Abstract
microRNAs comprise a few percent of animal genes and have been recognized as important regulators of a diverse range of biological processes. Understanding the biological functions of miRNAs requires effective means to identify their targets. Combined efforts from computational prediction, miRNA over-expression or depletion, and biochemical purification have identified thousands of potential miRNA-target pairs in cells and organisms. Complementarity to the miRNA seed sequence appears to be a common principle in target recognition. Other features, including miRNA-target duplex stability, binding site accessibility, and local UTR structure might affect target recognition. Yet computational approaches using such contextual features have yielded largely nonoverlapping results and experimental assessment of their impact has been limited. Here, we compare two large sets of miRNA targets: targets identified using an improved Ago1 immunopurification method and targets identified among transcripts up-regulated after Ago1 depletion. We found surprisingly limited overlap between these sets. The two sets showed enrichment for target sites with different molecular, structural and functional properties. Intriguingly, we found a strong correlation between UTR length and other contextual features that distinguish the two groups. This finding was extended to all predicted microRNA targets. Distinct repression mechanisms could have evolved to regulate targets with different contextual features. This study reveals a complex relationship among different features in miRNA-target recognition and poses a new challenge for computational prediction.
Keywords: Argonaute, gene regulation, RISC complex
Animal genomes contain hundreds of microRNA genes (miRBase 13.0). Recent estimates suggest that miRNAs comprise ≈1% of genes in Drosophila and Caenorhabditis elegans and 2–3% of genes in mouse and human. To date, functional analysis in vivo has revealed biological roles for only a small fraction of these (1–3). One issue limiting progress in understanding the miRNA functions is identification of the target mRNAs that they regulate. Computational target identification is primarily based on sequence complementarity to the miRNA (reviewed in ref. 2), but many computational strategies also make use of sequence context to predict miRNA targets. Comparisons of different methods show limited overlap among the predicted targets, although those that place more emphasis on pairing to the seed sequence at the 5′ end of the miRNA tend to produce similar results. Most of these methods identify many possible targets for each miRNA, often hundreds (e.g., refs. 4–9).
A growing body of experimental evidence shows that miRNAs can regulate many targets. Overexpression of miRNAs in heterologous cell types can affect the levels of hundreds of mRNAs with target sites (e.g., ref. 10). Conversely, depletion of miRNAs can lead to increased levels of a comparable number of target mRNAs (11, 12). Changes in target RNA stability can result from miRNA-induced deadenylation of the mRNA (13–15). mRNA up-regulation, combined with target prediction has helped to identify biologically relevant targets of specific miRNAs (e.g., ref. 16). Whole proteome analyses have shown that miRNA induced changes in protein expression correlate with changes in mRNA level, in trend if not in magnitude (17, 18). Yet, there are well-documented instances of miRNA-mediated regulation at the protein level that do not involve changes in mRNA level (14, 17, 18). Therefore, methods to identify targets by miRNA-induced changes in expression profile can only tell part of the story. This highlights the need for alternative means to identify miRNA targets.
One such alternative involves identification of microRNA targets by virtue of their physical association with miRNA-containing ribonucleoprotein complexes (19–24). In ref. 19, we reported a method based on Ago1 immunopurification (IP) that proved to be effective. Eleven new targets were identified for miR-1, including some that had not been predicted. Although the specificity was high, with all new targets experimentally validated, the method had limited sensitivity, identifying ≈1/10th of the expected number of targets. Here, we present an improved Ago1 IP protocol, which permits identification of hundreds of potential miRNA targets, and compare the contextual features of targets identified by IP to the targets destabilized at the mRNA level upon Ago1 depletion.
Results
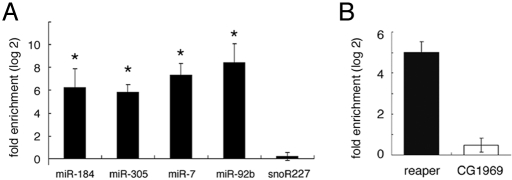
In an effort to improve the sensitivity of miRNA IP, with minimal loss of specificity, we tested a variety of antibody concentrations, incubation times and wash conditions (Fig. S1). Sensitivity was assessed by quantitative PCR to monitor miRNA levels (over a broad range of abundance: miR-184 comprises 17% of S2 cell miRNA; miR-305: 1.5%; miR-7: 0.1%; miR-92b: 0.01%; (25). The 4 miRNAs were enriched >50-fold in IP from cells expressing HA-tagged Ago1 compared with control cells not expressing the transgene (Fig. 1A, *, P < 0.05). The small nucleolar RNA (snoR227) showed no enrichment (log2 = 0.18). Thus, the IP protocol can recover miRNAs over a broad range of expression levels without enriching for unrelated small RNAs.
Fig. 1.
miRNP immunopurification. (A) Enrichment of miRNAs measured by Q-PCR. y axis: fold enrichment (log2 scale). Splicing RNA U27was used for normalization and snoR227 as a control. Results represent 3 independent IP experiments. (B) Enrichment of a known miR-2a target reaper by IP. Data normalized to rp49 for 3 independent experiments (t test, P < 0.05).
The association between miRNAs and targets must be stable for IP to be useful. To assess this association, we measured the recovery of reaper mRNA, a known target of miR-2a. The miR-2 family comprises >13% of S2 cell miRNAs. reaper mRNA was enriched ≈32-fold by IP. A control lacking known miRNA binding sites was not enriched (Fig. 1B). This compares favorably with the 2- to 3-fold enrichment of reaper obtained in the study in ref. 19.
Expression Profiling of mRNA Associated with Ago1.
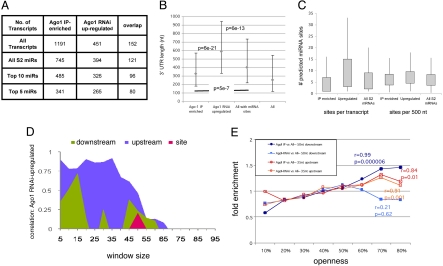
To examine the population of mRNAs associated with Ago1 complexes, total RNA was recovered by IP with anti-HA from S2 cells expressing HA-Ago1 and from untransfected control cells. After 2 rounds of linear amplification the RNA was used to generate probes for expression profiling. Approximately 7,000 transcripts can be detected reliably by expression profiling in S2 cells (11). A total of 1,621 mRNAs were significantly IP enriched with a P value of <0.05. Of these, 1,191 were enriched >1.4-fold, 464 were enriched >2-fold and 89 were enriched >4-fold (Fig. S2A). A considerable fraction of S2 cell transcripts are associated with Ago1 complexes, although for most the degree of enrichment is low in magnitude.
S2 cells miRNAs fall into 26 seed families with distinct target specificities (Table S1). The 5 and 10 most abundant seed families comprise 73% and 88% of S2 cell miRNA. IP-enriched targets for all S2 miRNAs were slightly more abundant than S2 cell transcripts in general (P < 1 × 10−10; Table S2 and Fig. S2B). IP-enriched targets for the top 5 and top 10 seed families were also slightly more abundant (P = 0.02; P = 3.5 × 10−6). Although statistically significant, these differences are small in magnitude and should not pose a concern for use of IP for target identification.
We next compared IP using the Ago1 transgene with IP of endogenous Ago1 using monoclonal anti-Ago1 [kindly provided by M. Siomi (Keio University School of Medicine, Tokyo, Japan)]. Enrichment was determined relative to a control IP with monoclonal anti-Myc after normalization to rp49. Sixteen targets were assayed by quantitative RT-PCR (Q-PCR). All showed significant enrichment, with a strong correlation between the two methods (Fig. S1C, correlation coefficient r = 0.71, P = 0.0018). Thus, the results are largely independent of the antibody used.
Experimental Validation of Target Enrichment.
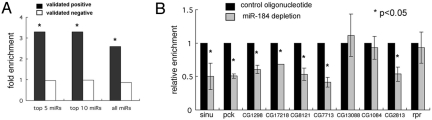
The performance of Ago1 IP was tested with reference to a nonredundant set of experimentally validated miRNA-target pairs, consisting of 67 validated positives and 29 validated negatives (Table S3). More targets were IP-enriched than expected after normalization for the number of transcripts in each category (Fig. 2A). Enrichment was >3-fold for targets of the 5 and 10 most abundant miRNA seed families (20 positives/489 IP-enriched with sites for the top 10 miRNAs; P = 6.0 × 10−5). The enrichment was slightly lower when all S2 cell miRNAs were considered (23 positives/745 IP-enriched transcripts with sites for all miRNAs), but remained significant (P = 3.6e-4). There was no enrichment for the 29 miRNA-target pairs that tested negative experimentally. For comparison with the analysis of reaper (Fig. 1B), the average magnitude of IP enrichment was 5.5-fold for all validated positives. Enrichment was greatest for abundant miRNAs (Fig. S1D). It is noteworthy that some validated targets were not enriched by IP. Different factors may contribute to this. Some in vivo validated pairs might not be functional in S2 cells. Among the validated targets for top 5 miRNAs that were not IP enriched, 28% were up-regulated at the RNA level upon Ago1 depletion (Table S3). Target turnover may also affect recovery.
Fig. 2.
Transcripts associated with Ago1 complexes. (A) Comparison of IP results with experimentally validated miRNA targets. Fold enrichment of the nonredundant target set (Table S3) for the 5 and 10 most abundant miRNA families and for all S2 miRNA families. y axis: enrichment calculated as the number of validated targets IP-enriched divided by the number not IP-enriched after normalization for transcript number in each category. P values: Fisher's exact two-tailed test. (B) Effect of miR-184 depletion on the recovery of predicted targets. y axis: Relative IP enrichment calculated by normalizing IP fold enrichment in miR-184 depleted cells to control cells. Transcript levels quantified by Q-PCR (normalized to rp49) and compared with control IP with empty beads. reaper lacks miR-184 sites and serves as a control. *, P < 0.05, Student's t test.
Antisense oligonucleotides were used to deplete specific miRNAs in S2 cells to ask if target recovery depends on the miRNA. miR-184 was chosen because many of its predicted targets were enriched by IP (Table S4). Cells were treated with anti-miR-184 or with a scrambled sequence control and subjected to IP with anti-Ago1. Nine of the thirty-two miR-184 targets were assayed by Q-PCR. IP recovery of 7 was lower in miR-184 depleted cells (Fig. 2B), indicating that binding to miR-184 contributes to IP enrichment. The partial reduction in recovery likely reflects incomplete depletion of miR-184. Other miRNAs might also contribute: 8/9 have predicted sites for miRNAs other than miR-184, including sites for 4 of the top 10 seed families. IP enrichment of specific targets can be attributed to association with a specific miRNA.
Additional Validation of New miRNA Targets Identified by IP.
From the set of computationally predicted miR-184 targets, we selected 17 that were IP enriched and 17 that were not. Transcript levels were measured by Q-PCR after miR-184 depletion. Twelve of seventeen IP enriched targets were up-regulated in cells depleted of miR-184 (P < 0.05; Fig. S3A and Table 1). Control mRNAs were not affected. For comparison, only 2 of a set of 17 miR-184 targets that were not IP-enriched were up-regulated in miR-184 depleted cells. Luciferase reporter assays were also used to assess miRNA-mediated regulation of the set of IP enriched miR-184 targets. Five of the twelve targets up-regulated on miR-184 depletion have been validated (7). We tested one of these as a positive control, 5 other up-regulated transcripts and 3 from the IP, but not up-regulated set (Table 1). Control UTRs lacking mir-184 sites were unaffected by miR-184 depletion. The 6 up-regulated transcripts showed modest but significant (P < 0.05) increases in luciferase activity on miR-184 depletion, but the other 3 IP positive transcripts did not (Fig. S3B).
Table 1.
Summary of IP target validation
| Predicted miR-184 target | IP enriched | IP miR-184 dependent | Up-regulated miR-184 depletion | UTR reporter assay* |
|---|---|---|---|---|
| sinu | + | + | + | +, K |
| pck | + | + | + | K |
| CG1298 | + | + | + | K |
| CG8121 | + | + | + | + |
| CG7713 | + | + | + | + |
| CG17218 | + | + | + | |
| CG2813 | + | + | + | |
| CG6583 | + | + | K | |
| CG9796 | + | + | + | |
| CG10620 | + | + | + | |
| CG13088 | + | − | + | + |
| CG1084 | + | − | + | K |
| CG6965 | + | − | − | |
| CG8010 | + | − | − | |
| CG3446 | + | − | − | |
| CG11059 | + | − | ||
| CG15154 | + | − |
Listed are the 17 genes with predicted miR-184 site that are enriched in the Ago1-IP microarray study with a minimum cutoff > 1.4 fold, P < 0.05. Seven out of 9 genes tested showed the dependence of IP enrichment on miR-184 (marked ″+″, blank = not tested). Twelve genes showed mRNA up-regulation upon miR-184 depletion (marked ″+″). Tested-positive and nonresponsive luciferase reporters in S2 cells are shown in column 5. *, K indicates UTR validation in ref. 7.
Table 1 summarizes tests used to classify the predicted miR-184 targets recovered by IP. Those up-regulated at the RNA level in miR-184 depleted cells were also shown to be regulated in 3′ UTR reporter assays. Five of seventeen IP enriched transcripts were not regulated at the RNA level. The 3 tested by 3′ UTR reporter assay were not up-regulated in miR-184 depleted cells. It may be that these were enriched by IP due to other miRNAs (all contain at least one site for other S2 miRNAs). Or, they might represent Ago1 associated RNAs that are not miRNA targets. In practice, transcripts that are selected by IP and up-regulated upon miRNA depletion can be considered high-confidence targets.
It is worth noting that 11 of 12 transcripts that were up-regulated upon miRNA depletion had at least 1 evolutionarily conserved target site, as did the 2 up-regulated mRNAs that were not recovered by IP. This distinguishes them from the 5 IP enriched transcripts that were not up-regulated on miR-184 depletion, in which target sites are not conserved. Evolutionary selection for function may favor sites that promote destabilization of the target RNA as a consequence of miRNA binding, as noted for targets downregulated at both mRNA and protein level by miRNA overexpression (10, 17). These correlations do not imply that nonconserved sites are not functional, but suggest differences in how they are regulated.
What Types of miRNA Target Sites Are Enriched by IP?
A growing body of evidence indicates that pairing to residues 2–8 of the miRNA, the “seed,” makes an important contribution to target site function (4–6, 10, 17, 18, 26, 27). Yet, some experimentally validated target sites include mismatches or G:U base pairs in the seed region (27–32).
The frequency of occurrence of particular types of sites was examined for IP enriched transcripts. These include 8-mer seed matches, which are sufficient to confer robust regulation, 7-mer seed matches which confer weak regulation unless supplemented by additional pairing to the miRNA 3′ end, and 6-mer matches that require additional base pairing (27). G:U base pairs and mismatches were also examined. For the top 5 miRNA seed families (73% of all miRNA reads), we observed statistically significant enrichment for 8-mer and 7-mer matches beginning in positions 1 or 2; and 6-mer matches beginning in position 2 (Fig. 3A; *, P < 0.05). Four of these were enriched for top 10 seed families (88% of miRNAs); three were enriched when the analysis was extended to all S2 cell miRNAs. There was no enrichment for seeds with G:U pairing or mismatches.
Fig. 3.
Features of IP enriched mRNAs. (A) miRNA target seed type enrichment analysis. “8-mer, 7-mer or 6-mer” indicates the length of the seed match. “Position” indicates the first paired position from the miRNA 5′ end. y axis: fold enrichment calculated as % sites of each type in IP-enriched transcripts divided by % of matching sites in nonenriched transcripts. P values: Fisher's Exact Test (*, P < 0.05). (B) Assessment of site openness. Surface plot showing Pearson correlation coefficient (y axis) vs. sequence window size upstream and downstream of miRNA sites (x axis, nucleotides). The correlation coefficient measures the correlation between IP enrichment and degree of local nucleotide openness, binned from 10% to 80%. IP fold enrichment is calculated as percentages of all miRNA sites in IP-enriched transcripts divided by percentages of all miRNA sites in all S2 cell transcripts for each bin (9). The red triangle indicates the correlation coefficient for openness of the miRNA site (arbitrarily placed at 50 nt on the x axis); upstream, sequences 5′ of the site; downstream, sequences 3′ of the site. Enrichment and correlation coefficients were calculated for windows from 5 to 95 nt (Table S6 and Table S7).
The presence of perfect 8-mer, 7-mer and 6-mer seed matches improved the chance that a transcript was recovered by IP. The degree of enrichment decreased as the set of miRNAs increased, perhaps because most miRNAs are expressed at low levels. Enrichment depends on the proportion of possible matches to each seed that is recovered by IP. Low abundance miRNAs can contribute less to the quantity of targets recovered by IP, but their inclusion increases the number of different seeds from 10 to 26 and so affects the degree of enrichment for each seed.
Other Features of IP Enriched Targets.
To be recovered by IP, target RNAs must remain bound to the Ago1 complex. The hybrid energy of base pairing between miRNA and target should contribute to the stability of binding. Table S5 compares the free energy of miRNA-target base pairing for transcripts enriched by IP with those not enriched (ΔGhybrid as in ref. 9). The distribution of binding energies was significantly shifted toward more stable duplexes in IP-enriched transcripts (P = 0.005 using a two-sample Kolmogorov–Smirnov test). The correlation between binding energy ΔGhybrid and degree of enrichment in the IP set was modest (r = −0.60), on the borderline of statistical significance (P = 0.0503). On this basis, IP does not appear to select strongly for more stable target site binding.
Local secondary structure in the UTR might affect function by making a miRNA site more or less accessible. Previous studies have developed ways to predict site “openness” and suggested that this could improve computational target prediction (7, 9, 33). We found a significant shift toward greater site openness (9) in IP-enriched targets compared with those not enriched (Kolmogorov–Smirnov two-sample, P = 4 × 10−6; Table S6). The degree of enrichment for openness correlated well with increasing site accessibility (r = 0.865, P = 0.006). A comparable correlation was found when all S2 mRNAs were used as the control set (r = 0.868, P = 0.005).
To examine the impact of nearby sequences, we tested windows from 5 to 95 nt in length upstream and downstream of the site (Table S7). Fig. 3B presents a surface plot showing the correlation coefficient between degree of openness and enrichment in the IP set compared with all S2 transcripts. Upstream of the site, the correlations were significant for windows of up to 50 nt. Downstream, the correlations were significant for windows from 10 to 70 nt with an optimum at 50 nt (Table S7 and Fig. 3B). A previous analysis using a different miRNP protein in C. elegans (AIN-IP, GW182) found that a 25-nt upstream window had the best correlation with IP enrichment, but did not find evidence for downstream openness (9). These studies may have sampled different miRNP complexes, since antibodies to different RISC components were used, but species differences in UTR composition cannot be ruled out.
Comparison of Targets Identified by Ago1 IP and Ago1 Depletion.
Expression profiling of cells depleted of miRNA function is gaining acceptance as a means of identifying potential targets (e.g., refs. 11, 12, 16, and 18). In this context, we asked whether similar target features are selected for by IP and Ago1 depletion. A priori we expect them to be similar, because both reflect activity of Ago1 complexes. To our surprise, we found <1/3 overlap in the RNAs identified by the two methods (Fig. 4A and Table S8 lists all transcript data). Recent reports using Ago-2 to pull down miRNA targets in mammalian cells over-expressing a miRNA also showed limited overlap with targets downregulated at the mRNA level (20, 23). To explore the basis for these differences we examined the two groups for distinct structural and molecular features, using all S2 mRNAs as a common control set.
Fig. 4.
A comparative analysis on Ago1 IP-enriched transcripts Vs Ago1 RNAi-up-regulated transcripts. (A) Numbers of mRNAs with target sites identified by IP or by Ago1 RNAi. IP-enriched used a cutoff >1.4-fold and P < 0.05. Ago1 RNAi set from (12): up-regulated >1.5-fold (P < 0.05). All: all S2 cell mRNAs; all S2 miRNA: those with sites for any S2 miRNA. (B) y axis: median UTR length (nt) represented by gray squares. The range between 1st and 3rd quartiles are indicated by bars. P values: Wilcoxon U test, Bonferroni corrected (4 tests per group). The median for the Ago1 RNAi set is 262 nt longer than the IP set (P = 6e-21) and 187 nt longer than for all RNAs with sites (P = 6e-13). The IP set was 75 nt shorter than all RNAs with sites (P = 5 × 10−7). (C) Site density profiles in IP-enriched vs. Ago1 RNAi-up-regulated targets and all S2 miRNAs. The up-regulated set has more sites/transcript than the IP set (median shift 4, P = 2 × 10−18) or than all RNAs with sites (median shift 3, P = 1 × 10−14). Median site density was 6.9/500 nt in the up-regulated set vs. 6.0 for the IP set and 5.6 for all RNAs. These differences were significant using a K-S test (two tail). (D) Assessment of site openness for the Ago1 RNAi up-regulated set vs. all RNAs with sites (as in Fig. 3B). (E) Fold enrichment for the optimal upstream and downstream windows in IP-enriched and Ago1 RNAi up-regulated sets. x axis: average nucleotide openness binned from 10% to 80%. y axis: fold enrichment as compared with all S2 transcripts with sites, which were used as the common control. Pearson's correlation coefficient, r values and associated P values are shown.
There was little difference in seed types enriched by the two methods (Fig. S4A). However, sites with stronger binding energy were under-represented in the up-regulated transcript set (P = 0.01, cumulative distribution plot in Fig. S4B). There was a significant anti-correlation between more stable hybrids and degree of enrichment in the up-regulated transcript set (r = 0.82, P = 0.004). Thus, Ago1 RNAi up-regulated transcripts contained energetically weaker binding sites. This contrasts with the evidence of marginal selection for stronger sites in the IP set (r = −0.60, P = 0.0503).
The presence of multiple sites should increase the stability of binding. The number of sites increases with UTR length (e.g., ref. 4), so it might be expected that IP would select for long UTRs. Instead, we found RNAs with shorter UTRs enriched in the IP set (Fig. 4B). There were fewer sites in IP enriched UTRs, but this difference disappeared when data were normalized for UTR length (P = 0.1; Fig. 4C). UTRs were longer in the Ago1 RNAi up-regulated set and site density was higher compared with the IP set (Fig. 4 B and C; P = 0.01) or all transcripts (P = 4.5 × 10−7). Although it is not evident why IP enriches for shorter UTRs, it has been suggested that sites located close together act synergistically to promote target destabilization (17, 18, 34). This could explain why the up-regulated transcript set is enriched for longer UTRs with a higher site density.
A more striking difference was found between the IP and up-regulated sets in the degree of openness of target sites and flanking sequences. In contrast to the IP set, there was no correlation between binding site openness and enrichment in the up-regulated transcript set (r = 0.2, P = 0.62; Table S6). Fig. 4D shows the correlation coefficients for the binding site and for upstream and downstream openness at various window sizes. Upstream of the site, the correlations were statistically significant for windows of up to 35 nt, but there was limited correlation for downstream openness (Table S7; compare with Fig. 3B). Fig. 4E shows the enrichment for openness of upstream and downstream optimal sequence windows for the IP and up-regulated sets, compared with all S2 RNAs. The difference between these two groups is striking for downstream sequence openness.
Functional Clustering Suggests Distinct Biological Functions in the Two Target Groups.
To explore what types of functions these groups of transcripts encode, we analyzed Gene Ontology annotations (http://david.abcc.ncifcrf.gov). Functional annotation clustering results for the 745 transcripts enriched in Ago1 IP, the 394 up-regulated by Ago1 RNAi and the 121 enriched in both groups are shown in Table S9 (transcripts lacking predicted S2 miRNA sites were excluded). Interestingly, the IP-enriched transcripts were over-represented for functions related to lipid, glucose and amino acid metabolism, mitochondrial function, and other enzymatic activities. The top clusters in the Ago1 RNAi up-regulated group involved signal transduction, cell differentiation, morphogenesis and other developmental processes. Bearing in mind that S2 cells are thought to be macrophage-like in their origin, the transcripts downregulated at the RNA level by miRNAs may be those encoding functions that could be detrimental in this cell type (4), whereas those reflected in the IP-enriched set might be targets whose activity levels are more finely tuned by miRNAs. It will be of interest to see whether a comparable segregation of functional classes is observed in datasets from tissue specific miRNAs in vivo.
Genome-Wide Analysis Shows miRNA Targets with Distinct Structural and Molecular Properties.
Our findings indicated that shorter UTR length was correlated with greater site and flanking sequence openness in the Ago1 IP-enriched transcript set, whereas longer UTRs with higher site density were enriched in the set up-regulated by Ago1 depletion. This prompted us to ask whether the two methods were revealing an underlying difference in overall UTR composition.
To address this we performed a similar analysis for target site density and openness on UTRs of all predicted miRNA targets. A clear trend of increasing miRNA site density as a function of UTR length was found, as reported in ref. 4. UTRs of 100–300 nt and 300–500 nt had lower site densities than the median of all UTRs with sites (Fig. S5A). For UTRs >500 nt, site density was higher than the median of all UTRs with sites. The increasing site density gradually flattened for UTRs >1Kb in length. The differences were statistically significant.
Intriguingly, the local structural properties flanking miRNA sites were also different for targets with short and long UTRs. As observed for the IP set, downstream sequences of all UTRs ranging from 100 nt to 700 nt showed greater accessibility (Fig. S5B and Table S10). The correlation dropped off sharply at >700–900 nt and longer UTRs in the 900- to 1,100-nt group became depleted for openness. Table S10 shows analysis of the openness of upstream sequences and the binding site with similar results.
These observations indicate that at least two features of miRNA target sites change as a function of the length of the UTR in which they are found. First, site density increases with UTR length, to a maximum level of ≈5/100 nt for all miRNAs. Second, the accessibility of sites measured by sequence openness is considerably higher in short UTRs.
Discussion
This study provides evidence that miRNP IP is a useful means to identify microRNA targets. Intriguingly, targets identified by IP differ in structural and molecular properties from the group identified by virtue of their effects on target mRNA stability after Ago1 depletion. We have found that this reflects an underlying genome-wide difference in site context as a function of UTR length. IP selects for sites with a greater degree of local structural openness around the binding site, which tend to be found in shorter UTRs. In contrast, transcripts up-regulated after Ago1 depletion do not appear to require favorable binding energy or local UTR accessibility, but are selected for higher site density, which tends to occur in longer UTRs. Many miRNA targets are known to be regulated primarily at mRNA level, others mainly at the protein level, yet it is not clear how the specificity is achieved. These findings raise the question of whether differences in the mode of target regulation might be reflected in the differences in UTR features reported here.
Looking beyond the impact that these features have on biochemical target site identification, these differences presumably reflect the impact of selection for target site function during evolution. Short UTRs tend to have fewer sites. Our findings suggest that there has been selection for greater accessibility of these sites. Selection against sequence changes that would allow for local mRNA base pairing would promote openness of the site and flanking sequences. This might facilitate interaction with the RISC complex. At the longer end of the UTR spectrum there appears to have been selection for more sites, flattening off at ≈5 sites per 100 nt for all miRNAs. Higher density might allow for more effective regulation by cooperation between different miRNAs or by having multiple sites for one miRNA in a UTR. Experimental evidence for synergistic regulation by multiple miRNAs is noted in refs. 17, 18, 26, 27, and 34.
Longer 3′UTRs have been shown to be preferentially targeted for regulation by adenine/uridine-rich element binding proteins in Drosophila (AUBPs, 35). AUBPs destabilize mRNAs by recruiting RNA degradation enzymes. Interestingly, ≈20% of Ago1 RNAi up-regulated RNAs have conserved AU-rich elements, a 2-fold over-representation (P = 7e-12) vs. 6.7% of the IP set (under-representation P = 0.02). It is tempting to speculate a fraction of Ago1 RNAi up-regulated targets might be influenced by both AUBPs and miRNA machineries. These findings contribute to an emerging picture of interplay between AUBPs and miRNA regulation (36, 37).
The differences in contextual features associated with target sites as a function of UTR length pose a new challenge for improving computational approaches to target prediction. Approaches that favor site context, overall binding energy or conservation are likely to favor one part of the UTR target spectrum over another. A systematic analysis of the interactions between contextual features, including synergistic regulation and structural openness, could improve the accuracy of prediction.
Materials and Methods
Immunopurification of Ago1 from S2 cells is described in Fig. S1A and SI Materials and Methods. UTR luciferase reporter assays were performed as described in refs. 27 and 38 with modifications described in SI Materials and Methods. miRNA and mRNA quantitative PCR was performed as described in SI Materials and Methods. Expression profiling was performed by the EMBL Gene Core using Affymetrix Drosophila 2.0 arrays (see SI Materials and Methods). miRNA target site prediction and statistical analysis is detailed in SI Materials and Methods. David bioinformatic tools were used for gene ontology. Functional annotation clustering was applied as described in ref. 39.
Supplementary Material
Acknowledgments.
We thank V. Benes and T. Bähr-Ivacevic of the EMBL GeneCore for expression profiling; S. Lim and E. Loeser for technical support; T. Sandmann for help with microarray and bioinformatic analysis; and J. Varghese, W. Ge, P. Verma for helpful suggestions. Work in the S.M.C. laboratory was supported by European Union Sixth Framework Programme Grant “Sirocco” LSHG-CT-2006-037900 and the Singapore Millennium Foundation via core funding to Temasek Life Sciences Laboratory. Work in the V.A. laboratory was supported by National Institutes of Health Grants F32GM087039 (to M.H.) and GM34028 and GM066826 (to V.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908149106/DCSupplemental.
References
- 1.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 8.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammell M, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Rehwinkel J, et al. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eulalio A, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 14.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. Rna. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 18.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. Rna. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karginov FV, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landthaler M, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. Rna. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nahvi A, Shoemaker CJ, Green R. An expanded seed sequence definition accounts for full regulation of the hid 3′ UTR by bantam miRNA. Rna. 2009;15:814–822. doi: 10.1261/rna.1565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon DJ, et al. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long D, et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 34.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cairrao F, Halees AS, Khabar KS, Morello D, Vanzo N. AU-rich elements regulate Drosophila gene expression. Mol Cell Biol. 2009;29:2636–2643. doi: 10.1128/MCB.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: Is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 39.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.