Abstract
Motivated by the long-term goal of theoretically analyzing long-range proton transfer (PT) kinetics in biomolecular pumps, a number of technical developments were made in the framework of QM/MM simulations. A set of collective reaction co-ordinates is proposed for characterizing the progress of long-range proton transfers; unlike previous suggestions, the new coordinates can describe PT along highly non-linear three-dimensional pathways. Calculations using a realistic model of carbonic anhydrase demonstrated that adiabatic mapping using these collective coordinates gives reliable energetics and critical geometrical parameters as compared to minimum energy path calculations, which suggests that the new coordinates can be effectively used as reaction coordinate in potential of mean force calculations for long-range PT in complex systems. In addition, the generalized solvent boundary potential was implemented in the QM/MM framework for rectangular geometries, which is useful for studying reactions in membrane systems. The resulting protocol was found to produce water structure in the interior of aquaporin consistent with previous studies including much larger number of explicit solvent and lipid molecules. The effect of electrostatics for PT through membrane protein was also illustrated with a simple model channel embedded in different dielectric continuum environments. The encouraging results observed so far suggest that robust theoretical analysis of long-range PT kinetics in biomolecular pumps can soon be realized in a QM/MM framework.
1 Introduction
Proton transfer (PT) is a crucial step in many biochemical processes. In many enzymes, local PT between the substrate and amino acids forms the basis for the general acid-base catalysis [1]. Long-range proton exchange between distant active sites has been recently proposed to be a mechanism for catalytic co-operativity [2]. In bioenergetics, protons are pumped across the lipid membrane through specific proteins [3–6] to create a proton concentration gradient, which may then be utilized in other energy consuming processes such as ATP synthesis [7]; similarly, a number of technical applications, such as fuel cells [8], require well-controlled long-range PTs.
With major efforts from both the experimental [9, 10] and theoretical community [11–15] in the past several decades, basic mechanisms for localized PT in enzyme active sites are rather well understood: e.g., it is accepted that the rate of the proton transfer is largely modulated by the electrostatics in the active site [16]. Proton donor-acceptor dynamics, which is likely coupled to the overall fluctuation of the enzyme, also plays an important role [17].
Long-range PTs, by contrast, are more challenging to understand at a quantitative level. It is generally accepted that long-range PT occurs through the help of hydrogen-bonded ”wires” formed by water molecules [18–20], in many cases with participation of titratable amino acid sidechains [21]. However, the precise transfer pathway(s) and rate-limiting factors are often difficult to unravel due to the large number of solvent and protein residues involved. Understanding these factors is crucial for the investigation of many fundamentally important issues such as efficiency of energy transduction in proton pumps.
From the experimental point of view, a large number of mutagenesis experiments and kinetic measurements for the corresponding mutants can in principle be used to probe the transfer pathway and important interactions that regulate the transfer kinetics. Yet the fact that mutation may introduce non-trivial perturbation to the solvent and protein structure always leaves some degree of ambiguity in the interpretation.
In carbonic anhydrase, a relatively small enzyme, for example, the precise nature of the rate-limiting event for an 8 Å long-range PT remains controversial even after many experimental studies over the last two decades [22–29]. Issues of debate include the role of water wire formation and the importance of water wire geometry, specifically, the number of water molecules and their orientations along the wire. Theoretical analysis, therefore, is highly desirable as a complementary technique and can, eventually, provide mechanistic insights on an atomistic level. Indeed, a large number of theoretical studies have been applied to several systems involving long-distance PT, such as carbonic anhydrase [24–29], gramicidin [30, 30–33], bacteriorhodopsin [34, 35], aquaporin [36–38], the synthetic LS2-channel [39] and cytochrome c oxidase [40–43]. These theoretical studies certainly provided valuable mechanistic information, but it is perhaps fair to state that a quantitative understanding has not been obtained in any of those systems. For instance, in gramicidin [30, 32, 44] and aquaporin [36–38], studies from a number of groups using different simulation protocols proposed rather different rate-limiting events.
These controversies reflect the fact that a number of technical challenges have to be overcome for generally robust theoretical analysis of long-range PT.
First, since a large number of bond-breaking and formation events are involved, an accurate description of the reaction energetics requires a quantum mechanical treatment of a large number of atoms. This is particularly true for PT coupled to fairly complex chemical reactions, such as in cytochrome c oxidase [45]. Most previous simulation studies employed empirical models for proton transfer along water wires, which may not be sufficiently accurate or flexible.
Second, since a large number of shuttling groups are explicitly involved in the long-range PT, sufficient conformational sampling of the protein and solvent atoms at a relevant temperature is required for quantitative estimate of the transfer kinetics. In many previous studies minimum energy paths (MEP) were computed using the nudged elastic band (NEB) [46] or conjugated path refinement (CPR) [47] algorithms. These MEPs are useful for qualitative analysis but inadequate for a more quantitative comparison with experiments. With more demanding computational cost, potential of mean force (PMF) was evaluated for the PT in several studies [30–33, 36–39]. In this way the effect of thermal fluctuations can be included. In addition to limitations in the potential functions used in those studies, another rather general problem concerns the choice of reaction coordinate in the PMF calculations; most previous choices either assume a specific pattern of transfer pathways (e.g., step-wise) or work best only for transfers across linear water chains.
Finally, long-range PT involves significant charge separation, thus careful treatment of electrostatics in the highly heterogeneous protein, solvent, and possible lipid environment is of great importance. Although this issue has been addressed in some studies using either Ewald summation [48] or Langevin dipole models [49], many studies, especially those involving QM/MM potentials, use very approximate electrostatics schemes that compromise the reliability of the results.
Our long-term goal is to understand the mechanism of vectorial proton pumping involved in biochemical energy transductions using QM/MM simulations. Here we describe a number of technical developments we have made towards this goal. Specifically, we introduce a set of new collective coordinates based on the center of excess charge [31,50] to characterize the PT process. Their collective nature allows the calculation of potential of mean force without assuming a priori the pathway of the transfer, and they are sufficiently flexible to cope with non-linear PT pathways. Moreover, the generalized solvent boundary potential approach [51] has been implemented in the QM/MM framework, which made it possible to study long-range PT in heterogeneous environments with reliable potential functions. These technical advances, in conjunction with the developments and benchmark of effective QM methods in our groups, will soon allow reliable theoretical analysis of proton transfer kinetics in complex biomolecular pumps.
In the following, we first describe the new reaction coordinates and compare them to earlier suggestions in Sect. 2, which is followed by a brief summary of the GSBP implementation in the QM/MM framework. We then test and illustrate these new protocols using a number of systems in Sect. 3, which include carbonic anhydrase, a model channel and aquaporin. Finally, we highlight a number of conclusions in Sect. 4.
2 Computational methods
As emphasized in the Introduction, a reliable description of long-range PT kinetics requires state-of-the-art treatment of electronic structure of the system as well as sufficient and effective sampling of the heterogeneous protein, solvent, and possibly lipid degrees of freedom. For the electronic structure, our general strategy is to use the hybrid QM/MM potential. In addition to the typical technical details concerning the QM/MM scheme [52–58], the major challenge is to develop and benchmark a QM method that is both fast and accurate for the problem at hand. An attractive method in this regard is the self consistent charge density functional tight binding (SCC-DFTB) method [52, 59], which has been successfully applied to a number of biological systems [60–67] including those involving proton or hydride transfer reactions. The method has been parameterized for C, N, O, H, P, S and Zn [61], and has been shown to often give more accurate results than commonly used semi-empirical methods such as AM1 and PM3 [60, 61, 68–71]. On-going efforts in our groups will further improve the accuracy of the SCC-DFTB approach for proton affinity and proton transfer reactions, which will not be discussed here. Instead, we focus here on two technical aspects that are related to the sampling of conformational space relevant to the long-range PT. The first issue concerns the definition of a reaction coordinate that characterizes the PT process, which is crucial in meaningful calculations of PMF in systems with complex PT pathways. The second aspect deals with the implementation of a boundary potential for QM/MM simulation of reactions in membrane proteins. Specifically, the generalized solvent boundary potential (GSBP) approach proposed by Roux and co-workers [51] has been implemented in the SCC-DFTB/MM framework. The GSBP approach treats the most important degrees of freedom at the microscopic level and describes effects due to other regions of the protein, solvent and lipid membrane with continuum electrostatics, making it possible to sample the reactive degrees of freedom in a very efficient manner. In the following, we first introduce the reaction coordinate and make comparisons to previous suggestions, then we briefly summarize the GSBP implementation in the QM/MM framework for rectangular boundary.
2.1 Reaction Coordinate for Long-range Proton Transfers
Although in some systems PT is fast and thus can be monitored with unbiased nanosecond MD simulations [72], most PT processes have significant barriers and require alternative methods for investigation. Powerful simulation methods such as the transition path sampling technique [73] have been proposed to study the real-time dynamics of rare events in the condensed phase. However their high computational cost limits their applicability. An alternative for studying rare events, e.g., PT, is to compute the PMF along a well-chosen reaction coordinate. By ”well-chosen”, we mean that the reaction coordinate captures the nature of the degree(s) of freedom that most strongly regulate the reaction kinetics. Due to charge migration in long-range PT, the protein and solvent environment undergo significant reorganizations; an appropriate reaction coordinate, therefore, is the energy gap between different diabatic states that correspond to localized proton coordinations, similar to the ”solvent coordinate” used to describe electron transfers in solution. This energy gap coordinate was indeed used extensively by Warshel and co-workers [11, 26, 74] in the study of long-range PT using EVB potentials [75], in which the diabatic states are well defined. With adiabatic QM/MM potentials, however, it is more difficult to define an energy gap coordinate; moreover, application of the energy gap coordinate to simultaneous (concerted) multiple proton transfers is less straightforward.
Therefore, we choose to define the reaction coordinate in terms of the geometrical and charge property of the PT reaction, i.e., the location of the center of excess charge (CEC) relative to the proton donor and acceptor groups. We note that although this type of coordinate is geometrical in nature, its dependence on the charge distribution of the ”proton wire” ensures that the environment appropriately reorganizes as its value varies. Indeed, previous comparison of the energy gap coordinate and a geometrical coordinate found very similar PMF results for short-range proton transfer reactions. [76]. Moreover, activated dynamics simulations obtained a transmission coefficient fairly close to unity (~ 0.4) for a number of studies of short-range PT reactions in enzyme systems using geometrical coordinate to define the reaction coordinate [77, 78]. Whether the success applies quantitatively to long-range PT processes remains to be investigated.
Generally speaking, when defining a geometric reaction coordinate, the task is to extract those degrees of freedom best describing the reaction. Specifically for PT involving water molecules, a rapid interchange takes place [79] between structures which are close to the ideal structures of Eigen- and Zundel ions [80,81]. The defect, i.e. the excess proton coordinated to water molecules, travels without any atom actually moving further than fractions of an Ångstrøm [32,79]. Hence the challenge for long-range PT is to separate these subtle fluctuations responsible for proton transfer from other motion in the immediate protein and solvent environment.
2.1.1 Previous Suggestions
For a short-range PT, in which the location of the transferring proton is well-defined, a commonly used reaction coordinate is the anti-symmetric stretch involving the donor (D), the transferring proton (H) and the acceptor (A),
| (1) |
This in principle can be generalized to multiple proton transfers by using a linear combination of anti-symmetric stretch coordinates for all sets of donor, transferring proton and acceptor atoms. Although such a linear combination was indeed found useful in the study of PT through two and three intervening water molecules in carbonic anhydrase [27], its use becomes cumbersome and less robust for more complicated pathways.
In the study of PT through the water wire in gramicidin A, a reaction coordinate based on the CEC was used by Roux and coworkers [31, 50], which involves the projection of the total dipole moment of the water wire on the z-axis (the axis of the water wire),
| (2) |
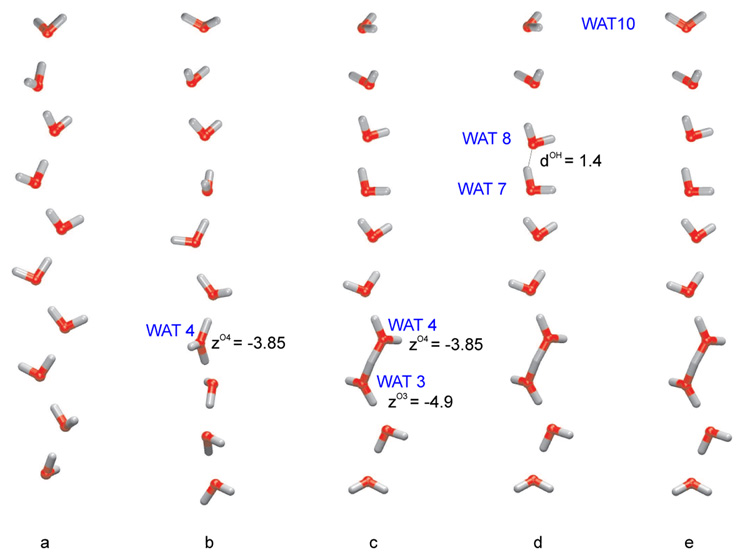
where NH and NO are the total number of hydrogen and oxygen nuclei in the water wire, rz’s are z coordinates and qH and qO are the partial charges of H (+1e) and O (−2e), respectively. The CEC coordinate is hence defined by µz/e, where e is the unit charge. For an unprotonated chain of water molecules (e.g. O10H20, Fig. 1a) the CEC gives the z component of the total dipole moment; in a protonated wire (e.g. Fig. 1b–e), on the other hand, it corresponds to the projection of the center of excess charge (proton defect) along the z axis.
Figure 1.
Water wire models for illustrating different reaction coordinates that describe long-range proton transfers. a) a water wire without any excess proton; b) a water wire with a H3O+-ion located at WAT4; c) a water wire with a Zundel -ion between WAT3 and WAT4; d) same as c) but with one water molecule (WAT8) displaced to mimic a close collision of water molecules. e) same as c) but with on water molecule (WAT10) rotated by a small angle to illustrate the contamination of µz/e. The values of different reaction coordinates discussed here are shown in Table 1. The distance cutoff for the bonds drawn was chosen to be 1.3 Å.
In contrast to the anti-symmetric stretch (δ), the CEC coordinate is a global, collective coordinate, meaning that it reflects not only the location of the excess proton in a water wire but also the configuration of all the water molecules in the wire. This sensitive dependence on the orientation of individual water molecules makes µz/e easily ”contaminated” by the fluctuation of the water wire (vide infra).
An alternative reaction coordinate suggested by Chakrabarti et al. [36] (denoted by v in the following), which takes a local view at the problem, counts the number (wOi) of protons coordinated to each oxygen atom Oi in the wire,
| (3) |
| (4) |
Here and in the following dA,B denotes the Cartesian distance between atoms A and B. The switching function fsw(d) is given in the following equation with suggested values of rsw =1.4 Å and dsw =0.05 Å [36]:
| (5) |
Although this coordinate in its optimum yields a more precise description of the location of the excess charge compared to µz/e, the limitation is that the information included is purely local. As a result, the functional form introduced can’t distinguish between three hydrogen atoms coordinated to an oxygen atom in an oxonium ion and three hydrogen atoms close to an oxygen atom due to a collision of two water molecules. Although this problem can be partially overcome either by choosing a better switching function or by a judicious choice of the parameters in the switching function (Eq. 5), it is difficult to fully eliminate fluctuations of the reactions coordinate due to water collisions. Even though close encounter of water molecules (r <1.4 Å) are not overwhelmingly frequent, the contamination effect of the reaction coordinate is significant as illustrated below.
2.1.2 A new reaction coordinate
To take into account the advantages of global and local considerations, we propose a reaction coordinate that unites the formulations discussed above. This modified CEC (mCEC) coordinate for a linear proton wire (along the z axis), which may include both water and protein groups, is defined as:
| (6) |
where Xj represents a coordinating atom for protons during the translocation, is the projection of the position vector of atom A on the z axis. wXj is the weight associated with the atom Xj and is defined as the number of protons coordinated to that atom in its reference state. The reference state is the least protonated state of the atom in both reactant and product. For instance, if atom Xk has two protons coordinated in the reactant but only one in the product, then wXk = 1. When the proton transfer is solely through water molecules as shown in Fig. 1, the oxygen atoms have a wX of +2 (also see Sect. 3.1 for a more general example). In the more general case, X can be any proton acceptor with a different weight (see below for more specific examples). The correction (third term in Eq. 6) consists of the sum over all contributions to the z-component of the bond vector from individual bonds. To establish the definition of a bond in this context, we use the same switching function fsw (d) as Chakrabarti et al. (Eq. 5) for the distance dXi,Hj. Parameters of rsw =1.3 Å and dsw =0.03 Å were found to give the best results.
2.1.3 Comparison of different coordinates for linear proton wires
To illustrate the difference between the newly proposed ξz coordinate and previous suggestions (µz/e, v), we compare their behavior with different situations of protonated and non-protonated water wires (Fig. 1). The coordinates were collected from molecular dynamics simulations of the model channels described below (Sect.3.2).
When there is no excess proton in the water wire, the value of the CEC coordinate (µz/e) is significantly different from 0 as expected (Table 1). The value of the mCEC coordinate (ξz), by contrast, is very close to zero; this demonstrates that the correction (third term in Eq. 6) appropriately removes contributions from any components irrelevant to the proton transfer as designed. The reaction coordinate of Chakrabarti et al. [36] (v) gives unstable results (large numerical value) for un-protonated water wire because the denominator in Eq. 3 approaches zero in this case; this is not a significant shortcoming because no such instability is anticipated for cases with excess proton(s).
TABLE 1.
Values for different reaction coordinates (in Å) for the water wires displayed in Fig. 1.
For the water wire with a localized hydronium ion (Fig. 1b), ξz gives the z coordinate of the oxygen atom in WAT4, which is the oxygen that bears the extra proton. Slight fluctuations in the bond lengths are reflected in v, which is slightly displaced towards WAT3 (Table 1). With a Zundel like ion in the wire (Fig. 1c), both v and ξz give values between the z coordinate of the oxygen atoms in WAT4 and WAT5. The different distances from the central proton to the two neighboring oxygen atoms (1.3 and 1.4 Å to O4 and O5, respectively) lead to slightly different values of the two reaction coordinates due to different functional forms. With the current set of parameters in fsw, both coordinates locate the excess proton closer to the WAT5 oxygen, which is consistent with the geometry.
The shortcoming of the v coordinate becomes clear with the case in Fig. 1d, in which one water molecule (WAT8) was displaced to simulate a close collision between water molecules. The mCEC coordinate is not affected by this, but the v coordinate changes significantly: the oxygen in WAT8 appears to have more than two bonded hydrogens, leading to a non-vanishing weight wOi that modifies the value of v. While configurations with such close collisions are not frequently sampled, the large impact on the value of v is devastating to PMF simulations. Moreover, the importance of this problem grows with the system size as the weight of even miniscule collisions increases as a function of its distance to the actual location of the excess proton.
As far as the original CEC coordinate (µz/e) is concerned, it does not give a value close to the actual location of the excess proton in any cases in Fig. 1. Although this feature by itself does not invalidate using µz/e for characterizing long range PTs, a problematic feature of µz/e is that it fails to distinguish between degrees of freedom essential for the PT and fluctuations in the environment. To illustrate this point, one water molecule (WAT10) at the end of the single file was manually rotated (Fig. 1e). While the coordinates v and the ξz were not affected by this change, the CEC coordinate showed a shift larger than that observed in the transition from an oxonium ion (Fig. 1b) to a Zundel ion (Fig. 1b). This gets even more problematic when larger systems with more water molecules are examined.
2.1.4 Generalization to complex proton wires in three dimensions
Test calculations in the last subsection demonstrated that the mCEC coordinate, ξz, is robust for describing proton transfer in a linear water chain. Although this is sufficient for long-range PT in some membrane channels with nearly ideal transfer geometry, it is useful to extend ξz to a three-dimensional vector such that PT along more complex pathways can be treated. The proposed functional form is,
| (7) |
where notations are similar to those in Eq. 6 except for the trivial vector extension.
The discussions thus far assumed that each group along the PT pathway can either accept or donate a proton, which is reasonable for water molecules. For long-range PT in proteins, titratable groups may participate in the PT pathway, which may give rise to more complex scenarios that further complicate the definition of the reaction coordinate. For example, in the sidechains of glutamate and aspartate, the protonation and deprotonation of the two titratable heavy atoms Xk,Xl (e.g., O∈ atoms in Glu) are coupled. In those cases, we propose further revision of the mCEC: the weights for the donor/acceptor pair in (Eq. 7) are set as: where wpair is the number of protons coordinated to the residue in its reference state (as described above for atoms). Moreover, the following term is added to for each donor-acceptor pair:
| (8) |
The term m(X,{H}) contains the information regarding whether there is at least one proton coordinated to the respective atom. It is a differentiable approximation to the maximum function for the switching functions (fsw (dX,Hi)) concerning distances between atom X and all protons along the PT pathway:
| (9) |
where a reasonably large integer (e.g., 15) works well for n.
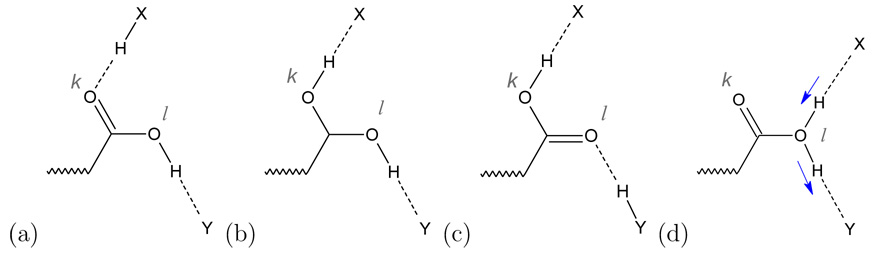
To illustrate the physical significance of the terms in Eq. 8,9, consider the sequence of structures depicted in scheme 1 a)–c). With idealized geometry in structures a,b and c, the weights m(Xk, {H}) and m(Xl, {H}) are (0,1), (1,1) and (1,0), respectively. In the transitional structure shown in 1b, the mCEC with the correction term included locates the excess proton at the midpoint between the two oxygen atoms. The switching functions in Eq. 8 ensure a smooth transition for the reaction coordinate during the proton transfer. The maximum like function m has to be used to allow an alternative PT mechanism as shown in Scheme 1d, which is conceivable in the study of the bacterial photosynthetic reaction center (König et al., work in progress). Here, m(Xk, {H}) and m(Xl, {H}) are 0 and 1 respectively.
Scheme 1.
Proton transfer through a protonated carboxylic acid as an example of a coupled donor-acceptor pair (Ok,Ol). Two mechanisms are shown: (a)–(c) sequence of steps, with Ok and Ol acting as proton acceptor and donor respectively. (d) Ol acts both as donor and acceptor; the arrows indicate the movement of the protons during the reaction.
Unlike ξz, the vectorial quantity can not be readily used in PMF calculations. For this purpose, we further define a collective reaction coordinate ζ for mapping the transfer of the mCEC (as reflected by ) between an initial donor atom D and a final acceptor atom A,
| (10) |
where dξ,D and dξ,A represent the distance from mCEC to the initial donor and the final acceptor atoms respectively; C is a coefficient defining the range of the reaction coordinate and is taken as 1 unless otherwise stated. Apparently, ζ is a switching function dominated by dξ,D and dξ,A at the lower and higher limits, respectively; as ζ evolves from negative to positive values, the mCEC displaces from the initial donor to the final acceptor. The choice of functional form in Eq. 10 is not unique and many other smooth switching functions could be used; e.g., another possible expression is [82],
| (11) |
The value of ζR of 0 and 1 corresponds to the mCEC localized to the initial donor and final acceptor, respectively.
Finally, we note that due to the collective nature of ζ and ζR, their values do not explicitly depend on the number of shuttling groups (i.e., water and titratable residues) along the PT pathway. Therefore, they are conveniently suited for studying PT in complex environments, in which there are either many possible transfer pathways with different numbers of shuttling groups existing at the same time or the number of shuttling groups fluctuates at a time-scale faster compared to that of the PT process. A good example is carbonic anhydrase, for which water wires of different lengths were consistently observed in different MD simulations [28, 29, 83] and the typical life-time for such water wires is on the order of pico-seconds, as compared to the µs time-scale for the PT.
2.2 Electrostatics in QM/MM simulations
As emphasized in several recent studies [66, 83–85], appropriate treatment of electrostatics is utterly important to QM/MM simulations. The issue is most serious for the simulation of processes that involve significant change in the charge distribution, such as oxidation-reduction reactions and long-range proton transfers. Inappropriate treatment of electrostatics may not only affect the quantitative aspect of the result, but also produce qualitative changes in the behavior of biological systems. For example, water structure in the active site of enzyme carbonic anhydrase was found to differ significantly with different electrostatic models used in QM/MM simulations [83].
The most robust electrostatic model up to date involves Ewald summation (or related numerical improvements) with the periodic boundary condition [48]. However, the necessity of including a large number of explicit solvent molecules to avoid artifacts in such simulations limits its use in QM/MM simulations of large systems [86, 87]. Since the systems of interest in the context of proton pumping are often rather large, it is desirable to adopt schemes that avoid periodic boundary condition. In this regard, the generalized solvent boundary potential approach proposed by Roux and co-workers [51] is very attractive. It treats a relatively small region of the protein-solvent-membrane system in microscopic details, with the contribution from the remaining degrees of freedom described at the continuum electrostatics level. This allows very efficient sampling of the most relevant configuration space for the reaction of interest, without significantly sacrificing important environmental effects. In a recent publication [83], we described extension of the GSBP approach to a QM/MM framework and implementation for spherical boundary condition. Here we further extended the implementation to rectangular boundary condition, which is useful for membrane systems. In the following, for the sake of completeness, we briefly review the GSBP approach in its original form [51] and then describe our implementation in the QM/MM framework.
2.2.1 Generalized solvent boundary potential
In a system composed of biomolecules in solution and membrane, interactions between charge distributions are shielded in a nontrivial way due to complex boundaries between different dielectric environments. In the generalized solvent boundary potential (GSBP) approach [51], the system is partitioned into an inner region and an outer environment, where the dielectric property can vary (e.g., containing both bulk solvent and a slab of membrane). Atoms in the inner region are allowed to move during the simulation whereas atoms in the outer region are fixed.
Accordingly, the electrostatic solvation free energy ΔWelec can be partitioned into contributions from the interactions of the outer-outer (oo), inner-outer (io), and inner-inner (ii) parts of the system:
| (12) |
The outer-outer contribution is a constant and therefore is not of interest. The contribution of the inner-outer part can be computed in a straightforward manner,
| (13) |
where is the reaction field due to the outer region atoms, which have to be computed only once (since the outer region is fixed) and saved on a set of grid points in the inner region. Note that actually the total electrostatic potential due to the outer region atoms, is saved instead of the reaction field potential so that direct Coulombic interaction between the inner and outer region atoms is included.
Since the inner region atoms move during the simulation, the electrostatic problem is solved by using a basis set expansion for the charge distribution in the inner region,
| (14) |
where the expansion coefficients cm can be calculated using The overlap matrix elements Omn of the basis functions are computed as and the vector of generalized multipole moments Qn are defined as
Correspondingly, the inner-inner contribution to the solvation free energy takes the following form:
| (15) |
where M is termed the reaction field matrix and is the numerical representation of the Green’s function for the Poisson-Boltzmann equation in the bm basis. Similar to , M only needs to be computed once via solving the Poisson-Boltzmann equation and does not depend on the instantaneous configuration of the inner region.
The choice of basis functions depends on the geometry of the problem. For membrane proteins, a rectangular boundary (see below) is appropriate. Accordingly, Legendre polynomials are taken as the basis functions.
2.2.2 Generalized solvent boundary potential in the SCC-DFTB/MM Framework
As described in recent work [83], the GSBP approach can be easily extended to a general QM/MM framework. It is natural to assume that all QM atoms are within the inner region. Then, it is straightforward to show that the QM/MM contribution to the solvation free energy has the form,
| (16) |
The elements of the vector QMM differ from the full charge vector by the fact that modifications required at the QM/MM frontier [53] are accounted for [83]. In the spirit of SCC-DFTB [59], the QM charge distribution is expressed in terms of Mulliken charges, which simplify the calculation of the corresponding GSBP terms. The generalized multipole moments QQM are computed as:
The QM/MM related GSBP contributions are solved in a self-consistent manner by including the GSBP terms in the Hamiltonian matrix elements Hµv for the QM region,
| (17) |
where Sµv is the overlap matrix element; and are defined as the following
| (18) |
| (19) |
The above formulation is exactly the same as that discussed in our recent work [83], in which GSBP was implemented in the SCC-DFTB/MM framework for spherical boundary condition. In the current work, we extended the implementation to rectangular boundary condition, which involves computation of and using the Legendre polynomials. The implementation is available in the latest developmental version of CHARMM (c32a2).
3 Test calculations
Several systems were chosen to systematically test the simulation protocols developed here. First, two PT pathways in a realistic enzyme model for carbonic anhydrase were studied to test the robustness of new reaction coordinates for long-range PT through non-linear water chains. Next, PT in a model channel embedded in different dielectric environments was studied. Finally, an aquaporin was used to further illustrate the reliability of the GSBP based QM/MM simulation setup for the study of more realistic membrane systems.
3.1 Proton Transfer in carbonic anhydrase II
Carbonic anhydrase II (CAII) is a zinc-enzyme that has been widely used as a prototypical model for studying long-range PT in proteins. Its biological function is to interconvert CO2 and which makes it important in respiration processes [88, 89].
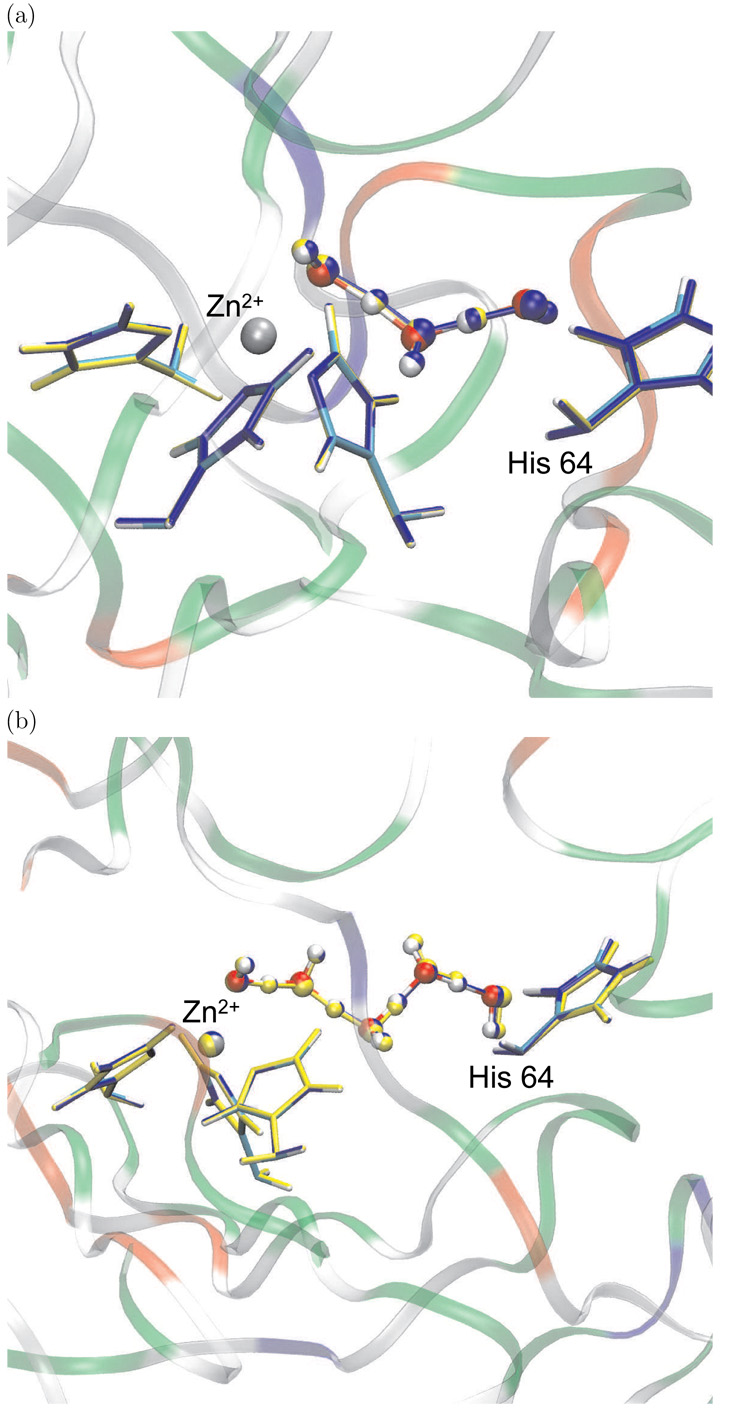
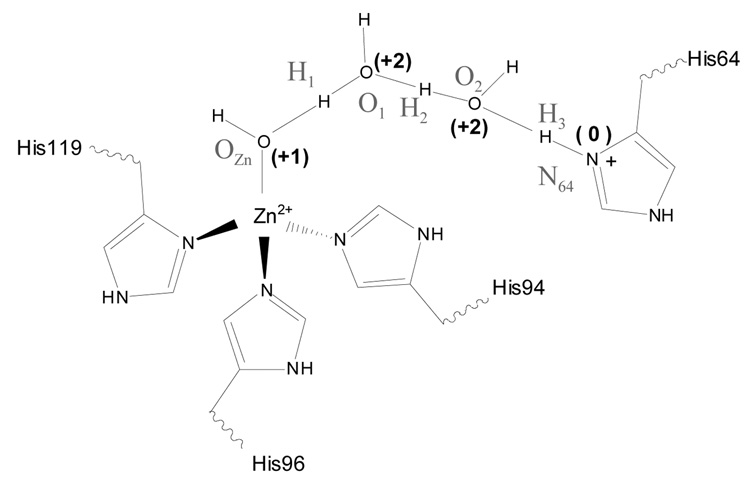
An important step during the catalytic cycle is the transfer of a proton between a zinc-bound water and a histidine residue (His 64) close to the surface of the protein; in the forward direction, the PT generates a zinc-bound hydroxide, which is the catalytic species that reacts with CO2. The distance between the zinc-bound water and His 64 is about 8 Å in the x-ray structure, which led to the suggestion that PT has to be mediated by water molecules in the active site [90–92]. Although two-water molecules have been observed in the x-ray structures that connect the zinc-bound water and His 64 through hydrogen bonds, simulation studies with different potential functions [28, 29, 83] have shown that the water structure in the active site is rather dynamical and water bridges connecting the zinc site and His 64 typically include from two to four water molecules. The on-going debate [22, 23, 25, 26, 28, 29] concerns whether formation of a specific type of water bridge contributes dominantly to the kinetic bottleneck of the PT and whether the PT proceeds through a concerted or step-wise mechanism [24–27, 93]. In the current work, we only use CAII to illustrate the new reaction coordinates via comparison to minimum energy path results. Detailed discussions of mechanistic issues are reported elsewhere [27] (Riccardi and Cui, work in progress).
3.1.1 Computational Setup
The enzyme model was set-up using the GSBP protocol: it contained a 16 Å microscopic spherical region centered around Zn2+ in which all atoms were allowed to move, and a 2 Å layer of Langevin region in which atoms were harmonically constrained. The remaining protein atoms and x-ray water molecules formed the ”outer” region were fixed in space. Spherical harmonics up through the 20th order were used as the basis functions for the electrostatics in GSBP. The QM region included the zinc ion and all of its ligands (His 94, 96, 119 plus a water), the His 64 sidechain and the bridging water molecules (2 and 4, see below); they were treated using SCC-DFTB, while the rest atoms were described with the CHARMM 22 force field for proteins [94]. For further details, refer to Ref. [27, 83].
Two configurations collected from MD simulations, as described in an earlier publication [83], were used as the starting geometry. The MD simulations were done in the ”COHH” state [28], in which the zinc-bound water is deprotonated and His 64 is doubly protonated (H64H+); thus the PT proceeds from H64H+ to the zinc-bound hydroxide in the present set of results. The two starting configurations contain two and four bridging water molecules, respectively, between the zinc-bound hydroxide and H64H+. Adiabatic mapping calculations were carried out using both ζ (Eq.10) and ζR (Eq.11) to map out the potential energy profile along these approximate reaction coordinates; the weights associated with the heavy atoms along the water wire for ζ and ζR are illustrated in Scheme I. Minimum energy pathways were determined using the conjugate peak refinement (CPR) algorithm [47] in CHARMM. The results of the adiabatic mapping and the MEP, both the energetics and critical geometrical parameters along the path, are compared in the following. In addition, adiabatic mapping calculations were carried out for strictly step-wise PTs for the two-water bridge case, to illustrate the importance of having a flexible reaction coordinate. These calculations used a series of anti-symmetric stretch (δ, Eq. 1) to drive the sequential PTs; three possible sequences of transfers were studied (see below). No mass-weighting was used in any calculations and no zero-point energy correction was included in the energetics.
3.1.2 Results
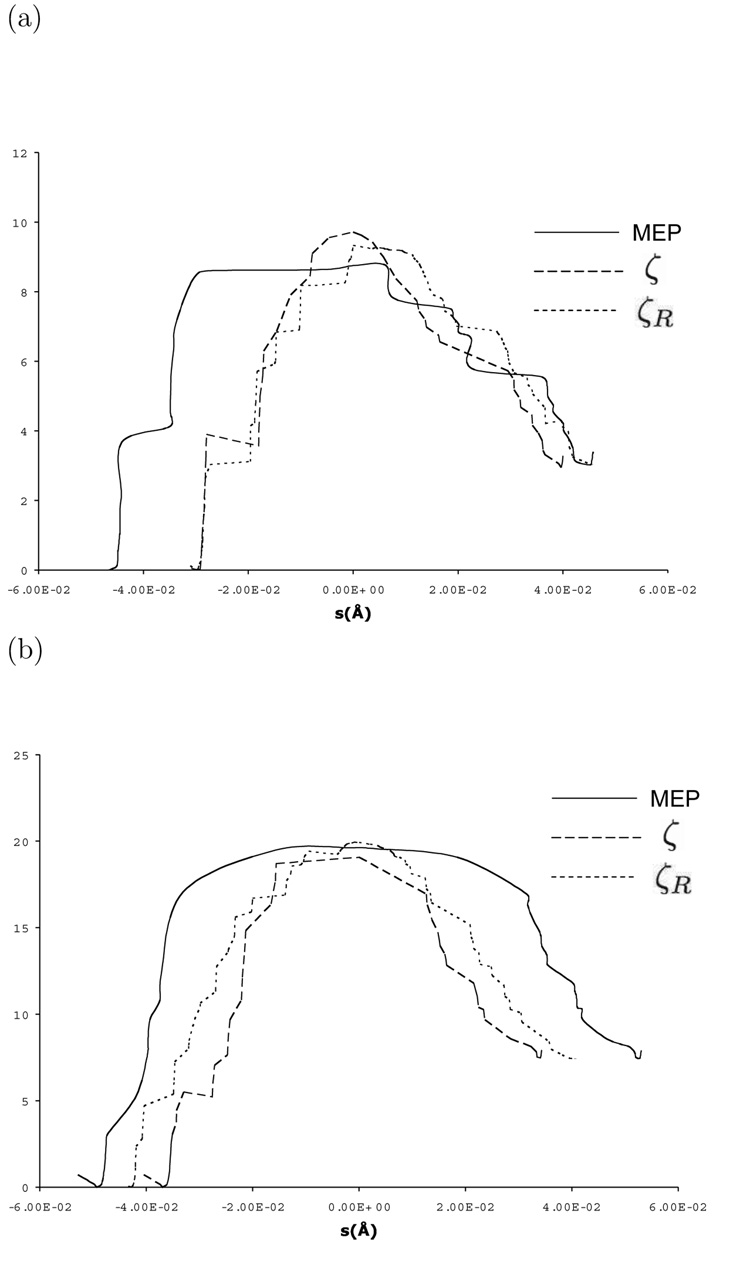
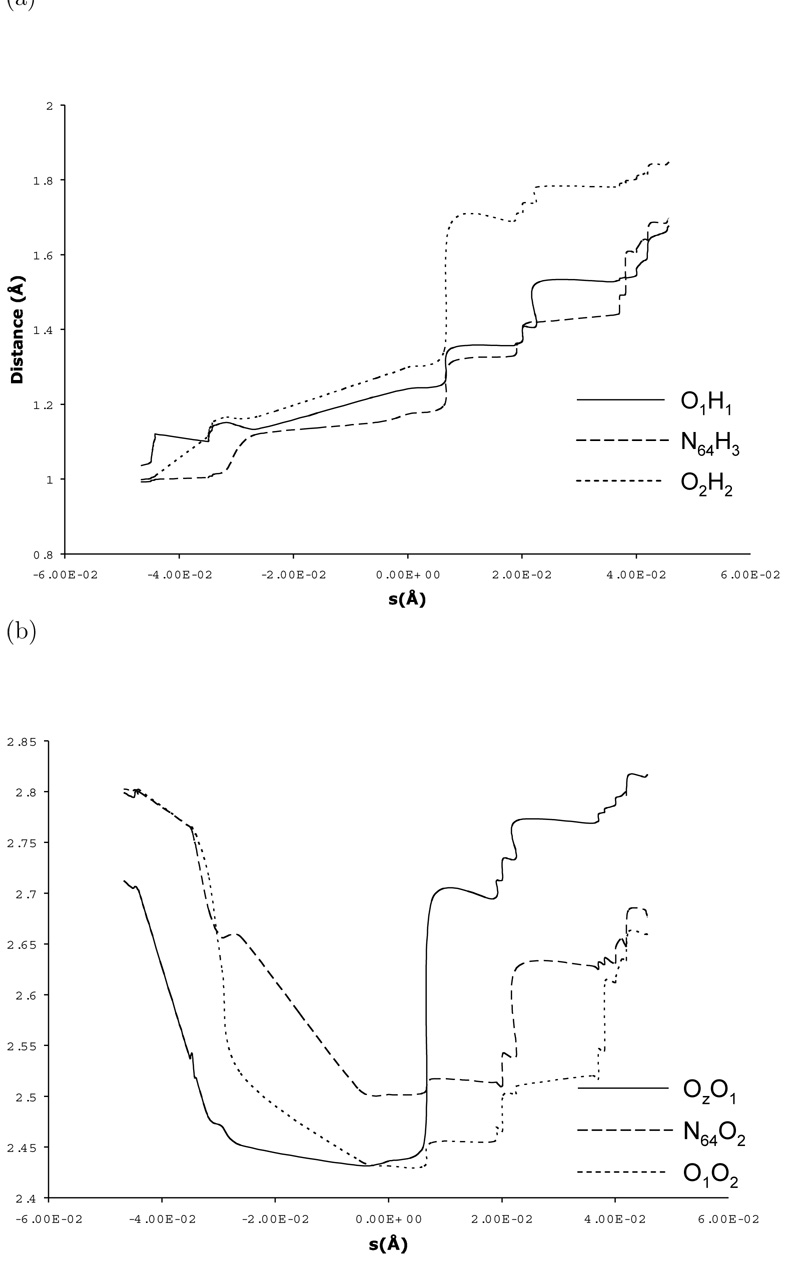
For the two-water bridge configuration chosen here, the MEP calculations identified only one unique saddle point (Fig. 2a) and the barrier height is approximately 8.7 kcal/mol. The transfer of the three protons occurred in a nearly concerted fashion, with the transfer of the middle proton (H2) somewhat ahead of the other two (Fig. 3a, Table 3). The distances between proton donor and acceptor atoms also varied significantly during the PT, which is a well-known result [95] (Fig. 3b).
Figure 2.
Energy profiles along different reaction paths ) for the proton transfer in carbonic anhydrase starting from a configuration with (a) two (b) four bridging water molecules. The horizontal axis is defined as the path length (in Å) measured relative to the highest energy structure along the path. The longer path lengths and broad features in the MEP results are due to the small cumulative variations in the MM degrees of freedom, which is a limitation in the conjugate peak refinement algorithm used here.
Figure 3.
Critical geometrical parameters along the minimum energy path for the proton transfer in carbonic anhydrase starting from a configuration with two bridging water molecules. The horizontal axis is defined as the path length (in Å) measured relative to the highest energy structure along the path. (a) The distances between transferring protons and the original oxygen donor atoms; (b) The distances between proton donor and acceptor atoms. For the atom labels, see Scheme I.
TABLE 3.
Comparison of critical distances (in Å) in the transition state for the proton transfer through two bridging water molecules in carbonic anhydrase obtained using different protocolsa
| MEP | ζ | ζR | |
|---|---|---|---|
| rOZnO1 | 2.43 | 2.44 | 2.48 |
| rO1O2 | 2.44 | 2.44 | 2.44 |
| rO2NH64 | 2.50 | 2.51 | 2.58 |
| rOZnH1 | 1.26 | 1.28 | 1.34 |
| rH1O1 | 1.17 | 1.16 | 1.14 |
| rO1H2 | 1.14 | 1.16 | 1.23 |
| rH2O2 | 1.30 | 1.28 | 1.20 |
| rO2H3 | 1.27 | 1.29 | 1.43 |
| rH3NH64 | 1.24 | 1.23 | 1.16 |
The transition state for MEP is the saddle point along the minimum energy path obtained using the conjugate peak refinement algorithm. For ζ (Eq.10) and ζR (Eq.11), the structure corresponding to the highest potential energy in the adiabatic mapping results were used. For atom labels, see Scheme I.
Results from adiabatic mapping using ζ and ζR are in very good agreement with the MEP data. As shown in Fig. 2a and Table 2, barrier heights, exothermicity of the reaction and qualitative shape of the energy profile from MEP calculations are well reproduced by both ζ and ζR based adiabatic mapping calculations; e.g., the barrier using ζ and ζR is 9.7 and 9.3 kcal/mol, respectively, compared to the value of 8.7 kcal/mol from MEP calculations. The evolution of critical geometrical parameters along the path (i.e., those presented in Fig. 3) are also well reproduced using both ζ and ζR based calculations (Table 3). This is quite satisfying considering that ζ and ζR are collective coordinates that do not specify, a priori, the sequence of proton transfers. Specifically, the structure of the saddle point from MEP calculations agrees well with the highest-energy structures from adiabatic mapping calculations using the two reaction coordinates (Fig. 4a). The critical distances shown in Table 3 further illustrated the concerted nature of the PT transition state.
TABLE 2.
Comparison of barrier heights E‡ and exothermicities ΔE (in kcal/mol) for the proton transfers in carbonic anhydrase obtained using different protocolsa
| E‡ | ΔE | |
|---|---|---|
| 2-water bridge | ||
| MEP | 8.7 | 3.0 |
| ζ | 9.7 | 3.2 |
| ζR | 9.3 | 3.0 |
| Step-wise (seq. 1)b | 22.7 | 5.9 |
| Step-wise (seq. 2)b | 37.1 | 5.9 |
| Step-wise (seq. 3)b | 29.5 | 7.3 |
| 4-water bridge | ||
| MEP | 19.6 | 7.4 |
| ζ | 19.0 | 7.4 |
| ζR | 19.9 | 7.4 |
The MEP results are from minimum energy path calculations using the conjugate peak refinement algorithm. Other results were obtained using adiabatic mapping calculations using specific reaction coordinates (ζ (Eq.10), ζR (Eq.11) or δ (Eq. 1)). The SCC-DFTB/CHARMM-GSBP protocol was used for the potential function (see text).
In the ”Step-wise” calculations, different sequences of PTs were followed in a strict step-wise manner using consecutive adiabatic mapping calculations with δ as the reaction coordinate. Following the notation in Scheme I, the PT sequences are: H1-H2-H3(seq. 1); H1-H3-H2 (seq. 2); H3-H2-H1 (seq. 3). The difference between the exothermicity in step-wise PTs and MEP/ζ,ζR results is due to the relaxation in the MM environment after a larger number of minimizations; the small value relative to the difference in barrier heights, however, does not complicate the comparison between different PT pathways.
Figure 4.
Overlay of transition state structures from adiabatic mapping and minimum energy path calculations for proton transfer through (a) two (b) four bridging water molecules in carbonic anhydrase. For precise values for critical geometrical parameters, see Table 3 and 4.
Adiabatic mapping calculations following a strictly step-wise transfer mechanism, by contrast, showed very different energetics. As shown in Table 2, the energetics depend very sensitively on the sequence of the transfers. Transfer sequences that leave localized the hydroxide-hydronium pair is highly unfavorable energetically with barriers on the order of 40 kcal/mol; a sequence that involves the formation of a localized hydroxide is less demanding although the barrier is still substantially higher (22 kcal/mol) than the concerted pathway (~ 9 kcal/mol) revealed in both MEP and adiabatic mapping calculations using ζ and ζR. Although it is not our intention to conclude based on these calculations that concerted PT in CAII is much preferable than a strictly step-wise mechanism, it is worth mentioning that reaction path calculations starting from multiple (~ 100) initial structures and PMF calculations do support this scenario [27].
For the four-water bridge configuration it is striking that only one saddle point was found in the MEP calculations as well. The broad feature of the MEP energy profile in the barrier region is due to the small variations of MM atoms rather than major changes in the reactive degrees of freedom, which is a limitation to the CPR algorithm in the absence of any special coordinate weighting. The barrier height of 19.6 kcal/mol in this specific reaction path is substantially higher than the barrier of 8.7 kcal/mol in the two-water-bridge case discussed above. We emphasize that these results alone do not suggest that the barrier of PT through a longer water bridge is necessarily higher than that through a shorter water bridge, because significant variations in the behavior was found when multiple protein configurations were considered (Riccardi, Cui, work in progress). The issue of interest here is whether ζ and ζR can reproduce MEP results when the length of the water wire is long. The results suggest that this is the case, as both energetics (Table 2) and transition state structures (Fig. 4b, Table 4) were well reproduced by adiabatic mapping calculations; the agreement is more impressive using ζR.
TABLE 4.
Comparison of critical distances (in Å) in the transition state for the proton transfer through four bridging water molecules in carbonic anhydrase obtained using different protocolsa
| 4-water bridge | MEP | ζ | ζR |
|---|---|---|---|
| rOZnO1 | 2.42 | 2.45 | 2.42 |
| rO1O2 | 2.44 | 2.52 | 2.49 |
| rO2O3 | 2.43 | 2.65 | 2.43 |
| rO3O4 | 2.42 | 2.57 | 2.43 |
| rO2NH64 | 2.49 | 2.45 | 2.42 |
| rOZnH1 | 1.16 | 1.15 | 1.15 |
| rH1O1 | 1.26 | 1.30 | 1.27 |
| rO1H2 | 1.20 | 1.15 | 1.20 |
| rH2O2 | 1.23 | 1.31 | 1.22 |
| rO2H3 | 1.17 | 1.04 | 1.18 |
| rH3O3 | 1.26 | 1.53 | 1.25 |
| rO3H4 | 1.14 | 1.03 | 1.14 |
| rH4O4 | 1.30 | 1.62 | 1.30 |
| rO4H5 | 1.22 | 1.15 | 1.22 |
| rH5NH64 | 1.27 | 1.37 | 1.16 |
In short, test calculations using CAII produced encouraging results suggesting that both collective coordinates introduced here, ζ and ζR, can be used to describe the energetics and mechanism of PT spanning a long-distance through water wires of rather complex geometries (Fig. 4).
3.2 A model proton channel
As a second example, PT along a chain of water molecules confined in a model channel was studied. The channel was embedded in different environments described by continuum models to explore the dielectric effect on the PT energetics. The effect of positioning permanent dipoles lining the channel was also briefly studied. This simple model was inspired by the recent heated discussions regarding why some membrane water channels conduct protons whereas others do not (see, e.g., a non-technical summary in Ref. [96]). For instance, PT through gramicidin is rather facile. In fact, this is the functional mechanism of this antibiotic peptide. By contrast, aquaporins, which are responsible for homeostasis in higher organisms, efficiently conduct water molecules [97, 98] yet exclude protons and other charged ions from passing through. Different factors have been suggested to determine proton conductance, which include structure of water wires inside the protein [98, 99], electrostatic interaction between the protein and the proton [100,101], the dielectric barrier [37] or combinations thereof [36]. Here, we constructed a simple model channel (Fig. 5) to study the influence of different dielectric environments on the energetics. However, the major goal is to illustrate the principles of the QM/MM/GSBP simulation protocol.
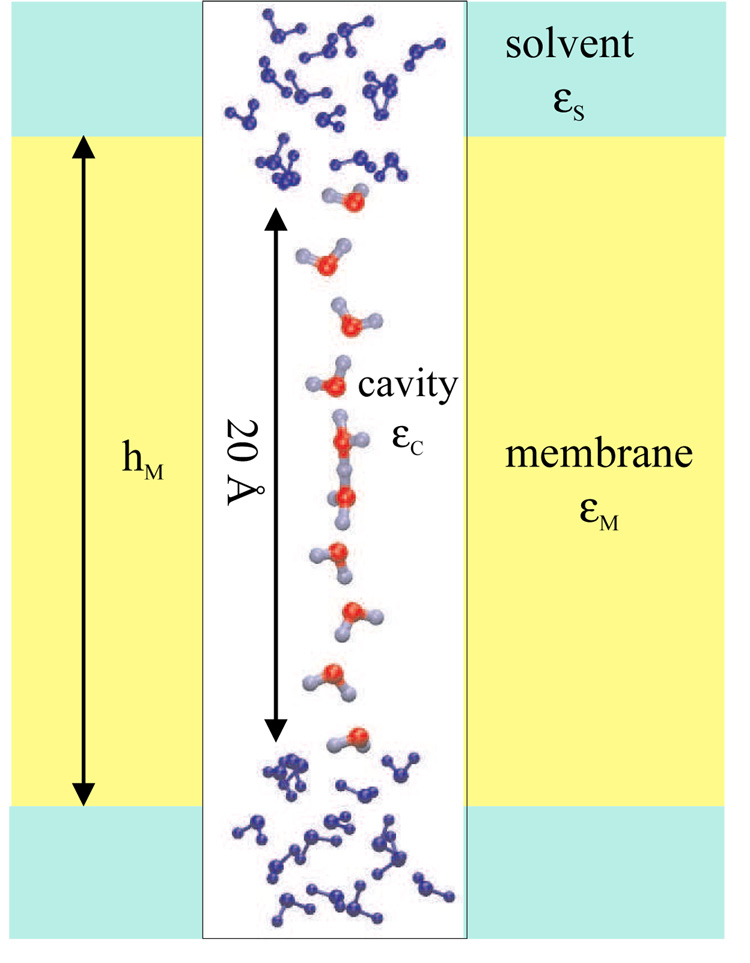
Figure 5.
Geometric and dielectric parameters for the model channel. See Table 5 for specific numerical values for the dielectric constants (∈C, ∈M, ∈S) and the membrane thickness hM.
3.2.1 Construct of the model
The model is similar to the one used in a previous study [50] and was constructed using the following procedure. A linear water chain consisting of 10 water molecules and one excess proton was built, in which the distance from the first to the last oxygen is 22.0 Å; the choice of the number of water molecules and the length of the chain were motivated by the structures of aquaporin and gramicidin A, in which the single-file part of the transmembrane water chain typically consists of 9–10 water molecules. The model ”channel” that holds the water wire was introduced by applying a cylindrical potential to each of the oxygen atoms in the water wire:
with kcyl =100 kcal mol−1 Å −2. The quantity r is the distance of the oxygen atom from the axis of the channel, and r0 is 0.5 Å; Θ is the Heaviside step function. To ensure appropriate solvation of the excess proton at the ends of the water chain, fifteen water molecules were added to each end of the chain. The coordinates of these water molecules were taken from an equilibrated box of water molecules at 300 K. The ”bulk” water molecules were subject to a set of quartic constraints to maintain a cubic shape of 3×3×2.5 Å.
This ”dumb-bell” model system was then embedded into different dielectric environments including vacuum, pure solution and membrane systems. Important geometrical parameters are illustrated in Fig. 5 and their values are summarized in Table 5 for different environments. In the membrane set-ups, the grids used in GSBP and midpoint of the membrane was centered on the model channel. The grid dimensions were 92×92×185, and a coarse grid spacing of 1.6 Å and a finer spacing of 0.4 Å were used in focusing Poisson-Boltzmann (PB) calculations required for the various quantities used in the GSBP computations. The dielectric constants for the membrane and bulk water were set to be 2.0 and 80.0, respectively; although the PB module in CHARMM allows the use of different dielectric constants for the lipid polar heads and non-polar tails, this was not pursued here and will be tested more systematically in a more quantitative study of pKa in membrane proteins in the future. The salt concentration was chosen to be 150 mM. Although Im et al. [102] showed that the artifacts encountered with trilinear interpolation should have a minor effect on molecular dynamics simulations, we used the slightly more involved cardinal B-spline method for interpolating between the gridpoints. In the GSBP calculations for the inner electrostatics, Legendre polynomials up to order 10 were used as basis functions, which was found sufficient in previous studies for systems of similar size [51, 103].
TABLE 5.
Geometric and dielectric parameters for the model channel shown in Fig.5: dielectric coefficients for cavity ∈C, membrane ∈M and solvent ∈S and membrane thickness hM.
| ∈C | ∈M | ∈S | hM [Å] | |
|---|---|---|---|---|
| vacuum | 1 | 1 | 1 | n/a |
| water | 1 | 80 | 80 | n/a |
| membrane 20 | 1 | 2 | 80 | 20 |
| membrane 30 | 1 | 2 | 80 | 30 |
To study the effect of polar residues on the PT energetics, dipoles were added in the lumen of the model channel embedded in the 30 Å model membrane. Specifically, four dipoles with rather modest dipole moments composed of two opposite charges (|q|=0.1e,d=1.5Å) were circularly arranged around the channel at a radius of 3.3 Å. Three of these annuluses were positioned at z=−1.4,1.2, 3.8 Å (Fig. 6b), which were motivated by the carbonyl groups in gramicidin A; two extreme orientations of the dipoles were considered.
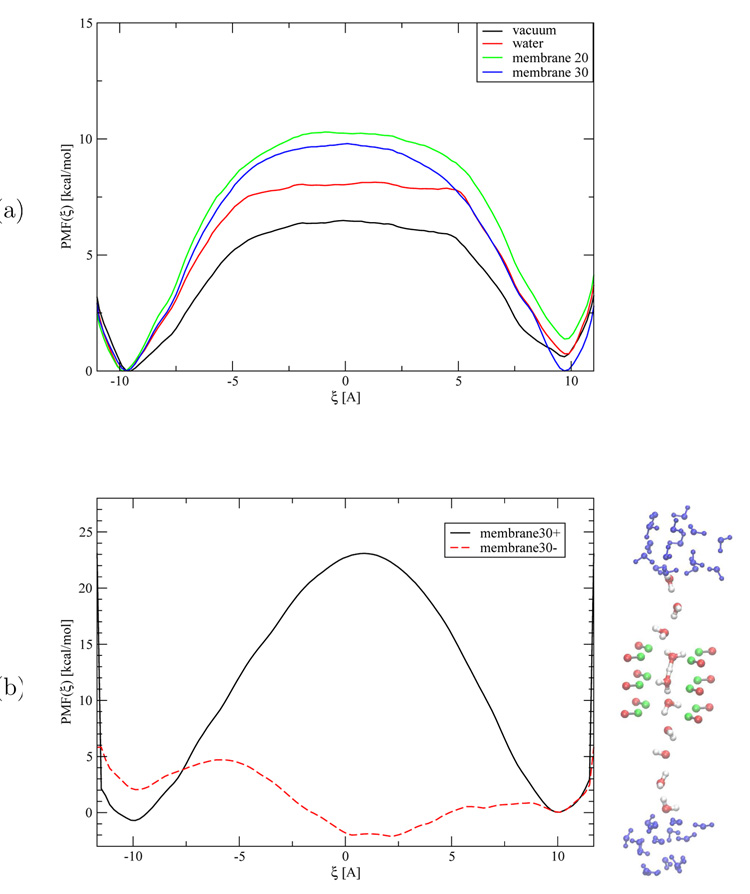
Figure 6.
(a) Potential of mean force for proton transfer through a purely non-polar model channel embedded in different dielectric environments. (b) Potential of mean force for proton transfer through the model channel with dipoles arranged in the channel. The solid (dashed) line refers to a setup where the positive (negative) charges of the dipoles point to the center of the channel.
The chain of water molecules and the excess proton were described with the SCC-DFTB method, while the rest explicit water molecules were described with the TIP3P model. PMF calculations were carried out using the standard umbrella sampling technique [104] with ξz as the reaction coordinate since the water wire here is highly linear. A total of ten windows was used, and each window contained 40 ps of equilibration and 100 ps of production calculations.
3.2.2 PT energetics in different environments
In vacuum, the proton transfer in the dumb bell model has a modest barrier of 6.5 kcal/mol (Fig. 6a); there is a barrier because the excess proton is preferentially stabilized by the explicit solvent molecules at the ends of the water wire. The barrier increases slightly to 8.1 kcal/mol, when the model is immersed in bulk water represented using a dielectric medium of ∈ = 80. With the dielectric model for the membrane, the effect of preferential solvation at the ends of the water wire becomes more significant, which causes the PT barrier to increase even further; with the membrane thickness (or low-dielectric part of the membrane) of 20 and 30 Å, respectively, the barrier is 10.3 and 9.8 kcal/mol, respectively.
The results shown here are in qualitative agreement with electrostatic calculations of Burykin et al. [37], who found a barrier of about 15 kcal/mol for transferring a proton through a water wire in a cylindrical pore of radius 4 Å through a 30 Å thick membrane. However, the barrier observed here includes only part of the dielectric barrier due to the membrane. Since the initial position of the excess proton is close to the low-dielectric region in the current model, the charge has already been partially desolvated. To include the entire dielectric barrier, the free energy necessary for transporting the proton from the bulk to the minimum in the PMF has to be computed [105].
The presence of the dipoles along the channel was found to have a major impact on the PT energetics, which was qualitatively expected. With the positive charges of the dipoles facing the center of the channel, the PT barrier is 23 kcal/mol (Fig. 6b). In contrast, the opposite polarity lowers the PT barrier dramatically from ~ 10 kcal/mol for the non-polar channel to nearly vanishing. In fact, the most stable configuration corresponds to the excess proton localized in the center of the channel close to one sets of dipoles (z ~ 1:2 Å).
In short, study of this simple model showed that the GSBP approach is able to capture the qualitative effect of different environments on the PT energetics along a chain of water molecules. The transition from a high dielectric medium to a low dielectric medium modeled in the slab model described above imposes a notable effect on the PT energetics, on the order of a few kcal/mol. However, the effect of the polar groups mimicked by a set of dipoles of rather small magnitude has a more significant effect. Hence, the results of our simple model support the claim of Warshel and co-workers that non-polar membrane channels per se do not conduct protons, unless the charged species can be stabilized within the channel as described recently [37].
3.3 Water structure in aquaporin
Finally, we study the water distribution in aquaporin as an illustration of the simulation protocol being developed here applied to a realistic membrane protein system. The ultimate goal is to understand proton blockage in aquaporin. As mentioned above, although several studies [36–38] have been carried out, there are still debates regarding the contribution of various factors to the high barrier for the PT. All previous calculations used empirical models describing the proton transfer process, thus it remains meaningful to carry out reliable QM/MM simulations to better understand the importance of various factors that have been proposed. In this work, however, we focus on the water structure, which has been proposed to contribute to the high barriers for PT in aquaporins and other proton-blocking proteins [98,99]; the PT energetics and mechanism of proton blockage will be discussed elsewhere (Hoffmann et al., work in progress).
Aquaporin water channels have been shown both experimentally [106] and through molecular dynamics simulations [36, 98, 99, 101, 107] to have a pronounced water structure. Similar observations were also made in gramicidin in simulation and experiment as reviewed in [108]. Here we chose to investigate the aquaporin channel since the embedded water structure has been confirmed by multiple simulations using different force fields (GROMACS, CHARMM), water models (TIP3P, SPC, PM6) as well as different simulation protocols [36, 98, 99, 101, 107]. For example, Tajkhorshid et al. [98] carried out simulations for a glycerol conducting aquaporin [106]: an aquaporin tetramer was embedded in an explicit membrane-water environment and the Ewald summation for long-range electrostatics was applied with periodic boundary conditions. The simulations showed distinct water structure around hydrogen-bond-forming moieties in the lumen region, and the locations of high water densities match well with those identified from high-resolution x-ray structures presented in the same paper. For our purpose, comparing results to those previous calculations using explicit membrane and bulk solvent molecules is expected to be a stringent test of the GSBP-QM/MM simulation protocol being developed here.
3.3.1 Simulation set-up
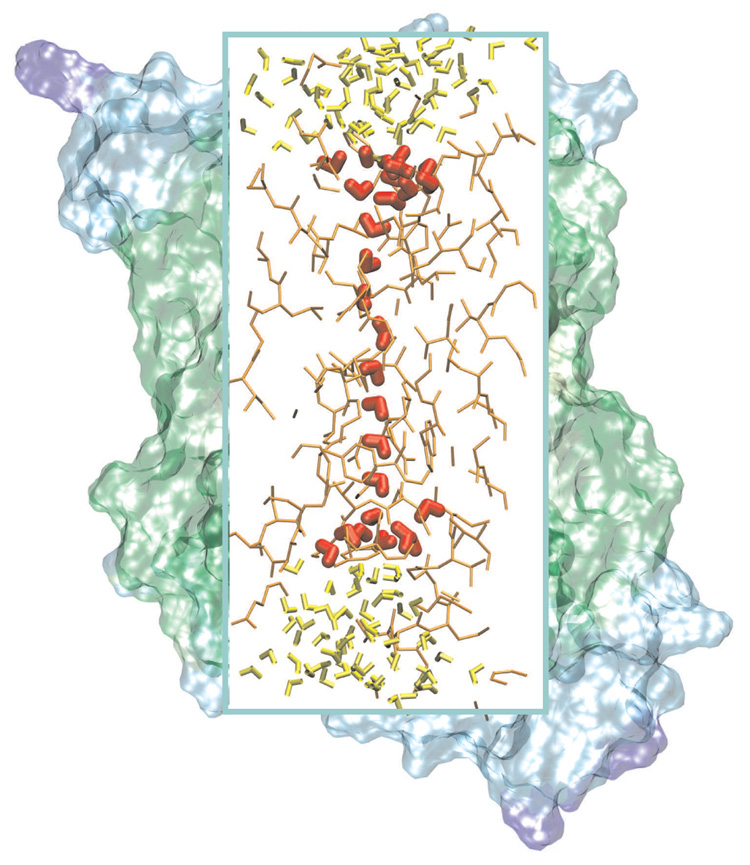
The QM/MM-GSBP simulations were initiated from a snapshot from the classical simulation of Tajkhorshid et al. [98] mentioned above. Only one monomer was selected for simulation and the rest proteins and explicit membrane were discarded. As shown schematically in Fig. 7, the GSBP set-up partitioned the system into inner and outer rectangular cavities that contain 2473 and 1828 atoms, respectively. The size of the inner region is 22×25×54 Å3, which is described by a set of grids with dimensions of 139×161×175 in the PB calculations for the various GSBP components; a coarse grid spacing of 1.6 Å and a finer spacing of 0.4 Å were used in focusing Poisson-Boltzmann (PB) calculations. The coordinate origin and midpoint of the membrane was set to a water molecule in the center of the channel, which corresponds to W522 in the x-ray structure [98]. The membrane, which was entirely treated with a dielectric model, was set to have a thickness of 35 Å the dielectric coefficients of membrane and bulk solvent were chosen to be 2.0 and 80.0, respectively, and the salt concentration was chosen as 150 mM. Similar to the model channel simulations discussed above, Legendre polynomials up to order 10 were used as basis functions for the GSBP inner region electrostatics. The protein-solvent boundary was set up using the atomic Born radii of Nina et al. [109]. The atoms outside the inner cavity were constrained to their initial position. Protein atoms at the boundary of inner and outer cavity were constrained according to the previously described protocol [51]. For the Coulombic interactions, extended electrostatics model [110] was used where interactions beyond 12 Å were treated using a multipolar expansions including dipole and quadrupole terms.
Figure 7.
Partitioning of the aquaporin system for QM/MM-GSBP simulations. The inner rectangular box shows the atoms located in the inner cavity in the GSBP set-up. The atoms treated using SCC-DFTB are highlighted. Atoms in the outer region were fixed during the simulation and their effect were taken into account through continuum electrostatics (see text).
The model contains a total of 154 water molecules, 27 of which were treated using SCC-DFTB. Treating these water molecules with a QM model provides a consistent and natural description for the PT process; the co-operativity among the water molecules, which is missing in previous classical treatments [36, 98, 99,101,107], is also taken into account. Current research in the group investigates the proton exclusion mechanism in aquaporins (Hoffmann et al. , work in progress) based on the model introduced here, which will provide a further inside in the impact of co-operativity in the water chain and polarization, which has been proposed to be potentially important [111]. All of these QM waters were in the interior of aqua-porin in the starting configuration. To prevent lateral diffusion of QM and MM waters, planar restraints were introduced at the interface between QM and MM water molecules. The QM/MM frontier and respective restraints were chosen such that additional QM water molecules can diffuse into and out of the pore. The reservoirs of QM water (red) can be seen on each side of the monofilar water chain in Fig. 7. This is in contrast to the setup in a previous study [36], which reproduced the location of the water molecules in the channel but was limited in its predictivity by restricting water from entering or exiting the pore.
Eight trajectories were calculated, each with a time period of 20 ps for equilibration and 100 ps of data collection. For the determination of relative water densities, the pore radius of the lumen was determined at each point along the channel axis using the Hole2 program [112].
In addition, a set of calculations in which the environment of the inner region was simply replaced by vacuum was also carried out for comparison with the GSBP results. The length of the MD simulations was the same as in the GSBP based simulations.
3.3.2 Results and discussions
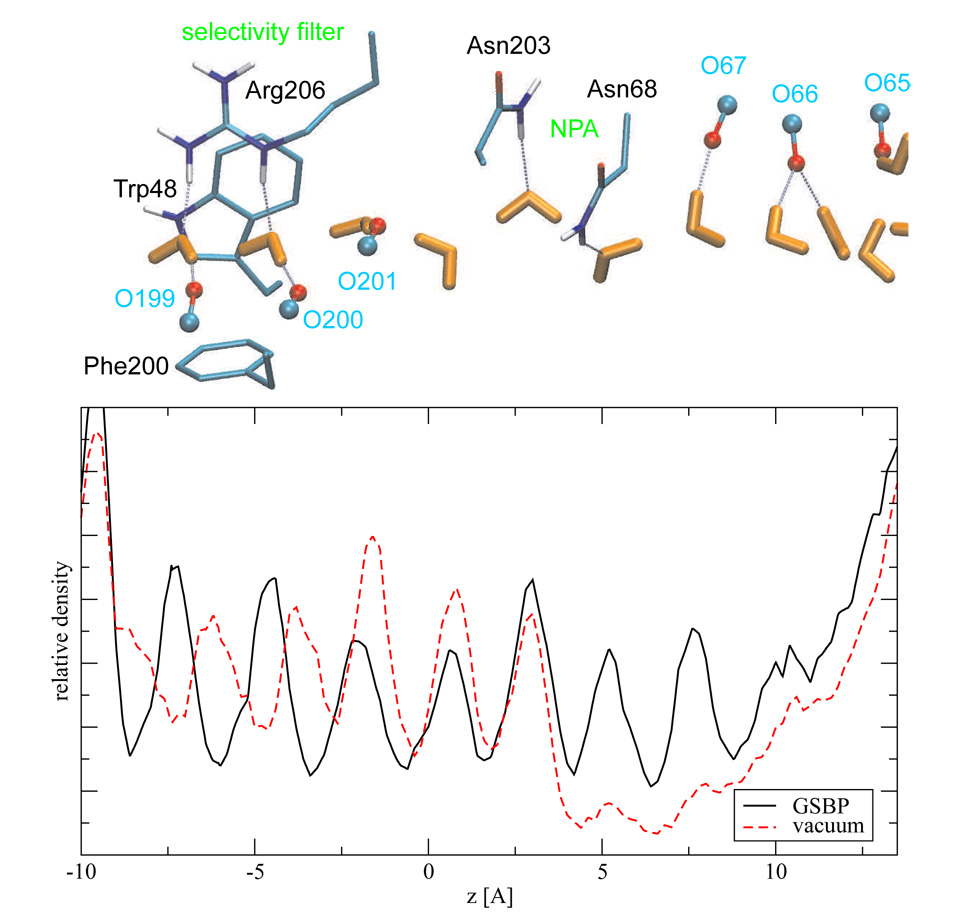
As shown in Fig. 8, the distribution of water molecules has distinct features inside the aquaporin. The peaks in the relative water density coincide well with the position of specific protein polar groups. These including the well-conserved NPA motif in the center of the channel and two half membrane spanning, short peptide sequences (65–68, 199–203) whose carbonyl groups line the channel; Arg 206 in the selectivity filter region also helps to position water molecules. The role of Trp48 and Phe200 is mainly attributed to narrowing the pore size. This might improve the interaction of the water molecules with Arg206 [99]. These observations are in good agreement with previously published data [98, 99] using models that are substantially larger in size. Moreover, the bifurcating orientation of the water wire centered around Asn 203 as observed in previous studies is also reproduced by the current simulation.
Figure 8.
Computed relative water densities in the GLPF-G channel along the membrane norm (z axis) for the vacuum- and the GSBP-based simulations, which were obtained through the frequency of observing the water oxygen atoms at a the respective position. The average positions for selected atoms from the GSBP simulations are also shown to illustrate the correlation between water density and position of polar groups along the channel, which include both protein sidechains (e.g., Arg 206, Asn 68, Asn 203) and main chains (e.g., a series of carbonyl groups).
In the vacuum simulations, the distribution of water agreed well with GSBP only in the central region of the channel. The agreement is visibly worse in regions beyond 5 Å from the center. In particular, the peak positions of the water molecules are shifted in the region of negative z value; these shifts can not be attributed to the shifts in the position of polar groups in the lumen, which are displaced by less than 0.1 Å in the vacuum simulations in comparison to the GSBP simulation. Moreover, disruptions of the water chain were observed between z= 5–10 Å, which is clearly visible in the average density profile. Previous periodic boundary based simulations [101] did notice disruption of the water wire inside aquaporin, although these events were very rare. Therefore, the observed breakdown of the well-ordered water structure in the vacuum simulation is unexpected for a channel with many polar interactions despite its narrow radius. In previous work, such pulsatory behavior was observed only in narrow and purely hydrophobic channels, such as a hydrophobic model of the aquaporin channel [101], carbon nanotube [113] and smooth hydrophobic pore [114] and cylinders [115]. In fact, since the pulsatory transport of water in narrow pores depends on collective oscillation of the bulk water [114], the lack of large body of bulk solvent in the current set-up is not expected to produce channel depletion.
In short, the QM/MM/GSBP simulation presented here agrees well with previous periodic boundary simulations with explicit lipid membrane in terms of water structures inside aquaporin. This is quite remarkable considering the much smaller number of atoms explicitly included in the present protocol and the fact that the membrane was replaced by a simple dielectric model. The comparison with vacuum simulation clearly demonstrated the importance of proper long-range electrostatic treatment for intrinsically heterogeneous systems such as membrane-bound proteins.
4 Concluding Remarks
Although localized proton transfer (PT) reactions have been generally considered as well-characterized, reaction mechanisms for long-range PTs in biological systems are often poorly understood. The large number of protein residues and water molecules involved in the PT process makes determination of the kinetic bottleneck a challenging task for both experimental and theoretical investigations. For example, the rate-limiting step of PT in the enzyme carbonic anhydrase remains controversial despite both experimental and theoretical studies over the past several decades.
In this paper we presented recent developments in our groups motivated by the long-term goal of theoretically characterizing kinetics of long-range PT in biomolecular pumps such as cytochrome c oxidase, which, as discussed in recent studies [40, 116] might exhibit interesting kinetic properties as required by their biological function. Specifically, we introduced a new set of collective coordinates for characterizing the progress of long-range PT. In contrast to earlier suggestions, which were also analyzed here, the new set of coordinates based on the modified center of excess charge (mCEC) works for PT along not only linear but also complex three-dimensional transfer pathways. These coordinates were verified by comparing the corresponding adiabatic mapping results (energetics and key geometrical parameters) to minimum energy path calculations. Excellent agreements were observed for PT through rather complex water wires with realistic model of carbonic anhydrase using a SCC-DFTB/CHARMM potential. These results suggest that the new coordinates can be used as the reaction coordinate for computing meaningful potential of mean force for long-range PT. We note in particular that the collective nature of the new coordinates makes it straightforward to consider the equilibration of different water-wire configurations in the PMF calculations, which is often found rapid compared to the time-scale of the PT process.
In addition, we extended the implementation of the generalized solvent boundary potential (GSBP) [51] in the QM/MM framework [83] to rectangular geometries, which is useful for the simulation of reactions in membrane systems. As discussed in previous studies [51, 83, 103], GSBP is a computationally effective approach for treating long-range electrostatics in heterogeneous macromolecules; it allows one to explicitly sample the phase space for only a small number of protein, solvent and possibly lipid atoms, while approximating the effect due to the rest of the system using continuum electrostatics. For example, the QM/MM-GSBP set-up here used a fairly small model for aquaporin (22×25×54 Å3, which contained ~2500 atoms) and treated both bulk solvent and the lipid membrane using continuum models; the simulation reproduced the water structure in the aquaporin channel compared to previous MD simulations using much larger number of explicit solvent and lipid molecules. Simulations of the same reduced system with the inner region surrounded by vacuum, by contrast, produced rather different water distributions, which further illustrated the importance of long-range electrostatics and effectiveness of the GSBP approach. The influence of electrostatics in long-range PT was also briefly investigated with PT in a model channel embedded in different dielectric continuum environments. Similar to previous work of Burykin et al. [37], the results indicate that PT through a purely hydrophobic channel embedded in membrane encounters a significant barrier, unless polar interactions present in the channel to stabilize the hydronium ion.
With the encouraging results obtained here, we anticipate that a set of robust QM/MM simulation protocols will soon be available for theoretical analysis of long-range PT kinetics in complex biomolecular pumps. Two important issues remain to be resolved. First, effective QM methods that produce reliable energetics for PT involving not only water molecules but also protein sidechains and possibly metal-bound ligands need to be developed; extension of the SCC-DFTB approach [59] is likely to be productive along this line. Second, although a continuum model for lipid membrane and bulk solvent was found satisfactory in the current model studies, whether this is generally applicable to membrane protein systems remains to be clarified. In this regard, quantitative studies such as pKa [85] or binding free energy simulations of membrane systems are particularly useful. Once these technical issues are resolved, theoretical analysis will play a major role in uncovering the secrets of biomolecular pumps.
Scheme 2.
Atom labels and weights (in parentheses) associated with the definition of the modified center of excess charge coordinate (Eq. 7).
Acknowledgements
PHK was partially supported by a stipend within the DFG Research Training Group GK-693 of the Paderborn Institute for Scientific Computation (PaSCo). PHK and MH were supported within the DFG research program 490. The work at UW-Madison (NG and QC) was partially supported by NSF (MCB-0313743 and CHEM-CAREER-0348649); the work at Illinois (ET) was supported by NIH (P41–RR05969 and R01–GM067887). QC is a Alfred P. Sloan Research Fellow. A part of the computations was carried out on the joint cluster PLING of theoretical physics and Paderborn center for parallel computing (PC2). Computational resources from the National Center for Supercomputing Applications at the University of Illinois are also greatly appreciated.
Further, the authors acknowledge helpful discussions with Patricia Schaefer, Demian Riccardi and Mark Formaneck. PHK and MH also thank Dr. Marc Amkreutz for stimulating discussions.
References and Notes
- 1.Fersht A. In: Structure and Mechanism in Protein Science. Guide to Enzyme Catalysis and Protein Folding. Freeman WH, editor. 1999. [Google Scholar]
- 2.Frank RAW, Titman CM, Pratap JV, Luisi BF, Perham RN. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- 3.Birge RR. Biochim. Biophys. Acta. 1990;1016:293. doi: 10.1016/0005-2728(90)90163-x. [DOI] [PubMed] [Google Scholar]
- 4.Okamura M, Paddock M, Graige M, Feher G. Biochimica et Biophysica Acta. 2000;1458:148. doi: 10.1016/s0005-2728(00)00065-7. [DOI] [PubMed] [Google Scholar]
- 5.Wikstrom M. Biochim. Biophys. Acta - Bioenergetics. 2004;1655:241–247. doi: 10.1016/j.bbabio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Brzezinski P. Trends in Biochem. Sci. 2004;29:380. doi: 10.1016/j.tibs.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Nicholls DG, Ferguson SJ. Bioenergetics. Vol. 3. Academic Press; 2002. [Google Scholar]
- 8.Kreuer K, Paddison SJ, Spohr E, Schuster M. Chem. Rev. 2004;104:4637–4678. doi: 10.1021/cr020715f. [DOI] [PubMed] [Google Scholar]
- 9.Kohen A, Klinman J. Acc. Chem. Res. 1998;31:397–404. [Google Scholar]
- 10.Liang Z, Klinman J. Curr. Opin. Struct. Biol. 2004;14:648–655. doi: 10.1016/j.sbi.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Hwang JK, Warshel A. J. Am. Chem. Soc. 1996;118:11745–11751. [Google Scholar]
- 12.Gao J, Truhlar DG. Annu. Rev. Phys. Chem. 2002;53:467. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- 13.Cui Q, Karplus M. J. Am. Chem. Soc. 2002;124:3093. doi: 10.1021/ja0118439. [DOI] [PubMed] [Google Scholar]
- 14.Kiefer PM, Hynes JT. J. Phys. Chem. A. 2003;107:9022–9039. [Google Scholar]
- 15.Hammes-Schiffer S. Biochem. 2002;41:13335–13343. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- 16.Warshel A. Annu. Rev. Biophys. Biomol. Struct. 2002;32:425. doi: 10.1146/annurev.biophys.32.110601.141807. [DOI] [PubMed] [Google Scholar]
- 17.Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 18.Nagle JF, Morowitz HJ. Proc. Nat. Acad. Sci. 1978;75:298–302. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagle JF, Mille M. J. Chem. Phys. 1981;74:1367. [Google Scholar]
- 20.Grotthus C. Ann. Chim. 1806;58:54. [Google Scholar]
- 21.Sham YY, Muegge I, Warshel A. Proteins: Struct., Funct., Genet. 1999;36:484–500. [PubMed] [Google Scholar]
- 22.Silverman DN. Biochim. Biophys. Acta. 2000;1458:88–103. doi: 10.1016/s0005-2728(00)00061-x. [DOI] [PubMed] [Google Scholar]
- 23.Fisher Z, Hernandez Prada JA, Tu C, Duda D, Yoshioka C, H A, Govindasamy L, Silverman DN, McKenna R. Biochemistry. 2005;44:1097. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 24.Aqvist J, Warshel A. J. Mol. Biol. 1992;224:7–14. doi: 10.1016/0022-2836(92)90572-2. [DOI] [PubMed] [Google Scholar]
- 25.Braun-Sand S, Strajbl M, Warshel A. Biophys. J. 2004;87:2221–2239. doi: 10.1529/biophysj.104.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schutz CN, Warshel A. J. Phys. Chem. B. 2004;108:2066–2075. [Google Scholar]
- 27.Xu D, Riccardi D, Ghosh N, Elstner M, Guo H, Cui Q. J. Am. Chem. Soc. Comm. Ed. 2005 Submitted. [Google Scholar]
- 28.Toba S, Colombo G, Merz KMJ. J. Am. Chem. Soc. 1999;121:2290–2302. [Google Scholar]
- 29.Lu D, Voth GA. Proteins: Struct., Funct., and Genet. 1998;33:119–134. [PubMed] [Google Scholar]
- 30.Pomès R, Roux B. Biophys. J. 2002;82:2304. doi: 10.1016/S0006-3495(02)75576-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomès R, Roux B. Biophys. J. 1996;75:33–40. doi: 10.1016/S0006-3495(98)77492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomès R, Roux B. Biophys. J. 1996;71:19. doi: 10.1016/S0006-3495(96)79211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumaker MF, Pomès R, Roux B. Biophys. J. 2000;79:2840. doi: 10.1016/S0006-3495(00)76522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bondar A, Fischer S, Smith JC, Elstner M, Suhai S. J. Am. Chem. Soc. 2004;126:14668. doi: 10.1021/ja047982i. [DOI] [PubMed] [Google Scholar]
- 35.Bondar A, Elstner M, Suhai S, Smith JC, Fischer S. Structure. 2004;12:1281. doi: 10.1016/j.str.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Chakrabarti N, Tajkhorshid E, Roux B, Pomès R. Structure. 2004;12:65. doi: 10.1016/j.str.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Burykin A, Warshel A. Biophys. J. 2003;85:3696. doi: 10.1016/S0006-3495(03)74786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarti N, Roux B, Pomès R. J. Mol. Biol. 2004;343:493. doi: 10.1016/j.jmb.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Voth GA. Biophys. J. 2003;85:864. doi: 10.1016/S0006-3495(03)74526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popovic DM, Stuchebrukhov AA. J. Am. Chem. Soc. 2004;126:1858–1871. doi: 10.1021/ja038267w. [DOI] [PubMed] [Google Scholar]
- 41.Cukier RI. Biochim. Biophys. Acta. 2005;1706:134–146. doi: 10.1016/j.bbabio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Olkhova E, Huter MC, Lill MA, Helms V, Michel H. Biophys. J. 2004;86:1873–1889. doi: 10.1016/S0006-3495(04)74254-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wikstrom M, Verkhovsky MI, Hummer G. Biochim. Biophys. Acata. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- 44.Braun-Sand S, Burykin A, Chu ZT, Warshel A. J. Phys. Chem. B. 2005;109:583–592. doi: 10.1021/jp0465783. [DOI] [PubMed] [Google Scholar]
- 45.Wikstrom M. Curr. Opin. Struct. Biol. 1998;8:480. doi: 10.1016/s0959-440x(98)80127-9. [DOI] [PubMed] [Google Scholar]
- 46.Johannessón GH, Jónsson H. J. Chem. Phys. 2001;115:9644. [Google Scholar]
- 47.Fischer S, Karplus M. Chem. Phys. Lett. 1992;194:252. [Google Scholar]
- 48.Sagui C, Darden TA. Annu. Rev. Biophys. Biomol. Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- 49.Florian J, Warshel A. J. Phys. Chem. B. 1997;101:5583–5595. [Google Scholar]
- 50.Pomès R, Roux B. Biophys. J. 1998;75:33. doi: 10.1016/S0006-3495(98)77492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Im W, Berneche S, Roux B. J. Chem. Phys. 2001;114:2924. [Google Scholar]
- 52.Cui Q, Elstner M, Kaxiras E, Frauenheim T, Karplus M. J. Phys. Chem. B. 2001;105:569. [Google Scholar]
- 53.König PH, Hoffmann M, Frauenheim T, Cui Q. J. Phys. Chem. B. 2005 doi: 10.1021/jp0442347. in press. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Lee T, Yang W. J. Chem. Phys. 1999;110:46. [Google Scholar]
- 55.Zhang Y, Liu H, Yang W. J. Chem. Phys. 2000;112:3483. [Google Scholar]
- 56.Murphy RB, Philipp DM, Friesner RA. J. Comp. Chem. 2000;21:1442. [Google Scholar]
- 57.Antes I, Thiel W. J. Phys. Chem. A. 1999;103:9290. [Google Scholar]
- 58.Gao J, Amara P, Alhambra C, Field MJ. J. Phys. Chem. A. 1998;102:4714. [Google Scholar]
- 59.Elstner M, Porezag D, Jungnickel G, Elsner J, Haugk M, Frauenheim T, Suhai S, Seifert G. Phys. Rev. B. 1998;58:7260. [Google Scholar]
- 60.Elstner M, Frauenheim T, Suhai S. THEOCHEM. 2003;632:29. [Google Scholar]
- 61.Elstner M, Cui Q, Munih P, Kaxiras E, Frauenheim T, Karplus M. J. Comp. Chem. 2003;24:565. doi: 10.1002/jcc.10201. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X, Harrison DHT, Cui Q. J. Am. Chem. Soc. 2002;124:14871. doi: 10.1021/ja027063x. [DOI] [PubMed] [Google Scholar]
- 63.Formaneck MS, Li G, Zhang X, Cui Q. J. Theor. Comp. Chem. 2002;1:53. [Google Scholar]
- 64.Cui Q, Elstner M, Karplus M. J. Phys. Chem. B. 2002;106:2721. [Google Scholar]
- 65.Li G, Cui Q. J. Am. Chem. Soc. 2003;125:15028. doi: 10.1021/ja0367851. [DOI] [PubMed] [Google Scholar]
- 66.Li G, Zhang X, Cui Q. J. Phys. Chem. B. 2003;107:8643. [Google Scholar]
- 67.Li G, Cui Q. J. Phys. Chem. B. 2003;107:14521. [Google Scholar]
- 68.Elstner M, Jalkanen KJ, Knapp-Mohammady M, Frauenheim T, Suhai S. Chem. Phys. 2001;263:203–219. [Google Scholar]
- 69.Elstner M, Jalkanen KJ, Knapp-Mohammady M, Frauenheim T, Suhai S. Chem. Phys. 2000;256:15–27. [Google Scholar]
- 70.Hu H, Elstner M, Hermans J. Proteins: Struct. Funct. Gene. 2003;50:451–463. doi: 10.1002/prot.10279. [DOI] [PubMed] [Google Scholar]
- 71.Cui Q, Elstner M, Kaxiras E, Frauenheim T, Karplus M. J. Phys. Chem. B. 2001;105:569. [Google Scholar]
- 72.Smondyrev AM, Voth GA. Biophys. J. 2002;83:1987–1996. doi: 10.1016/S0006-3495(02)73960-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Annu. Rev. Phys. Chem. 2002;53:291. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 74.Shurki A, Warshel A. Adv. Prot. Chem. 2003;66:249. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]
- 75.Aaqvist J, Warshel A. Chem. Rev. 1993;93:2523. [Google Scholar]
- 76.Mo YR, Gao J. J. Comput. Chem. 2000;21:1458–1469. [Google Scholar]
- 77.Neria E, Karplus M. Chem. Phys. Lett. 1997;267:23. [Google Scholar]
- 78.Nam K, Prat-Resina X, Garcia-Viloca M, Devi-Kesavan LS, Gao J. J. Am. hem. Soc. 2004;126:1369–1376. doi: 10.1021/ja039093l. [DOI] [PubMed] [Google Scholar]
- 79.Marx D, Tuckerman ME, Hutter J, Parrinello M. Nature. 1999;397:601. [Google Scholar]
- 80.Eigen M. Angew. Chem. 1963;75:489–508. [Google Scholar]
- 81.Zundel G, Metzger H. Z. Phys. Chemie. 1968;58:225. [Google Scholar]
- 82.Elber R. J. Chem. Phys. 1990;93:4312–4321. [Google Scholar]
- 83.Schaefer P, Riccardi D, Cui Q. J. Chem. Phys. 2005 doi: 10.1063/1.1940047. In press. [DOI] [PubMed] [Google Scholar]
- 84.Nam K, Gao J, York DM. J. Chem. Theo. Comp. 2005;1:2–13. doi: 10.1021/ct049941i. [DOI] [PubMed] [Google Scholar]
- 85.D. Riccardi D, Schaefer P, Cui Q. J. Phys. Chem. B. 2005 doi: 10.1063/1.1940047. Submitted. [DOI] [PubMed] [Google Scholar]
- 86.Kuwajima S, Warshel A. J. Chem. Phys. 1988;89:3751. [Google Scholar]
- 87.Weber W, Hunenberger PH, McCammon JA. J. Phys. Chem. B. 2000;104:3668–3675. [Google Scholar]
- 88.Backman L. Eur. J. Biochem. 1981;120:257. doi: 10.1111/j.1432-1033.1981.tb05697.x. [DOI] [PubMed] [Google Scholar]
- 89.Silverman DN. Methods in Enzymology. 1995;249:479–503. doi: 10.1016/0076-6879(95)49046-9. [DOI] [PubMed] [Google Scholar]
- 90.Christianson DW, Fierke CA. Acc. Chem. Res. 1996;29:331. [Google Scholar]
- 91.Tu C, Silverman DN, Forsman C, Jonsson B-H, Lindskog S. Biochem. 1989;28:7913. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 92.Eriksson AE, Jones TA, Liljas A. Proteins: Struct., Funct., and Genet. 1988;4:274. doi: 10.1002/prot.340040406. [DOI] [PubMed] [Google Scholar]
- 93.Cui Q, Karplus M. J. Phys. Chem. B. 2003;107:1071. [Google Scholar]
- 94.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. of Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 95.Borgis D, Hynes JT. Chem. Phys. 1993;170:315–346. [Google Scholar]
- 96.Yarnell A. Chem. Eng. News. 2004;81:42. [Google Scholar]
- 97.Borgnia M, Nielsen S, Engel A, Agre P. Annu. Rev. Biochem. 1999;68:425. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 98.Tajkhorshid E, Nollert P, Jensen MO, Miercke LJW, O’Connell J, Stroud RM, Schulten K. Science. 2002;296:525. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- 99.de Groot BL, Grubmüller H. Science. 2001;294:2353. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 100.de Groot BL, Frigato T, Helms V, Grubmüller H. J. Mol. Biol. 2003;333:279. doi: 10.1016/j.jmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 101.Jensen MO, Tajkhorshid E, Schulten K. Biophys. J. 2003;85:2884. doi: 10.1016/S0006-3495(03)74711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Im W, Beglov D, Roux B. Comput. Phys. Comm. 1998;111:59–75. [Google Scholar]
- 103.Woo HJ, Dinner AR, Roux B. J. Chem. Phys. 2004;121:6392–6400. doi: 10.1063/1.1784436. [DOI] [PubMed] [Google Scholar]
- 104.Torrie GM, Valleau JP. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 105.Warshel A, Sussman F, King G. Biochemistry. 1986;25:8368–8372. doi: 10.1021/bi00374a006. [DOI] [PubMed] [Google Scholar]
- 106.Stroud RM, Miercke LJW, O’Connell J, Khademi S, Lee JK, Remis J, Harries W, Robles Y, Akhavan D. Curr. Opin. Struct. Biol. 2003;13:424. doi: 10.1016/s0959-440x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 107.Ilan B, Tajkhorshid E, Schulten K, Voth GA. Proteins. 2004;55:223. doi: 10.1002/prot.20038. [DOI] [PubMed] [Google Scholar]
- 108.Roux B, Karplus M. Annu. Rev. Biophys. Biomol. Struct. 1994;23:731–761. doi: 10.1146/annurev.bb.23.060194.003503. [DOI] [PubMed] [Google Scholar]
- 109.Nina M, Beglov D, Roux B. J. Phys. Chem. B. 1997;101:5239. [Google Scholar]
- 110.Stote RH, States DJ, Karplus M. J. Chim. Phys. 1991;88:2419–2433. [Google Scholar]
- 111.Lu D, Li Y, Rotkin SV, Ravaioli U, Schulten K. Nano Lett. 2004;4:2383. [Google Scholar]
- 112.Smart OS, Goodfellow JM, Wallace BA. Biophys. J. 2003;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hummer G, Rasaiah JC, Noworyta JP. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 114.Beckstein O, Sansom MSP. Proc. Natl. Acad. Sci. USA. 2003;100:7063–7068. doi: 10.1073/pnas.1136844100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Allen R, Melchionna S, Hansen JP. Phys. Rev. Lett. 2002;89:175502-1–175502-4. doi: 10.1103/PhysRevLett.89.175502. [DOI] [PubMed] [Google Scholar]
- 116.Cui Q. Theor. Chem. Acc. 2005 Submitted. [Google Scholar]