Abstract
Nucleic acid polymerases catalyze the formation of DNA or RNA from nucleoside-triphosphate precursors. Amino acid residues in the active site of polymerases are thought to contribute only indirectly to catalysis by serving as ligands for the two divalent cations required for activity or substrate binding. Two proton transfer reactions are necessary for polymerase-catalyzed nucleotidyl transfer: deprotonation of the 3′-hydroxyl nucleophile and protonation of the pyrophosphate leaving group. Using model enzymes representing all four classes of nucleic acid polymerases, we show that the proton donor to pyrophosphate is an active site amino acid residue. The use of general acid catalysis by polymerases extends the mechanism of nucleotidyl transfer beyond that of the well-established two-metal-ion mechanism. The existence of an active-site residue that regulates polymerase catalysis may permit manipulation of viral polymerase replication speed and/or fidelity for virus attenuation and vaccine development.
Nucleic acid polymerases catalyze the formation of DNA or RNA from 2′-deoxyribonucleotides or ribonucleotides, respectively. Polymerases are therefore required for the reproduction, maintenance and expression of the genomes of all organisms, including viruses. Nucleotidyl transfer, the chemical reaction catalyzed by polymerases is illustrated in Fig. 1. Nucleophilic attack on the α-phosphorous atom of the (2′-deoxy)ribonucleoside triphosphate by the primer 3′-hydroxyl leads to formation of a phosphodiester bond and release of pyrophosphate. All polymerases require two divalent cations, usually Mg2+, for activity and employ a two-metal-ion mechanism for nucleotidyl transfer (Fig. 1) 1,2. Metal A lowers the pKa of the primer 3′-hydroxyl, thus facilitating deprotonation of this moiety for in-line nucleophilic attack 3. Metal B orients the triphosphate for catalysis, stabilizes the negative charge that arises during formation of the pentavalent transition state and has been suggested to assist pyrophosphate release 1,3. Although it is clear that deprotonation of the primer 3′-hydroxyl is required for catalysis 4,5, only recently has it been suggested that protonation of the pyrophosphate leaving group occurs prior to its release from the enzyme 4. The acceptor of the 3′-hydroxyl proton and donor of pyrophosphate proton are not known. It is now quite clear that chemistry can be at least partially rate limiting for nucleotide addition by all classes of nucleic acid polymerases 4,6, as well as serve as a fidelity checkpoint 6-8. Identification of the acceptor and donor for these proton-transfer reactions may inspire new mechanistic hypotheses for how catalytic efficiency can be tuned by the nature (correct versus incorrect) of the bound nucleotide.
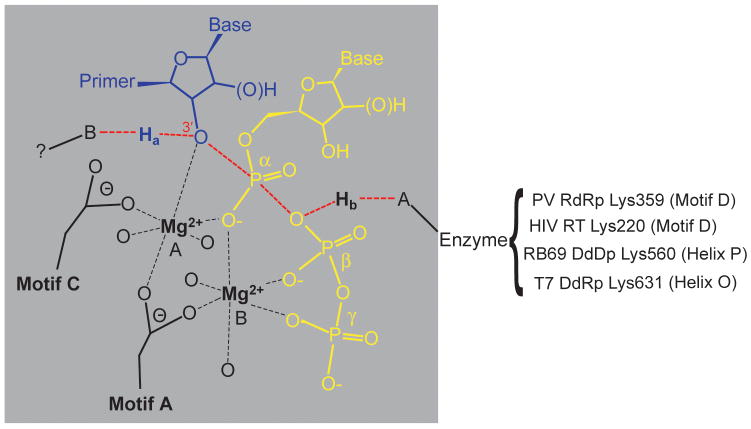
Figure 1. Extending the two-metal-ion mechanism of nucleotidyl transfer to include general acid catalysis.
Nucleoside triphosphate (yellow) enters the active site with a divalent cation (Mg2+, metal B). This metal ion is coordinated by the phosphates of the nucleotide, an Asp residue located in structural motif A of all polymerases, and likely water molecules (indicated as oxygen ligands to metal without specific designation). Metal B orients the triphosphate in the active site and may contribute to charge neutralization during catalysis. A second divalent cation binds (Mg2+, metal A) that is coordinated by the 3′-OH of primer terminus (blue), the nucleotide α-phosphate, as well as Asp residues of structural motifs A and C. Metal A lowers the pKa of the 3′-OH facilitating deprotonation and subsequent nucleophilic attack at physiological pH. As the transition state of nucleotidyl transfer is approached (indicated by dashed red lines), the primer 3′-OH proton, Ha, is transferred to an unidentified base (B), and we propose that the pyrophosphate leaving group is protonated (Hb) by a basic amino acid on the enzyme. The positions of the candidate general acids of the model polymerases employed in this study are shown.
Four classes of template-dependent nucleic acid polymerases exist: RNA-dependent RNA polymerase (RdRp); RNA/DNA-dependent DNA polymerase, the so-called reverse transcriptase (RT); DNA-dependent DNA polymerase (DdDp); and DNA-dependent RNA polymerase (DdRp). In this study, we have employed the RdRp from poliovirus (PV), the RT from human immunodeficiency virus type 1 (HIV), the DdDp from bacteriophage RB69, and the DdRp from bacteriophage T7 as representatives of the four classes of polymerases because of the wealth of mechanistic and/or structural information available for these enzymes 9-21. The objective of this study was to identify the proton donor to the pyrophosphate leaving group, as analysis of high-resolution structures of several polymerases poised for or undergoing catalysis implicated a basic amino acid residue rather than a water molecule as the proton donor. In addition, analysis of the pH dependence of nucleotide addition by PV RdRp was consistent with catalysis being dependent on a residue with a pKa value of 10.5 4.
Results
A general acid in nucleic acid polymerase catalysis
Analysis of the structural model for the RdRp from Norwalk virus (NV) in complex with primed template RNA and nucleotide showed that Lys374 of conserved structural motif D was located in the vicinity of the triphosphate moiety of incoming nucleotide (Supplementary Figs. 1a,b) 22. Structural motif D is conserved between RdRps and RTs and has no defined catalytic function 23. Analysis of structure-based sequence alignments showed only one conserved lysine residue in motif D of RdRps and RTs, including the telomere RT, telomerase (TERT) (Supplementary Fig. 1c). These sequence alignments identified Lys359 of PV RdRp and Lys220 of HIV RT in conserved structural motif D as candidates for the proton donors in these systems (Supplementary Fig. 1c). Lys219 of HIV RT was ruled out as the putative general acid because of the absence of this residue in the RT from human immunodeficiency virus type 2 (HIV-2) (Supplementary Fig. 1c). It is well established that the DNA polymerase activity of HIV-2 RT is on par with that observed for HIV RT 24. Unlike Lys374 of NV RdRp, Lys359 of PV RdRp and Lys220 of HIV RT were not oriented in a position to interact readily with the triphosphate moiety of the incoming nucleotide (Fig. 2). Structural motif D is one of the most dynamic elements of the palm subdomain of RdRps and RTs, with the position of this motif varying by as much as 6 Å when structures are compared (Supplementary Fig. 1b). Solution of the structure of HIV RT in complex with primed template DNA and nucleotide required crosslinking of the enzyme to DNA that could have influenced the orientation of motif D 14. This possibility is supported by a recently published model for the TERT elongation complex that positions the motif-D lysine in a position to serve as a general acid (Fig. 2) 25. Therefore, only biochemical studies could test the possibility that the conserved lysine in motif D functions as a general acid. Identification of candidates for the general acid in DNA-dependent DNA and RNA polymerases was more straightforward. Structural models show clearly that Lys560 in helix P of RB69 DdDp, a B-family polymerase 12, and Lys631 in helix O of T7 DdRp, an A-family polymerase 21, are positioned to serve as proton donors (Fig. 2) for these enzymes.
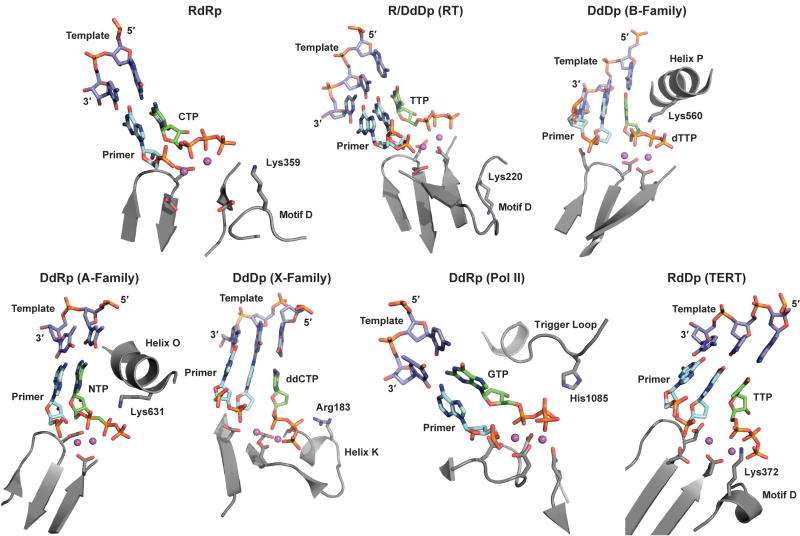
Figure 2. Interactions of NTP in the active sites of various polymerase families.
In all ternary complexes, the NTP (green C-atoms), primer/template (cyan and purple C-atoms, respectively), and key active site residues (grey C-atoms) are shown as sticks; the metal ions are displayed as spheres colored in magenta. RdRp: The RdRp from PV (pdb 1RA6) 19 with primer/template and CTP bound to the active site in the presence of two Mn2+ ions. Primer-template, CTP and Mn2+ ions were extracted from the structural complex of Norovirus (NV) polymerase (pdb 3BS0) 22 and modeled into the active site of PV RdRp. R/DdDp (RT): HIV RT with TTP bound to the active site in the presence of two Mg2+ ions (pdb 1RTD) 14. DdDp (B-Family): The RB69 DNA polymerase with the dTTP nucleotide bound in the presence of Ca2+ ions (pdb 1IG9) 12. DdRp (A-Family): The T7 RNA polymerase with the modified nucleotide, αβ-methylene-ATP bound in the presence of Mg2+ (pdb 1S76) 21. DdDp (X-Family): Pol β polymerase with the ddCTP bound to the active site in the presence of Mg2+ ions (pdb 1BPY) 50. DdRp (Pol II): Multi-subunit RNA polymerase II from yeast (pdb 2E2H) 31 in complex with nucleic acids and GTP substrate in the presence of Mg2+. RdDp (TERT): The catalytic subunit of telomerase with TTP bound to the active site in the presence of two Mg2+ ions (pdb 3DU5) 25. Primer-template, TTP and Mg2+ ions were extracted from the structural complex of HIV RT (1RTD) 14 and modeled into the active site of TERT as described previously 25. Residues equivalent to Lys359 of PV RdRp predicted to function as general acids, are labeled. The designations of the structural elements on which the general acid is located are also indicated explicitly.
We produced derivatives of each of the polymerases described above that contained a leucine residue instead of the candidate lysine proton donor (Supplementary Methods). Leucine was selected instead of alanine to prevent water from binding to the site and serving as a proton donor (Supplementary Fig. 2). For all polymerases tested, the maximal rate constant for nucleotide incorporation (kpol) was reduced by 50-2000 fold for the Leu derivative relative to the wild-type, lysine-containing (wt) enzyme (Table 1). This observation is consistent with a role for the lysine in the rate-limiting step for nucleotide incorporation. For all of the wt polymerases employed here, chemistry is at least partially rate limiting 4. With the exception of the leucine derivative of T7 DdRp, the kpol values derive from experiments that included a saturating nucleotide concentration (5×Kd,app) (Supplementary Methods). In the case of the leucine derivative of T7 DdRp, the highest concentration of nucleotide attainable was 2×Kd,app (Supplementary Methods). This circumstance increases the error on the kpol value measured; however, our ability to reach conclusions for the leucine derivative is not affected because the observed 100-fold reduction in kpol value for this derivative is much greater than the error of the measurement.
Table 1. Kinetic analysis of PV RdRp, HIV-1 RT, RB69 DdDp and T7 DdRp supports general acid catalysis in nucleotidyl transfer.
| Parameter Measured | PV RdRp | HIV RT | RB69 DdDp | T7 DdRp | ||||
|---|---|---|---|---|---|---|---|---|
| WT | K359L | WT | K220L | WT | K560L | WT | K631L | |
| kpol (s−1) | 50 ± 5 | 1 ± 0.1 | 60 ± 5 | 0.3 ± 0.1 | 200 ± 10 | 0.10 ± 0.01 | 60 ± 5 | 0.6 ± 0.1 |
| Kd,app (μM)a | 200 ± 20 | 700 ± 80 | 7 ± 1 | 5 ± 2 | 40 ± 5 | 1000 ± 100 | 300 ± 30 | 5.0 ± 1.0 × 104c |
| SDKIEb | 3.0 ± 0.3 | 2.5 ± 0.3 | 2.2 ± 0.4 | 1.8 ± 0.4 | 4.2 ± 0.2 | 1.8 ± 0.2 | 5.2 ± 0.5 | 2.6 ± 0.5c |
| PId | 2 | 1 | 2 | 1 | 2 | 1 | 2 | nde |
Kd,app is for (d)ATP (Supplementary Methods).
SDKIE is solvent deuterium kinetic isotope effect 28, calculated as kobs in H2O/kobs in D2O at saturating [(d)ATP].
Kd,app, kpol and SDKIE values listed for T7 K631L were obtained by using data collected with 80 mM ATP, a sub-saturating concentration.
PI is proton inventory 29, calculated from a plot of kn/k0 as a function of n. The data were fit to a modified Gross-Butler equation for either a two-proton-transfer model (eq. 1) or for a one-proton-transfer model (eq. 2). The value reported is the proton-transfer-model that best fits the data.
not determined
A substantial difference was not observed in the apparent dissociation constant (Kd,app) for the nucleotide substrate measured for the leucine derivatives of PV RdRp or HIV RT (Table 1). This observation is consistent with residues in conserved, structural motif (F) of these enzymes functioning independently in triphosphate binding 26. In contrast, the Kd,app value for nucleotide substrate measured for the leucine derivatives of RB69 DdDp and T7 DdRp increased 25-fold and 167-fold, respectively (Table 1). This observation is consistent with structural studies which show that these lysines interact with the nucleotide substrate (Fig. 2) 18,20.
Unlike many other polymerases, the stability of PV RdRp on its nucleic acid primer-template is not affected by pH, permitting the pH dependence of the kinetics of nucleotide incorporation to be interpreted (Supplementary Methods) 4. Consistent with Lys359 of PV RdRp functioning as a proton donor during nucleotidyl transfer, the descending limb of the pH rate profile observed for wt PV RdRp (Fig. 3a) 4 was lost for the K359L derivative (Fig. 3a). The ionization observed for the K359L derivative likely reflects deprotonation of the 3′-hydroxyl (Fig. 3a). Theoretical studies of nucleotidyl transfer by rat DNA polymerase beta have suggested that the primer 3′-hydroxyl has a pKa in the 8-9.5 regime 7. Because the pKa values measured are kinetic pKa values, the lack of equivalence in the pKa value for the K359L derivative to any measured for wt enzyme likely reflects a change in the rate-determining step for nucleotidyl transfer 4. Chemistry should be solely rate limiting for the K359L derivative whereas a conformational-change step and chemistry are equally rate limiting for wt enzyme 10.
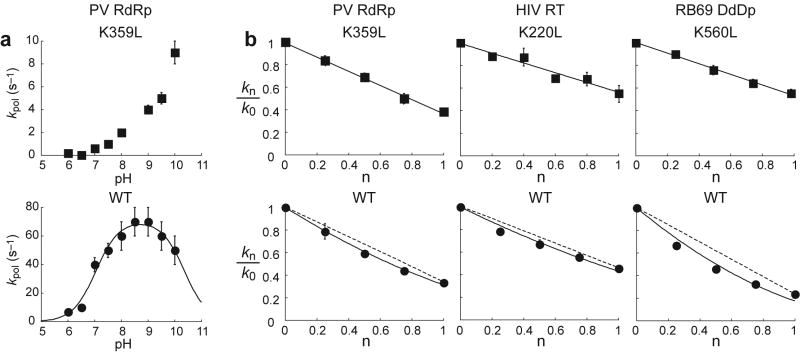
Figure 3. A conserved basic amino acid in the active site of multiple classes of polymerases participates directly in nucleotidyl transfer and functions as a general acid.
(a) Values for kpol plotted as a function of pH for K359L PV RdRp and WT PV RdRp. Kinetic data were obtained for AMP incorporation into S/S-+1 using the stopped-flow assay 4 in the presence of 5 mM MgCl2 (Supplementary Methods). In the case of WT PV RdRp, the solid line shows the fit of the data to a model describing two ionizable groups, Eq. 3 (Supplementary Methods), yielding pKa values of 7.0 ± 0.1 and 10.5 ± 0.1 4. Error bars indicate the standard deviation. Plots of the logkpol as a function of pH are provided as supplemental information (Supplementary Fig. 3). (b) Proton inventory plots for PV RdRp, HIV RT and RB69 DdDp with either the leucine derivative or lysine-containing polymerase (Supplementary Methods). Values for kn/k0 were plotted as a function of n. n is the mole fraction of D2O, kn is the observed rate constant for nucleotide incorporation in a reaction containing a given mole fraction of D2O and k0 is the observed rate constant for nucleotide incorporation in H2O. The solid lines represent the fit of the data to a one-proton-transfer model for the leucine derivative (filled squares) and to a two-proton-transfer model for the wild-type polymerase (filled circles) (Supplementary Methods). The dashed lines represent the predicted line for a one-proton-transfer model. Each data point represents the average of two or three independent experiments. Error bars indicate the standard deviation. The standard deviation was <10% in all cases. The panels for wt enzymes are adapted from Castro et al (2007) 4; more rigorous analysis of the two-proton model is reported therein.
Loss of the general acid leads to single proton-transfer
Theoretical studies of the free energy landscape for phosphoryl transfer almost always show proton transfer in the transition state 27 that would manifest experimentally as a solvent deuterium kinetic isotope effect 4. The solvent deuterium kinetic isotope effect is defined as the ratio of kpol values obtained when the reaction is performed in H2O relative to values obtained when the reaction is performed in D2O (Supplementary Methods) 28. The observation of a solvent deuterium kinetic isotope effect supports a proton-transfer reaction representing one of the microscopic steps reflected in the macroscopic rate constant, kpol. Each wt polymerase exhibited a solvent deuterium kinetic isotope effect of 2-5 (Table 1). This observation is consistent with chemistry contributing to the rate-limiting step(s) reported by the nucleotide-incorporation assay as observed for the wt enzymes 4. Interestingly, the solvent deuterium kinetic isotope effect measured for each leucine derivative was smaller than observed for the corresponding wt enzyme (Table 1). This observation could be interpreted in one of two ways. First, it is possible that the extent to which chemistry limits nucleotide addition by the leucine derivatives is less than occurs for the wt enzymes. Second, it is possible that fewer protons are being transferred. This latter possibility would be consistent with the candidate lysine residues contributing one proton transfer during nucleotide incorporation.
In order to count the number of protons being transferred during the nucleotidyl-transfer reaction, we performed a proton-inventory experiment (Supplementary Methods) 4,29. This experiment obtains kpol values in the presence of different mole fractions of D2O (kn). The ratio of kn/k0 (k0 is kpol in H2O) is plotted as a function of mole fraction of D2O (n). If a single proton is transferred during the reaction, then the data should fit to a line. This experiment was performed for all leucine derivatives except for T7 DdRp derivative, which could not be saturated with nucleotide. The data obtained for all of the leucine derivatives evaluated fit well to a straight line (Fig. 3b), consistent with a single proton transfer reaction occurring during nucleotidyl transfer for these enzymes. In contrast, the data for the corresponding wt enzymes failed to fall on a line defined by kn/k0 values at n=0 and n=1, consistent with a model for more than one proton transfer reaction occurring during nucleotidyl transfer (Fig. 3b). More extensive analysis of the proton-inventory experiment for the wt enzymes, including statistical analysis, was reported previously, convincingly demonstrating two-proton transfer reactions in the transition state for nucleotidyl transfer by the wt enzymes employed here 4. These data provide compelling evidence for the use of an active-site amino acid residue as a general acid to protonate the pyrophosphate leaving group to facilitate nucleotidyl transfer.
General acid catalysis in the presence of Mn2+
All of the experiments described to this point have been performed in the presence of Mg2+ because this divalent cation is considered to be the biologically relevant cofactor. In the presence of Mg2+, chemistry is only partially rate limiting for PV RdRp at pH 7.5 10. In addition, the extent to which chemistry contributes to the observed rate constant for nucleotide addition changes as a function of pH 4. In the presence of Mn2+, however, chemistry is the primary determinant of the rate constant for nucleotide addition by PV RdRp from pH 6.0 to pH 9.0 4. Unfortunately, experiments cannot be performed above pH 9.0 in the presence of Mn2+ due to precipitation of the metal hydroxide.
We evaluated the activity of the K359L derivative of PV RdRp in the presence of Mn2+ (Supplementary Fig. 4). The observed rate constant for nucleotide incorporation was reduced by 10 fold relative to wt enzyme to 1 s−1 without a substantial change in the apparent dissociation constant for nucleotide (Supplementary Fig. 4a). The phosphorothioate effect observed for the K359L derivative (6 ± 1) was essentially the same as measured for wt enzyme (7 ± 1), consistent with chemistry remaining as the rate-limiting step (Supplementary Fig. 4a). The solvent deuterium kinetic isotope effect observed for the K359L derivative of PV RdRp was 3 ± 1, a value that is more than two fold lower than the value of 7 ± 1 observed for wt enzyme (Supplementary Fig. 4a). These data are consistent with the loss of a proton-transfer reaction as observed in the presence of Mg2+. As expected, the dependence of the rate constant for nucleotide incorporation on pH was now essentially identical within the experimentally accessible pH range (Supplementary Fig. 4a). These data are consistent with chemistry serving as the rate-limiting step for both the K359L derivative and the wt enzyme in the presence of Mn2+. The reduced rate constant for nucleotide incorporation and solvent deuterium kinetic isotope effect observed for the K359L derivative relative to wt enzyme provide additional support for Lys359 being employed as a general acid. We conclude that PV RdRp, and likely the other polymerases, employ general acid catalysis regardless of the divalent cation cofactor employed.
Arg and His substitute for Lys as the general acid
In order to test further the use of a general acid for nucleotidyl transfer, we produced a PV RdRp derivative with Lys359 changed to arginine. In addition to testing our model for general acid catalysis, the ability of an arginine to function at this position as a general acid would be consistent with the use of arginine as a general acid in X-family polymerases like DNA polymerase beta (polβ) (Fig. 2) 30. An arginine at this position should produce a derivative that functions better than the K359L derivative but still worse than wt enzyme, exhibits unique pH dependence relative to wt enzyme and shows two proton-transfer reactions during nucleotidyl transfer. At pH 7.5, the K359R derivative bound nucleotide well and turned over ∼3-fold faster than the K359L derivative and 20-fold slower than wt enzyme (Fig. 4a). At pH 10, the kpol value for the K359R derivative had risen to within 2-fold of the maximum value observed for wt enzyme at pH 8.5 and continued to rise (Fig. 4b). In contrast, the value for the wt enzyme was falling at pH 10 (Fig. 4b). This difference can be explained by the ΔpKa value of 2 between lysine and arginine. The descending limb of the pH rate profile for the K359R derivative should appear at pH values greater than 11 (dashed line in Fig. 4b). Unfortunately, experiments cannot be performed at pH values higher than 10 because of the insolubility of metal hydroxide. Finally, the solvent deuterium kinetic isotope effect was 3.4 ± 0.1, essentially the same as wt enzyme (Table 1) and proton inventory data fit well to a two proton model (Fig. 4c). These data strongly support the use of a general acid for nucleotidyl transfer and suggest that both lysine and arginine can serve this function.
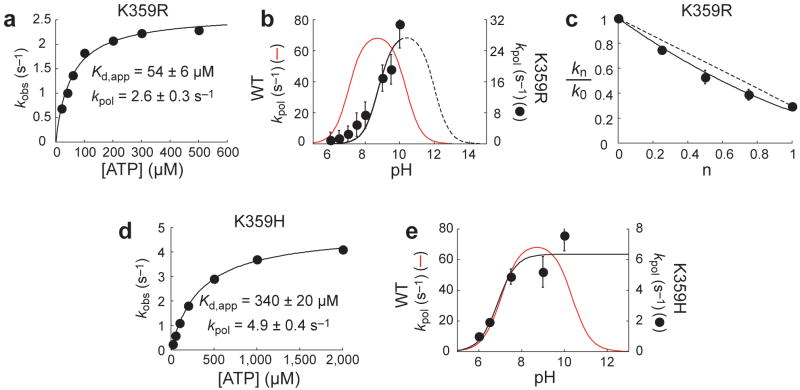
Figure 4. Altering nucleotidyl transfer kinetics by changing the amino acid employed as the general acid.
(a) Lys359 of PV RdRp was changed to Arg. Kinetic analysis of AMP incorporation by K359R PV RdRp at pH 7.5 fit to a hyperbola with a Kd,app for ATP of 54 ± 6 μM and a kpol of 2.6 ± 0.1 s−1. (b) pH rate profile for K359R PV RdRp fit to a model describing one-ionizable group, yielding a pKa value of 8.8 ± 0.3. The dashed line shows the predicted curve for a second ionizable group with a pKa of 12.0. The red line shows the curve of kpol as a function of pH for WT PV RdRp with pKa values of 7.0 and 10.5. Error bars indicate the standard deviation. (c) Proton inventory plot for K359R PV RdRp fit to a two-proton-transfer model. The dashed line represents the predicted line for a one-proton-transfer model. Each data point represents the average of two independent experiments. Error bars indicate the standard deviation. The standard deviation was <10%. (d) Lys359 of PV RdRp was changed to His. Kinetic analysis of AMP incorporation by K359H PV RdRp at pH 7.5 fit to a hyperbola with a Kd,app for ATP of 340 ± 20 μM and a kpol of 4.9 ± 0.4 s−1. (e) pH rate profile for K359H PV RdRp fit to a model describing one-ionizable group, yielding a pKa value of 6.9 ± 0.3. The red line shows the curve of kpol as a function of pH for WT PV RdRp with pKa values of 7.0 and 10.5. Error bars indicate the standard deviation.
Multi-subunit RNA polymerases contain a histidine in the trigger loop that may serve as the general acid for these enzymes (Fig. 2) 31. Histidine can function at this position of the PV RdRp (Fig. 4d). A 10-fold reduction in the kpol value was observed relative to wt enzyme with only a modest (50%) increase in the Kd,app value for nucleotide at pH 7.5 (Fig. 4d). The pKa value for the ascending limb of the pH rate profile for the K359H derivative was identical to wt enzyme; the descending limb was absent, consistent with a pKa value of 6-7 for histidine (Fig. 4e).
Discussion
We conclude that all four classes of nucleic acid polymerases employ general acid catalysis for nucleotidyl transfer. Importantly, the general acid is not absolutely essential but provides a 50-2000-fold rate enhancement depending upon the polymerase evaluated. The value of the pKa for pyrophosphate in the polymerase active site is not known. The absence of an absolute requirement for a general acid in polymerase-catalyzed nucleotidyl transfer suggests that the value of the pKa for the tetraanionic form of pyrophosphate is not sufficiently high to preclude it from serving as a leaving group. The differences in rate enhancement conferred by the general acid for the different polymerases may reflect the extent to which the general acid also contributes to the overall neutralization of the negative charge in the active site. RdRps and RTs have evolved an independent structural motif (F) to bind the triphosphate 26. In contrast, the other polymerase families have the general acid connected directly (helix O, helix K, trigger loop) or indirectly (helix P) to the structural elements that bind the triphosphate (Fig. 2). There is no doubt that neutralization of the negative charge that forms during the transition state will facilitate nucleotidyl transfer and likely also contributes to the reduced catalytic efficiency of the leucine derivatives 12,32,33.
Protonation of the pyrophosphate leaving group in the mechanism of nucleotidyl transfer has been essentially ignored until recently. Most structures of polymerases poised for or undergoing catalysis reveal an interaction between a basic amino acid of the polymerase and the beta phosphate of the nucleotide substrate (Fig. 2) that has been interpreted as a nucleotide binding determinant. However, the Kornberg laboratory proposed that the interaction between the beta phosphate of the incoming nucleotide and His1085 (trigger loop) of yeast RNA polymerase II could link substrate recognition to catalysis if this histidine served as a general acid for protonation of the pyrophosphate leaving group 31. Our recent studies of nucleotidyl transfer confirmed that protonation of the pyrophosphate leaving group does occur 4, thus encouraging the identification of the proton donor.
Nature has employed all three basic amino acids as the general acid (Fig. 2). The residue chosen may reflect the need to achieve a balance between the rate of polymerization and the fidelity of nucleotide incorporation. Enzymes that employ a lysine as a general acid, for example PV RdRp, HIV RT, RB69 DdDp and T7 DdRp elongate nucleic acids with rate constants on the order of 100 s−1 (Table 1). In contrast, rat DNA polymerase beta (Arg as general acid) and yeast RNA polymerase II (His as a general acid) synthesize nucleic acid an order of magnitude slower at physiological pH 8,30,34. That a difference in the general acid leads to changes in elongation rate is supported by the reduced rate of RNA synthesis observed for the K359R and K359H derivatives of PV RdRp (Fig. 4). In addition, changing His1085 of yeast RNA polymerase II to Tyr reduces the rate of catalysis by an order of magnitude without changing the observed affinity for the nucleotide substrate 34. Our study would suggest that the reduced rate of catalysis for the H1085Y mutant at physiological pH likely reflects the increased pKa value for the tyrosine hydroxyl proton relative to the histidine imino proton.
It has often been suggested that polymerase translocation occurs concomitant with or after pyrophosphate release 21,35. The work of Beese and co-workers has provided the best structural description of a polymerase undergoing catalysis by using a fragment of DNA polymerase I from Bacillus stearothermopholis, an A-family polymerase 36. In this system, movement of helix O is coupled to catalysis and translocation. Importantly, the general acid of all polymerases appears to be associated with exceptionally dynamic structural elements (Supplementary Fig. 1 and Supplementary Fig. 5), and the movement of these elements have been implicated in translocation 12,18,21,23,31,34,36. It is possible that the protonation-deprotonation cycle of the general acid during catalysis toggles helix O (and its structural equivalents) between the closed (protonated) and open (deprotonated) states, thus driving the translocation reaction. Consistent with this possibility is the observation that changing the general acid to Leu leads to substantial reductions in processivity for all nucleic acid polymerases employed in this study (Supplementary Fig. 6).
The use of a general acid by polymerases for nucleotidyl transfer has important implications for reaction reversal, pyrophosphorolysis. Because pyrophosphate in solution should be protonated, deprotonation of pyrophosphate should not occur readily for enzymes that employ lysine or arginine as a general acid as these residues should be rapidly reprotonated by solvent after pyrophosphate release. In contrast, enzymes like the multi-subunit RNA polymerases that employ a histidine should catalyze pyrophosphorolysis more efficiently, an observation that has been reported 37-39. The presence of histidine in these polymerases may have been an early solution to deal with the problem of polymerase arrest caused by backtracking, misincorporation, or template damage that is now dealt with by using editing factors 40.
Finally, it is worth noting that, where studied, the element harboring the general acid has been shown to contribute not only to incorporation speed but also incorporation fidelity 41-46. RNA virus pathogenesis and virulence are closely linked to replication speed and fidelity 47,48. It has recently been shown that a PV mutant encoding an RdRp with increased fidelity is attenuated 47,48, and this attenuated virus serves as a very effective vaccine strain 49. The identification of a single amino acid in the polymerase active site that can tune replication speed and perhaps modulate incorporation fidelity suggests the provocative hypothesis that a universal strategy for viral attenuation may exist that can be applied to the rational design of virus vaccine strains.
Methods
Materials
All general experimental materials, buffers, salts, etc., were of the highest grade available from Sigma, Fisher or VWR. A complete list of any specialty reagents employed is provided (Supplementary Methods).
Construction, expression and purification of polymerases and their derivatives
We constructed derivatives of all polymerases: PV RdRp, RB69 DdDp, T7 DdRp and HIV RT by using standard recombinant DNA protocols as described in the methods section provided as supplemental information. Briefly, we mutated DNA sequences by using the polymerase chain reaction. Forward and reverse primers employed for amplification were selected based on the presence of unique restriction sites suitable for subcloning of the mutated DNA fragment into the expression plasmid for the wild-type polymerase.
We expressed in E. coli and purified all polymerases as described previously 4. Briefly, we transformed cells with the appropriate plasmid and used these cells to produce an inoculum for large-scale growth. We induced gene expression during exponential growth by addition of isopropyl-β-D-thiogalactopyranoside. We lysed induced cells in appropriate buffers and we purified the enzymes to apparent homogeneity by using standard column chromatography resins and protocols. Details are provided (Supplementary Methods).
Nucleotide incorporation experiments
We performed nucleotide incorporation experiments as described previously 4. In general, we formed elongation complexes in the appropriate buffer by incubating polymerase with the appropriate primed template followed by rapid mixing with nucleoside triphosphate substrate, generally ATP. Rapid mixing was performed by using either a chemical quench flow (CQF) or stopped flow (SF) instrument (both from KinTek Corp., Austin, TX). For the CQF experiments, we employed 32P-labeled primers. We monitored primer extension by phosphorimaging of polyacrylamide gels. For the SF experiments, we employed templates containing 2-aminopurine ribonucleoside monophosphate on the 5′ side of the templating nucleotide. Primer extension causes a fluorescence change that could be monitored by using the SF instrument. All details, including modifications, are provided in the methods section provided (Supplementary Methods).
Solvent deuterium kinetic isotope effect (SDKIE) and proton inventory (PI) experiments
We performed SDKIE and PI experiments by monitoring pre-steady state nucleotide incorporation either using the chemical quench flow or stopped-flow instruments. We prepared enzymes, substrates and buffers in 100% water or 100% D2O followed by mixing at the appropriate ratio to obtain 0, 25, 50, 75 or 100% D2O. Deuterated glycerol was used in all solutions in D2O. The pD was used instead of pH for the solutions in D2O and was adjusted according to pD = pH + 0.4. SDKIE values were calculated as the ratio of kpol values obtained in H2O divided by that obtained in D2O. PI plots consisted of kn/kH20 as a function of n where kn is the observed rate constant for nucleotide incorporation at a particular mole fraction of D2O, kH2O is the observed rate constant for nucleotide incorporation in H2O and n is the solvent mole fraction of D2O. Proton-inventory data were fit to the modified Gross-Butler equation for a two-proton-transfer model 29:
| (1) |
or for a one-proton-transfer model 29:
| (2) |
where kn is the observed rate constant at the different percentages of D2O, kH2O is the observed rate constant in water, n is the mole fraction of D2O and Φ is the inverse of the isotope effect for each ionizable group.
Data analysis
Observed rate constants (kobs) for nucleotidyl transfer at various concentrations of nucleotide were obtained by fitting product-versus-time data to an equation defining a single exponential. Values for Kd,app and kpol were obtained by fitting kobs-versus-[NTP] data to an equation defining a hyperbola. Data were fit by nonlinear regression using the program KaleidaGraph (Synergy Software, Reading, PA). Specific equations employed are provided (Supplementary Methods).
Supplementary Material
Acknowledgments
We thank Stephen J. Benkovic, Philip C. Bevilacqua, J. Martin Bollinger, Katsuhiko Murakami, Kevin D. Raney and Joseph C. Reese for comments on the manuscript. This study was supported by a grant (AI45818) from NIH to CEC.
Footnotes
Author contributions C.E.C., J.J.A., C.C. and E.D.S designed research; C.C., E.D.S., K.R.M., J.J.A., I.M. and A.U. performed research; M.G. and W.K. contributed new reagents/analytic tools; C.E.C., C.C., E.D.S. and J.J.A. analyzed data; and C.E.C., E.D.S., C.C. and J.J.A. wrote the paper.
References
- 1.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–2. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 2.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Fothergill M, Goodman MF, Petruska J, Warshel A. Structure-energy analysis of the role of metal ions in phosphodiester bond hydrolysis by DNA Polymerase I. J Am Chem Soc. 1995;117:11619–11627. [Google Scholar]
- 4.Castro C, et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:4267–72. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florian J, Goodman MF, Warshel A. Computer simulation of the chemical catalysis of DNA polymerases: discriminating between alternative nucleotide insertion mechanisms for T7 DNA polymerase. J Am Chem Soc. 2003;125:8163–77. doi: 10.1021/ja028997o. [DOI] [PubMed] [Google Scholar]
- 6.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–6. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 7.Florian J, Goodman MF, Warshel A. Computer simulations of protein functions: searching for the molecular origin of the replication fidelity of DNA polymerases. Proc Natl Acad Sci U S A. 2005;102:6819–24. doi: 10.1073/pnas.0408173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sucato CA, et al. DNA polymerase beta fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry. 2008;47:870–9. doi: 10.1021/bi7014162. [DOI] [PubMed] [Google Scholar]
- 9.Anand VS, Patel SS. Transient state kinetics of transcription elongation by T7 RNA polymerase. J Biol Chem. 2006;281:35677–85. doi: 10.1074/jbc.M608180200. [DOI] [PubMed] [Google Scholar]
- 10.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–37. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer-Orta C, et al. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci U S A. 2007;104:9463–8. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–67. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 13.Gohara DW, Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry. 2004;43:5149–58. doi: 10.1021/bi035429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–75. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 15.Pomerantz RT, Temiakov D, Anikin M, Vassylyev DG, McAllister WT. A mechanism of nucleotide misincorporation during transcription due to template-strand misalignment. Mol Cell. 2006;24:245–55. doi: 10.1016/j.molcel.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarafianos SG, et al. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. Embo J. 2002;21:6614–24. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–93. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Temiakov D, et al. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–91. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 19.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. Embo J. 2004;23:3462–71. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–34. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 21.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 22.Zamyatkin DF, et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283:7705–12. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 23.Canard B, Chowdhury K, Sarfati R, Doublie S, Richardson CC. The motif D loop of human immunodeficiency virus type 1 reverse transcriptase is critical for nucleoside 5′-triphosphate selectivity. J Biol Chem. 1999;274:35768–76. doi: 10.1074/jbc.274.50.35768. [DOI] [PubMed] [Google Scholar]
- 24.Hizi A, Tal R, Shaharabany M, Loya S. Catalytic properties of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J Biol Chem. 1991;266:6230–9. [PubMed] [Google Scholar]
- 25.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–7. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 26.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–56. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosta E, Kamerlin SC, Warshel A. On the interpretation of the observed linear free energy relationship in phosphate hydrolysis: a thorough computational study of phosphate diester hydrolysis in solution. Biochemistry. 2008;47:3725–35. doi: 10.1021/bi702106m. [DOI] [PubMed] [Google Scholar]
- 28.Schowen RL. Mechanistic deductions from solvent isotope effects. Progr Phys Org Chem. 1972;9:275–332. [Google Scholar]
- 29.Venkatasubban KS, Schowen RL. The proton inventory technique. CRC Crit Rev Biochem. 1984;17:1–44. doi: 10.3109/10409238409110268. [DOI] [PubMed] [Google Scholar]
- 30.Kraynov VS, Showalter AK, Liu J, Zhong X, Tsai MD. DNA polymerase beta: contributions of template-positioning and dNTP triphosphate-binding residues to catalysis and fidelity. Biochemistry. 2000;39:16008–15. doi: 10.1021/bi0008480. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–54. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang Y, Oelschlaeger P, Florian J, Goodman MF, Warshel A. Simulating the effect of DNA polymerase mutations on transition-state energetics and fidelity: evaluating amino acid group contribution and allosteric coupling for ionized residues in human pol beta. Biochemistry. 2006;45:7036–48. doi: 10.1021/bi060147o. [DOI] [PubMed] [Google Scholar]
- 33.Yang G, Lin T, Karam J, Konigsberg WH. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry. 1999;38:8094–101. doi: 10.1021/bi990653w. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–56. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchand B, Gotte M. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J Biol Chem. 2003;278:35362–72. doi: 10.1074/jbc.M304262200. [DOI] [PubMed] [Google Scholar]
- 36.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci U S A. 2003;100:3895–900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erie DA, Yager TD, von Hippel PH. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- 38.Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci U S A. 1994;91:8057–61. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Hawley DK. Identification of a 3′-->5′ exonuclease activity associated with human RNA polymerase II. Proc Natl Acad Sci U S A. 1993;90:843–7. doi: 10.1073/pnas.90.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–73. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 41.Bebenek A, et al. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics. 2002;162:1003–18. doi: 10.1093/genetics/162.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll SS, Cowart M, Benkovic SJ. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991;30:804–13. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 43.Johnson VA, et al. Update of the Drug Resistance Mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–8. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 44.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. Embo J. 1995;14:4609–21. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki M, Yoshida S, Adman ET, Blank A, Loeb LA. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J Biol Chem. 2000;275:32728–35. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, et al. The L561A substitution in the nascent base-pair binding pocket of RB69 DNA polymerase reduces base discrimination. Biochemistry. 2006;45:2211–20. doi: 10.1021/bi052099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–16. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–8. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–61. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 50.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–15. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.