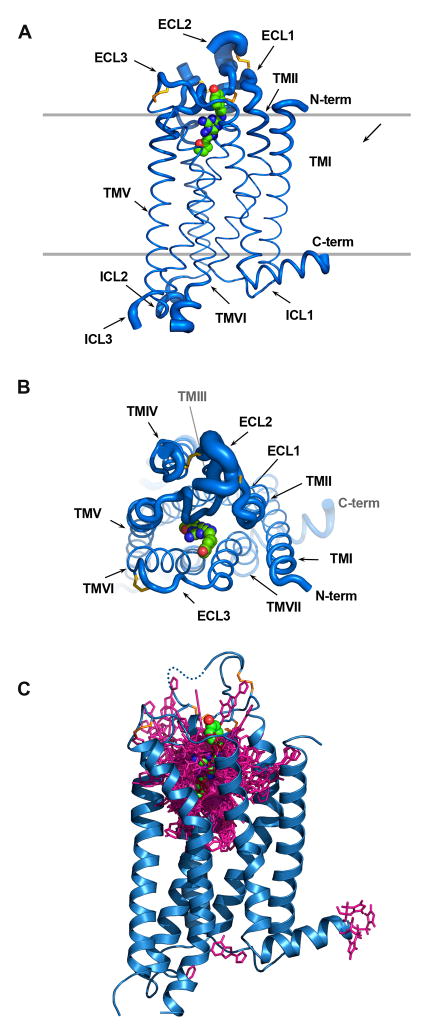
Figure 3.
Superposition of all 206 submitted models to the crystal structure of the human adenosine A2A receptor (PDB ID: 3EML without the T4-lysozyme). Protein Cα atom superposition between model and crystal structure was done using the align command in PyMOL (version 1.0r2, www.pymol.org). The receptor is shown as two orthogonal views of Cα traces (A, B) with tube thickness being proportional to the RMSD about each Cα position clearly showing how well the TM regions were modeled and how much uncertainty are in the loop regions. (C) A superposition of stick diagrams of the ligand (ZM241385) from 169 models, a CPK model is used to delineate the observed position in the crystal structure. The C-terminus (residue numbers >306) is removed from all models.
Figure 4.
Distribution of Cα RMSDs for TM helices I to VII (helix I: 6–34; helix II: 40–67; helix III: 73–107; helix IV: 117–142; helix V: 173–205; helix VI: 222–258; helix VII: 266–291), and the loop regions (ICL1: 35–39; ICL2: 108–116; ECL1: 68–72; ECL2:143–172 excluding 149–155 that are missing in the crystal structure; ECL3: 259–265).
Model examples
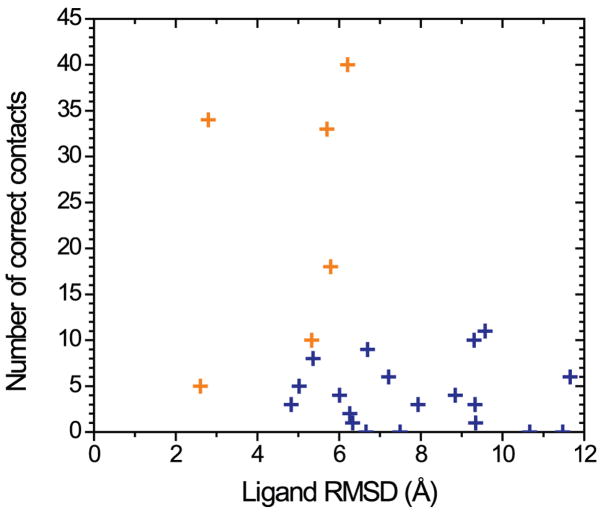
Despite the apparent difficulty in accurately predicting the receptor-ligand interactions, some models had consistent features with the crystal structure, although model ranking continues to be one of the most challenging areas of development. Here, we focus on the predictions from the top ten groups, ranked according to the combined z-score, and assess the model quality at greater details (Figure 5). Note that, with predictions for only one target, the statistical significance of the group ranking cannot be judged, as is typically done in CASP experiments by a head-to-head comparison on common targets between the top groups23. To further support our selection of the best predictions, we ranked all models using an alternative metric, binding site contactRMSD, which treats all ligand-binding residues with equal weight, and is an RMSD of receptor-ligand contact distance over all ligand-binding residues. We found that both the z-score ranking and the contactRMSD ranking agree on the selection of the best model, and the majority of other top predictions.
Figure 5.
A scatterplot of the number of correct contacts (y axis) versus ligand RMSD (x axis) for the best predictions from all groups. The best predictions from the top six groups are marked as gold crosses.
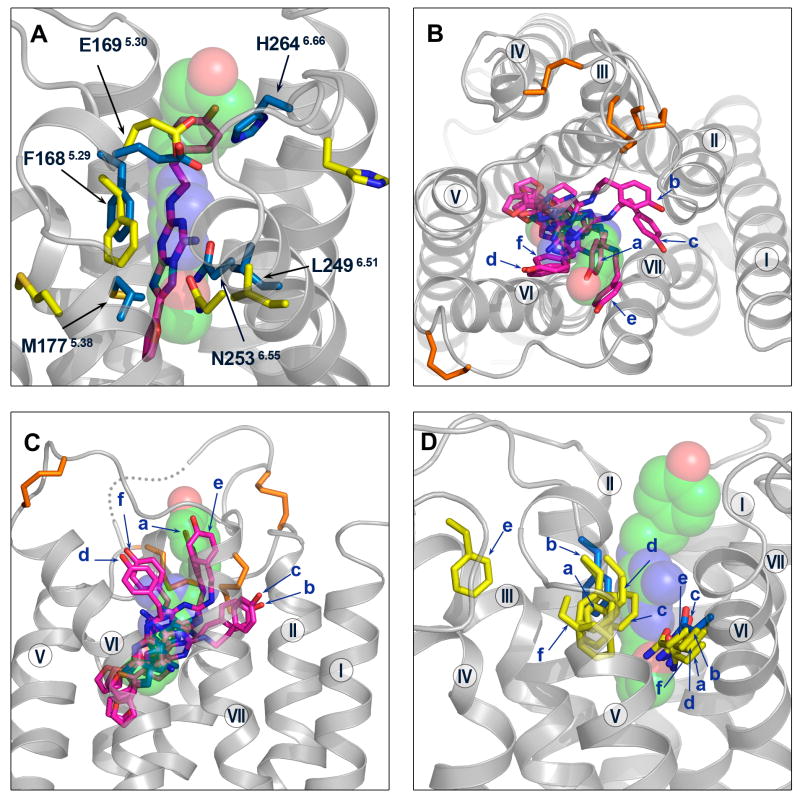
The best model (Costanzi) among all submitted models has a ligand RMSD of 2.8 Å RMSD and 34 of 75 correct contacts (Figure 6A and Table I). The ligand is modeled in a native-like binding pose, with an extended conformation and a nearly perpendicular orientation to the membrane plane. The model accurately predicts some of the key receptor-ligand interactions: it captures the hydrogen bonding interaction between the N2536.55 sidechain and the exocyclic amino group (N15 atom) of the ligand, and the aromatic stacking interaction between the F1685.29 sidechain and the bicyclic triazolotriazine core of the ligand. Compared to the crystal structure, the ligand in the model is positioned deeper in the binding pocket, bringing the furan ring closer to TM helices III and V. The inaccuracy in the ligand position is most likely due to errors in the sidechain positions of the two crucial ligand-binding residues (F1685.29 and E1695.30) in ECL2 (extracellular loop 2) and the sidechain orientation of M1775.38 at the extracellular end of TM helix V: the aromatic ring of F1685.29, which interacts with the bicyclic ring, is positioned too deeply; the adjacent E1695.30 forms a hydrogen bonding interaction with the hydroxyl group in the phenolic substituent instead of the exocyclic N15 atom near the bicyclic ring; the sidechain of M1775.38 is not oriented toward the binding cavity. In addition, the family conserved disulfide bond C773.25 – C1665.27 is predicted accurately, but the disulfide bond in ECL3 C2596.61 – C2626.64 is not, presumably contributing to the inaccuracy in the sidechain orientation of H2646.66, which is not pointed toward the binding site.
Figure 6.
Comparison between the best models and the crystal structure around the ligand-binding site. (A) The ligand and the ligand-binding residues F1685.29, E1695.30, M1775.38, L2496.51, N2536.55, H2646.66 are shown for the best model (Costanzi) and the crystal structure. The ligand is shown as sticks for the model (magenta colored carbon atoms) and semitransparent spheres (green colored carbon atoms) for the crystal structure; the ligand-binding residues are shown as yellow sticks for the model, and blue sticks for the crystal structure. Extracellular (B) and side views (C) of the ligand in the binding pocket for the best predictions from the top six groups (magenta sticks for models, and green spheres for the crystal structure). The receptor crystal structure is in gray ribbons. The disulfide bonds are shown in orange sticks (D) The ligand-binding residues F1685.29 and N2536.55 are shown as sticks for the best predictions from the top six groups (yellow for models, and blue for the crystal structure). In (B), (C) and (D) the models are labeled as a – Constanzi; b - Katritch; c - Lam; d - Davis; e - Maigret; f – Jurkowski.
Table I.
Summary of results for the best models from the top ranking groups.
| Group Name | Rank (Total Num Models) | Ligand RMSD (Å) | Ligand RMSD w/o phenoxy group (Å) | Number of Correct Contacts | Binding site residues RMSD (Å) | Protein Cα RMSD (Å) | TM I-VII Cα RMSD (Å) | ECL2 Cα RMSD (Å) | Combined z-score (avg ± s.d.) |
|---|---|---|---|---|---|---|---|---|---|
| Costanzi | 2 (4) | 2.8 | 2.7 | 34 | 3.4 | 3.0 (266) | 2.5 (212) | 3.8 (8) | 3.02 (0.86 ± 1.48) |
| Katritch/Abagyan | 1 (10) | 6.2 | 4.0 | 40 | 3.5 | 4.0 (283) | 2.7 (214) | 8.9 (23) | 2.76 (1.89 ± 1.13) |
| Lam/Abagyan | 1 (3) | 5.7 | 3.6 | 33 | 3.3 | 4.1 (283) | 3.6 (214) | 7.3 (23) | 2.42 (0.88 ± 1.34) |
| Davis/Barth/Baker | 4 (5) | 5.8 | 5.4 | 18 | 4.0 | 3.5 (283) | 2.1 (214) | 8.4 (23) | 1.46 (0.16 ± 0.86) |
| Maigret | 8 (10) | 2.6 | 2.1 | 5 | 7.3 | 5.1 (283) | 4.1 (214) | 9.1 (23) | 1.23 (0.05 ± 0.57) |
| Jurkowski/Elofsson | 2 (8) | 5.3 | 5.2 | 10 | 3.9 | 6.2 (283) | 2.9 (214) | 12.7 (23) | 1.04 (−0.02 ± 0.98) |
| Kanou | 7 (10) | 5.4 | 5.5 | 8 | 6.9 | 3.5 (279) | 2.8 (214) | 7.1 (23) | 0.91 (0.66 ± 0.11) |
| Goddard | 8 (10) | 5.0 | 3.9 | 5 | 4.8 | 4.3 (284) | 2.5 (214) | 10.7 (23) | 0.78 (0.16 ± 0.37) |
| Bologa | 3 (10) | 6.7 | 2.8 | 9 | 3.9 | 3.4 (278) | 2.5 (213) | 7.2 (19) | 0.72 (−0.14 ± 0.39) |
| Olson | 1 (9) | 4.8 | 4.7 | 3 | 5.8 | 3.5 (284) | 2.3 (214) | 7.5 (23) | 0.69 (−0.14 ± 0.58) |
Participants were allowed to submit up to 10 models, Rank indicates the ranking that the participant assigned to their best model as determined in the GPCR Docks 2008 study with the total number of models submitted by that participant in parentheses. The RMSD values are calculated for the heavy atoms of the ligand ZM241385 (all 25 atoms, and partially without the phenoxy group), heavy atoms of the binding site residues (F1685.29, E1695.30, M1775.38, W2466.48, L2496.51, H2506.52, N2536.55, H2646.66, M2707.35), Cα atoms of all residues, Cα atoms of residues in the TM helices I to VII (helix I: 6–34; helix II: 40–67; helix III: 73–107; helix IV: 117–142; helix V: 173–205; helix VI: 222–258; helix VII: 266–291), and Cα atoms of resides in ECL2 (143–172 excluding 149–155 that are missing in the crystal structure). All RMSD values are obtained after the models are superimposed to the crystal structure using the protein Cα atoms in PyMOL (version 1.or2, www.pymol.org). The assignment of residues in the ligand-binding site and the secondary structure elements is from the PDB header section (PDB ID: 3EML). The number of residues used in the RMSD calculation is in brackets. The combined z-score value for the best model, as well as the average and standard deviation values for all models submitted by each group, are shown.
The best predictions from the top six groups (Costanzi, Katritch/Abagyan, Lam/Abagyan, Davis/Barth/Baker, Maigret, Jurkowski/Elofsson) highlight the successes and difficulties in accurately predicting the ligand binding pose and receptor-ligand interactions (Figures 6B,C,D and Table I). The extended ligand conformation is accurately predicted in all six models, and the nearly perpendicular orientation is captured in four of the six models. The hydrogen bonding interaction between the N2536.55 sidechain and the exocyclic N15 atom of the ligand is correctly modeled in four models; however, in one of the four, the ligand makes no interaction with residues in ECL2. The aromatic stacking interaction between the F1685.29 sidechain and the bicyclic ring of the ligand is correctly modeled in four models; however, in all four models, the ligand is positioned too deeply in the binding pocket, and the M1775.38 sidechain is not oriented toward the binding cavity. There is one model that does not accurately capture either the hydrogen bonding interaction with N2536.55 or the aromatic stacking interaction with F1685.29, while five of the six models accurately predict the family conserved disulfide bond C773.25 – C1665.27. None of the six models capture the hydrogen bonding interaction between E1695.30 in ECL2 and the exocyclic N15 atom of the ligand.
Other models that ranked near the top (Kanou, Goddard, Bologa, Olson) are slightly less accurate but show similar trends as the top six models in their ability to accurately predict the ligand binding mode (Table I). The ligand is modeled in a native-like extended conformation in three of the four models. The hydrogen bonding interaction between the N2536.55 sidechain and the exocyclic N15 atom of the ligand is modeled accurately in three of the four models; whereas, the aromatic stacking interaction between the F1685.29 sidechain and the bicyclic ring of the ligand is modeled accurately in only one of the four models. The family conserved disulfide bond C773.25 – C1665.27 is captured in two models. Remarkably, one of the models (Goddard) accurately places the E1695.30 sidechain proximal to the exocyclic N15 atom of the ligand, and almost captures the hydrogen bonding interaction, even though the overall ECL2 conformation is inaccurate.
The best predictions were generally not ranked as the best models by the predictors at the time of model submission prior to the release of the crystal structure (Table I). Only two of the six best models were ranked first, and three of the six groups show a weak correlation between their model ranking and the model quality as assessed by the combined z-score for the accuracy around the ligand-binding site. Furthermore, the additional models submitted by the six groups are generally of lower quality than the best predictions (Table I). Only one of the six best models has a z-score that is within one standard deviation of the group average z-score.
Status of GPCR structure modeling and docking
The assessment of the submitted models shows that the best participating methods have the ability to predict close native-like ligand binding, but with limitations in capturing all of the key receptor-ligand interactions and correctly estimating model quality by ranking. The majority of the submitted models are quite far from achieving a native-like ligand binding pose. The most challenging aspect of GPCR structure prediction highlighted in this assessment seems to be in accurately modeling the ligand interactions with residues in the extracellular loop regions. This result is not surprising given the lack of structural homology in the loops among the known GPCR structures24, and the generally difficult task of modeling loops25, 26.
The most successful prediction methods relied on homology modeling approaches based on the template structures of β-adrenergic receptors, and in some cases with the additional template structures of rhodopsin, (PDB IDs: 2RH1 (β2AR), 2VT4 (β1AR), 1U19 (bovine Rhod), 2Z73 (squid Rhod)) to generate models of the receptor, followed by docking of the ligand to one or more receptor models using small-molecule docking programs such as Glide27, ICM28, GOLD29, and AutoDock30 (see Supplementary Information for description of prediction methods). The alignment of the human A2A sequence to the template structure seemed to have been straightforward, given the family conserved motifs and residues in the TM helices31. The ECL2 was modeled by de novo approaches in many of the top predictions (Katritch/Abagyan, Lam/Abagyan, Davis/Barth/Baker, Jurkowski/Elofsson, Goddard), but only partially modeled in the best prediction (Costanzi) for a short segment of eight residues, located N-terminus to TM helix V, that includes the disulfide-bond-forming C1665.27. Some of the criteria used to select and rank the final receptor-ligand complex models were: docking scores, conformational energy of the complex, agreement with mutagenesis and structure-activity relationship data, and binding selectivity studied by virtual ligand screening or by modeling other subtypes of adenosine receptor.
The reliability of the homology modeling approach depends on the availability of suitable templates32. The results of the current assessment show that the structures of β-adrenergic receptors alone or together with rhodopsin were suitable transmembrane templates in predicting the general structure of the adenosine A2A receptor. However, given the expected structural diversity in class A GPCRs, it is unclear whether the current set of techniques applied to the structure prediction of the A2A–ZM241385 complex would result in a similar level of accuracy for the prediction of other GPCRs, especially for those belonging to subfamilies that are phylogenetically distant from the amine and the opsin receptor clusters33. We believe the database of GPCR structures needs to expand further to provide suitable templates for accurate modeling of those other receptors.
The inaccuracies in homology models can arise from errors in side-chain packing, main-chain shifts in aligned regions, errors in unaligned loop regions, misalignments, and incorrect templates34. These errors relate to the issue of “adding value” to the template structure, which was addressed in the recent CASP experiment35, and also seems to be applicable to GPCR modeling. Indeed, ligand interactions with residues located in structurally divergent regions from the templates are consistently not modeled accurately in all of the six best predictions: hydrogen bonding interaction between E1695.30 in ECL2 and the exocyclic N15 atom of the ligand is not captured, and the sidechains of H2646.66 in ECL3 and M1775.38 in the extended bulge structure unique to A2A at the extracellular end of TM helix V are not oriented toward the binding site. An exception is the aromatic stacking interaction between F1685.29 in ECL2 and the bicyclic ring of the ligand, which is correctly modeled in some of the predictions. F1685.29 is located in the loop, but it is structurally homologous to F1935.32 which interacts with the carbazole heterocycle of the ligand carazolol in the β2AR structure, hence modeling of this interaction may have been guided by homology. Interestingly, F1685.29 is modeled more accurately than E1695.30 even though mutagenesis data shows mutation of E1695.30 to alanine reduces the affinity for both antagonists and agonists36, and no data is available for F1685.29. The inaccuracy in the orientation of the ligand binding pose, e.g. the parallel orientation with the phenolic substituent positioned close to TM helices II and III, may in part be due to the inaccurate modeling of the helical shifts in TM helices I, II, and III. The helical shifts alter the location of the binding pocket and redefine the pocket size and shape19; thus, it is expected that accurately modeling the helical shifts would contribute to a better prediction of the ligand binding pose. The helical shifts were most accurately modeled by an effective use of multiple template structures of rhodopsin and β-adrenergic receptors (Pogozheva/Lomize), or an all-atom refinement approach implemented by the ROSETTA program using a physically realistic model that recapitulates protein interatomic and protein-solvent interactions in the membrane environment37 (Davis/Barth/Baker).
Other sources of error include not modeling the water molecules that are either structurally important or directly involved in ligand binding interactions3. The ligand binding cavity in the A2A–ZM241385 structure has four ordered water molecules19, yet none of the submitted predictions included water molecules. We tried re-docking the ligand to the crystal structure using ICM28 and found that a native-like binding pose (within 1 Å heavy atom RMSD for the bicyclic ring and the furanyl substituent of the ligand, and <3.0 Å overall ligand RMSD) can be recovered without any water molecules, which suggests that water may not be critical for accurately predicting the ligand interactions. However, modeling water molecules together with the ligand may contribute in general to a better prediction of the ligand binding pose or affinity. Additional re-docking studies with the docking protocols used by the participating methods would help assess the effect of the water molecules, and the accuracy of the docking methods separately from that of the receptor modeling methods.
Lastly, it is interesting that the best model was from the Costanzi group who worked previously with adenosine receptor modeling and docking. Their domain knowledge on the adenosine receptor may have helped with the interpretation of the mutagenesis and ligand interaction data and such domain knowledge is almost certainly critical.
Conclusion
Accurate prediction of GPCR structure and ligand interactions clearly remains a challenge despite the increase in the number of experimentally solved GPCR structures within the last year. Assessment of these predictions highlights similar issues addressed by the CASP predictions for template-based modeling targets, i.e. the difficulty in loop modeling, refinement and improvement over the best available template, and model ranking. Accurate modeling of the structurally divergent region such as the extracellular loops, disulfide bond formation affecting helix residue registry, and the helical shifts in the TM region seems to be particularly critical for accurately predicting the key ligand interactions in GPCRs, and this area is perhaps the most in need of technological development. Progress in GPCR modeling and docking will require further improvements in the current prediction methods to “add value” to the best available templates and generate models that will be more useful for applications in structure-based drug design.
Supplementary Material
Acknowledgments
We thank the participating predictors for making this assessment possible. We thank Mike Hanson, V.-P. Jaakola, Chris Roth, and Vadim Cherezov for help with the analysis and comments on the manuscript, and Katya Kadyshevskaya and Vadim Cherezov for figure preparation. We are grateful to the Goddard group for providing the script to calculate the binding site contactRMSD. We thank Angela Walker for data tracking and assistance with the manuscript, and Josh Kunken for IT help during the assessment. This work was supported in part by the NIH Roadmap grant P50 GM073197 (JCIMPT) and Protein Structure Initiative grant U54 GM074961 (ATCG3D).
GPCR Assessment Participants
Arthur Olson (Department of Molecular Biology, The Scripps Research Institute)
Wiktor Jurkowski and Arne Elofsson (Center of Biomembrane Research, Department of Biochemistry & Biophysics, Stockholm University)
Slawomir Filipek (Laboratory of Biomodelling, International Institute of Molecular and Cell Biology)
Irina Pogozheva and Andrei Lomize (Peptide Synthesis and Molecular Recognition Laboratory, University of Michigan)
Bernard Maigret (Orpailleur team, LORIA, Nancy University)
Jeremy Horst1, Ambrish Roy 2, Brady Bernard1, Shyamala Iyer1, Yang Zhang2, and Ram Samudrala1 (1 Computational Biology Group, University of Washington; 2 Department of Molecular Biosciences, Center for Bioinformatics, University of Kansas)
Osman Ugur Sezerman (Sabanici University)
Gregory V. Nikiforovich1 and Christina M. Taylor2 (1 MolLife Design LLC; 2 Department of Biochemistry and Molecular Biophysics, Washington University)
Stefano Costanzi (Laboratory of Biological Modeling, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health)
Y. Vorobjev2, N. Bakulina2, and V. Solovyev1,2 (1 Department of Computer Science, Royal Holloway, University of London; 2 Softberry Inc.)
Kazuhiko Kanou1, Daisuke Takaya1,2, Genki Terashi1, Mayuko Takeda-Shitaka1,2 and Hideaki Umeyama1,2 (1 School of Pharmacy, Kitasato University; 2 RIKEN Systems and Structural Biology Center)
William A. Goddard III, Youyong Li, Soo-Kyung Kim, Bartosz Trzaskowski, Ravinder Abrol, and Adam Griffith (Materials and Process Simulation Center, California Institute of Technology)
Vsevolod Katritch, Manuel Rueda and Ruben Abagyan (Molsoft LLC)
Ian Davis, Patrick Barth, David Baker (Department of Biochemistry, University of Washington)
Michael Feig (Department of Biochemistry & Molecular Biology, Michigan State University)
Michal Brylinski, Hongyi Zhou, Seung Yup Lee and Jeffrey Skolnick (Center for the Study of Systems Biology, Georgia Institute of Technology)
Liliana Ostopovici-Halip and Cristian Bologa (Division of Biocomputing, University of New Mexico)
Polo Lam and Ruben Abagyan (Department of Molecular Biology, The Scripps Research Institute)
Eric S. Dawson, Kristian Kaufmann, Nils Woetzel, and Jens Meiler (Center for Structural Biology, Vanderbilt University)
Feng Ding, Adrian Serohijos, Shuangye Yin, and Nikolay V. Dokholyan (Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill)
David Rodriguez and Hugo Gutiérrez-de-Teràn (Fundación Pública Galega de Medicina Xenómica. Complejo Hospitalario Universitario de Santiago, A Choupana, s/n. Santiago de Compostela)
Henri Xhaard (Center for Drug Research, Faculty of Pharmacy, University of Helsinki)
References
- 1.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–8. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 2.Richon AB. Current status and future direction of the molecular modeling industry. Drug Discov Today. 2008;13:665–9. doi: 10.1016/j.drudis.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–49. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 4.Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–4. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 5.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–44. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Becker OM, et al. G protein-coupled receptors: in silico drug discovery in 3D. Proc Natl Acad Sci U S A. 2004;101:11304–9. doi: 10.1073/pnas.0401862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballesteros J, Palczewski K. G protein-coupled receptor drug discovery: implications from the crystal structure of rhodopsin. Curr Opin Drug Discov Devel. 2001;4:561–74. [PMC free article] [PubMed] [Google Scholar]
- 8.Bu L, Michino M, Wolf RM, Brooks CL., III Improved model building and assessment of the Calcium-sensing receptor transmembrane domain. Proteins. 2008;71:215–26. doi: 10.1002/prot.21685. [DOI] [PubMed] [Google Scholar]
- 9.Henin J, et al. Probing a model of a GPCR/ligand complex in an explicit membrane environment: the human cholecystokinin-1 receptor. Biophys J. 2006;90:1232–40. doi: 10.1529/biophysj.105.070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler CB, Pogozheva ID, LeVine H, 3rd, Mosberg HI. Refinement of a homology model of the mu-opioid receptor using distance constraints from intrinsic and engineered zinc-binding sites. Biochemistry. 2004;43:8700–10. doi: 10.1021/bi036067r. [DOI] [PubMed] [Google Scholar]
- 11.Evers A, Klabunde T. Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the alpha1A adrenergic receptor. J Med Chem. 2005;48:1088–97. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 12.Manivet P, et al. The serotonin binding site of human and murine 5-HT2B receptors: molecular modeling and site-directed mutagenesis. J Biol Chem. 2002;277:17170–8. doi: 10.1074/jbc.M200195200. [DOI] [PubMed] [Google Scholar]
- 13.Archer E, Maigret B, Escrieut C, Pradayrol L, Fourmy D. Rhodopsin crystal: new template yielding realistic models of G-protein-coupled receptors? Trends Pharmacol Sci. 2003;24:36–40. doi: 10.1016/s0165-6147(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 14.Gershengorn MC, Osman R. Minireview: Insights into G protein-coupled receptor function using molecular models. Endocrinology. 2001;142:2–10. doi: 10.1210/endo.142.1.7919. [DOI] [PubMed] [Google Scholar]
- 15.Ballesteros JA, Shi L, Javitch JA. Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol Pharmacol. 2001;60:1–19. [PubMed] [Google Scholar]
- 16.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya S, Hall SE, Li H, Vaidehi N. Ligand-stabilized conformational states of human beta(2) adrenergic receptor: insight into G-protein-coupled receptor activation. Biophys J. 2008;94:2027–42. doi: 10.1529/biophysj.107.117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenakin T. Efficacy at G-protein-coupled receptors. Nat Rev Drug Discov. 2002;1:103–10. doi: 10.1038/nrd722. [DOI] [PubMed] [Google Scholar]
- 19**.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. The human adenosine A2 receptor crystal structure served as the experimental template for comparison for this modeling and docking assessment. This is the second human GPCR structure to be experimentally determined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5:247–64. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moult J, et al. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. The very successful CASP (Critical Assessment of Protein Structure) project started in 1994, served as the model to conduct the reported GPCR modeling and docking assessment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lensink MF, Mendez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69:704–18. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 23.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69 (Suppl 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 24.Kobilka B, Schertler GF. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol Sci. 2008;29:79–83. doi: 10.1016/j.tips.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson MP, et al. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–67. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 26.Rohl CA, Strauss CE, Chivian D, Baker D. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55:656–77. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 27.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 28.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;(Suppl 1):215–20. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 29.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–23. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 30.Morris G, et al. Automated docking using a lamarkian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 31.Mirzadegan T, Benko G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 2003;42:2759–67. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–6. doi: 10.1126/science.1065659. The use of protein models and docking is dependent on how such data will be used. In this paper, Baker and Sali present an excellent presentation of where models are useful, in particular as hypothesis generators with the application dependent on the resolution of the structure. [DOI] [PubMed] [Google Scholar]
- 33.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 34.Marti-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 35.Read RJ, Chavali G. Assessment of CASP7 predictions in the high accuracy template-based modeling category. Proteins. 2007;69 (Suppl 8):27–37. doi: 10.1002/prot.21662. [DOI] [PubMed] [Google Scholar]
- 36.Kim J, et al. Glutamate residues in the second extracellular loop of the human A2a adenosine receptor are required for ligand recognition. Mol Pharmacol. 1996;49:683–91. [PMC free article] [PubMed] [Google Scholar]
- 37.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci U S A. 2007;104:15682–7. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.