Abstract
PDZ-RhoGEF is a member of the regulator of G protein signaling (RGS) domain-containing RhoGEFs (RGS-RhoGEFs) that link activated heterotrimeric G protein α subunits of the G12 family to activation of the small GTPase RhoA. Unique among the RGS-RhoGEFs, PDZ-RhoGEF contains a short sequence that localizes the protein to the actin cytoskeleton. In this report, we demonstrate that the actin-binding domain, located between amino acids 561–585, directly binds to F-actin in vitro. Extensive mutagenesis identifies isoleucine 568, isoleucine 569, phenylalanine 572, and glutamic acid 573 as necessary for binding to actin and for co-localization with the actin cytoskeleton in cells. These results define a novel actin-binding sequence in PDZ-RhoGEF with a critical amino acid motif of IIxxFE. Moreover, sequence analysis identifies a similar actin-binding motif in the N-terminus of the RhoGEF frabin, and, as with PDZ-RhoGEF, mutagenesis and actin interaction experiments demonstrate a motif of LIxxFE, consisting of the key amino acids leucine 23, isoleucine 24, phenylalanine 27, and glutamic acid 28. Taken together, results with PDZ-RhoGEF and frabin identify a novel actin binding sequence. Lastly, inducible dimerization of the actin-binding region of PDZ-RhoGEF revealed a dimerization-dependent actin bundling activity in vitro. PDZ-RhoGEF exists in cells as a dimer, raising the possibility that PDZ-RhoGEF could influence actin structure independent of its ability to activate RhoA.
Keywords: G protein, Rho, guanine-nucleotide exchange factor, actin, localization
Rho family GTPases are members of the Ras super family of monomeric G-proteins (1). The Rho family comprises six small GTPase subfamilies including Rho, Rac, Cdc42, Rnd, RhoBTB and RhoT/Miro (2). Out of these, the most well studied members include Rho (A, B and C), Rac (1 and 2) and Cdc42 proteins. Their roles in cell regulation include modulation of cytoskeletal structure, motility, cell division, gene transcription, vesicular transport and various enzymatic activities. As key regulators of the actin cytoskeleton, in fibroblasts, RhoA induces the formation of actin stress fibers and focal adhesions, Rac1 stimulates the protrusion of lamellipodia and membrane ruffles and Cdc42 promotes extension of filopodia and actin microspikes (3–5). Rho GTPases are found in all eukaryotic cells and so far twenty-two mammalian genes encoding Rho GTPases have been described (6).
Rho GTPases act as molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state. They are converted from an inactive (GDP-bound) state to an active state (GTP-bound) by guanine nucleotide exchange factors (GEFs) that function downstream of ligand bound integrins, growth factor receptors and heterotrimeric G-protein coupled receptors (GPCRs). The RhoGEFs are the best understood regulators of Rho activation in response to upstream stimuli (7), and so far about 70 RhoGEFs have been identified in the human genome (8, 9). The RhoGEFs are characterized by tandem Dbl homology (DH) and pleckstrin homology (PH) domains. In addition to the signature DH-PH domains, the RhoGEFs also contain a variety of other signaling domains that mediate interaction with numerous proteins resulting in a multitude of events.
Unique among the large family of RhoGEFs is a subfamily that contains a regulator of G protein signaling (RGS ) domain and is hence named RGS-RhoGEF. The RGS-RhoGEFs in mammals consist of p115-RhoGEF, PDZ-RhoGEF (PRG) and leukemia-associated Rho-GEF (LARG). These form an important group of proteins as they provide a direct link for activation of RhoA by cell surface receptors coupled to heterotrimeric G-proteins. Further, these exchange factors are specific for RhoA and do not activate Rac or Cdc42 (10–14). In addition to their RGS, DH and PH domains, which are characteristic of all three mammalian RhoGEFs, PRG and LARG contain an N-terminal PDZ domain.
PDZ domains of PRG and LARG have been shown to interact with plexins B1 and B2, LPA receptors (LPA1 and LPA2), insulin-like growth factor (IGF-1) receptor, and light chain 2 (LC2) of microtubule-associated protein (MAP) (14–19). Moreover, PRG and LARG are phosphorylated on tyrosine residues by focal adhesion kinase (FAK) in response to GPCR agonist stimulation of G proteins (20), and it has been shown that FAK, PRG and ROCK II cooperate to induce Rho/ROCK II dependent focal adhesion movement and trailing-edge retraction in response to LPA in fibroblasts (21). p115-RhoGEF, PRG and LARG have also been shown to form homo- and hetero-oligomers, and the C-terminal regions of the proteins are involved in their oligomerization (22, 23).
PDZ-RhoGEF (PRG), alternatively known as KIAA0380, ArhGEF11 or GTRAP48, is unique among the other RhoGEFs, LARG and p115Rho-GEF. While p115-RhoGEF and LARG are substantially activated by Gα13, PRG displays little or no activation of its Rho exchange activity when combined with Gα13 in vitro (24–27). PRG, a predominatly brain specific Rho-GEF (28), is able to induce neurite retraction in Neuro 2a cells. It has been shown that PRG promotes cell rounding and an increase in cortical actin in Swiss 3T3 cells (12), whereas, p115-RhoGEF induces stress fibers but not cortical actin reorganization and cell rounding.
Previously, we demonstrated that PRG co-localizes with the actin cytoskeleton in cultured cells and binds to actin complexes in cell lysates through a unique 25 amino acid region (amino acids 561–585) that is located between the RGS and DH-PH domains of the protein (29). PRG mutants that fail to interact with actin displayed an enhanced Rho-dependent signaling compared to wild type PRG. In the previous study, it was not determined whether PRG directly binds actin, or whether the interaction with actin was mediated by other actin-binding proteins. Here, we now demonstrate that the actin-binding region of PRG directly binds F-actin in vitro. In addition, we further characterize the actin-binding region of PRG by point mutation analysis, immunofluorescence localization, co-immunoprecipitation and actin co-sedimentation assays. We have identified the importance of four specific amino acids (I567, I568, F573 and E574) in the actin-binding domain of PRG that are directly responsible for in vitro actin-binding as well as in vivo colocalization with the actin cytoskeleton. Moreover, our studies demonstrate that a similar actin-binding motif exists in frabin, a RhoGEF that is not a member of the RGS-RhoGEF sub-family. Lastly, we report here that dimerization of the actin-binding region of PRG reveals an in vitro F-actin bundling activity.
MATERIALS AND METHODS
Plasmid construction
The N-terminal Myc epitope (EQKLISEED) tagged PRG and (Δ25)PRG in pCDNA3 have been described previously (29). Myc epitope-tagged frabin and GST-Dead FAB-PH1 frabin DNA (30) was kindly provided by Y. Takai (Osaka University Graduate School of Medicine, Suita, Japan).
GST(541–605)PRG and GST(541–605,Δ25)PRG were constructed by PCR amplification with Myc-PRG and Myc-(Δ25)PRG as templates respectively and subcloning into EcoRI-Sal1 restriction sites of pGEX5X-1. Frabin(1–150)GFP was generated by PCR amplification using Myc-frabin as template and subcloning into the EcoRI-Sal1 restriction sites of pEGFP-N1. Furthermore, GST(1–150)frabin was made by PCR amplification with GST-Dead FAB-PH1 frabin as a template and subcloning into EcoRI-Sal1 restriction sites of pGEX5X-1.
Stratagene QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to replace amino acids with alanine to create Myc(N567A)PRG, Myc(I568A)PRG, Myc(I569A)PRG, Myc(Q570A)PRG, Myc(H571A)PRG, Myc(F572A)PRG, Myc(E573A)PRG, Myc(N574A)PRG and Myc(N575A)PRG using Myc-PRG as a template. GST(541–605, I568A)PRG, GST(541–605, I569A)PRG, GST(541–605, F572A)PRG, GST(541–605, E573A)PRG and GST(541–605, N574A)PRG were obtained using GST(541–605)PRG as a template. Frabin(1–150, D22A)GFP, frabin(1–150, L23A)GFP, frabin(1–150, I24A)GFP, frabin(1–150, S25A)GFP, frabin(1–150, H26A)GFP, frabin(1–150, F27A)GFP, frabin(1–150, E28A)GFP, frabin(1–150, G29A)GFP and frabin(1–150, G30A)GFP were generated using frabin(1–150)GFP as a template, and GST(1–150, L23A)frabin, GST(1–150, I24A)frabin, GST(1–150, F27A)frabin and GST(1–150, E28A)frabin were made using GST(1–150)frabin as a template.
FKBP from pC4-Fv1E (Ariad Pharmaceuticals, Cambridge, MA), was amplified with forward and reverse primers containing a 5’ Xho1 and a 3’ Not1 site for subcloning into GST(541–605)PRG to make GST(541–605)PRG-FKBP. The correct sequence of the mutants were confirmed by DNA sequencing of the entire open reading frame (Kimmel Cancer Center Nucleic Acid Facility, Philadelphia, PA).
Cell culture and Transfection
COS-7 cells were propagated in DMEM (Mediatech, Herndon, VA) containing 10% fetal bovine serum and penicillin and streptomycin. Unless otherwise mentioned, cells were plated in six-well plates at 7.0 × 105 cells per well and grown for 24 h before transfection. One microgram of total expression plasmid was transfected into the cells by using FuGENE 6 (Roche Diagnostics, Indianapolis, IN).
Immunofluorescence Microscopy
COS-7 cells were grown on coverslips in six-well plates and transfected with appropriate plasmids for 24 h. Fixation and staining have been described previously (29). Briefly, cells were fixed with 3.7% formaldehyde in phosphate buffer saline (PBS) for 15 min, washed and then incubated in blocking buffer containing 2.5% non-fat milk in Tris-buffered saline (TBS)/1% Triton X-100. Cells were incubated with 1 µg/ml 9E10, anti-Myc mouse monoclonal antibody (Covance, Berkeley, CA) for 1h and then Alexa 594 goat anti-mouse secondary antibody (Molecular Probes, Eugene, OR) at 1: 250 dilution for 45 min. For green fluorescent protein (GFP)-tagged mutants, incubation with antibodies was omitted. For detection of actin, phalloidin conjugated to Alexa 488 or 594 (Molecular Probes, Eugene, OR) was used at a dilution of 1: 100 for 45 min. Thereafter, coverslips were washed with TBS/1% Triton X-100, rinsed in distilled water, and mounted on glass slides with 10 µl of Prolong Antifade Reagent (Molecular Probes, Eugene, OR).
Images were acquired using an Olympus BX-61 upright microscope with a 60 × 1.4 NA oil immersion objective and an ORCA-ER cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ) controlled by Slidebook vesion 4.0 (Intelligent Imaging Innovations, Denver, CO).
F-actin Co-Immunoprecipitation Assay
COS-7 cells grown in 10 cm plates were transfected with 7 µg of indicated constructs. 24 h after transfection, cells were washed twice with cold PBS and lysed using 500 µl lysis buffer as described previously (29). Briefly, cells were lysed for 45 min and lysates were incubated with anti Myc monoclonal or anti GFP polyclonal (Rockland, Gilbertsville, PA) antibodies for 3 h. The immunocomplexes were then recovered using 30 µl of Protein A/G Plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and analyzed by immunoblotting.
Preparation of recombinant proteins
Glutathione S-transferase (GST) fusion proteins of various fragments of PRG and frabin were expressed in transformed E. coli cells (BL 21). Cells were grown in 2YT medium at 37°C to A600 ~0.7, induced with 0.5 mM isopropyl-1-thiogalactopyranoside (Fisher Scientific, Fair Lawn, NJ) and grown for 3 h. Subsequently, cells were pelleted and lysed by sonication in STE (20 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1 mM EDTA pH 8.0) containing 5 mM DTT, 2% Triton X-100, 1.5% Sarkosyl, 2 mg/ml lysozyme, 1 mM PMSF, 10 µg/ml leupeptin, 10 µg/ml aprotinin and complete protease inhibitor cocktail tablets (Roche Diagnostics, Indianapolis, IN). The suspension was then centrifuged at 27,000g (Sorvall SS-34 rotor) for 30 min at 4°C. The supernatants were incubated with glutathione sepharose beads (GE Healthcare, Sweeden) for 2 h at 4°C. Beads were washed thrice with lysis buffer. Following washes, the GST-fused proteins were eluted off the beads with buffer (50 mM Tris-HCl, pH 8.0 and 1mM PMSF) containing 10 mM reduced glutathione (Sigma-Aldrich, St. Louis, MO). The eluted protein was subjected to dialysis in buffer containing 20 mM NaCl, 20 mM Tris-HCl (pH 8.0) and 2.5% glycerol. For F-actin bundling assays, proteins were cleaved off GST using Factor Xa Protease (New England Biolabs, Ipswich, MA); removal of GST was >90% routinely.
F-actin co-sedimentation assay
Purified GST-fused fragments were precleared by centrifugation at 200,000g (TLA 100.2 rotor; Beckman Instruments, Palo Alto, CA) for 20 min at 4°C to remove any aggregates and resultant supernatant was used for the experiment. Actin co-sedimentation assay was done essentially as described by the manufacturer (Cytoskeleton, Denver, CO). Briefly, recombinant proteins were incubated with 40 µg of freshly polymerized actin (F-actin) for 30 min at room temperature. After incubation, protein plus F-actin solution was subject to high speed centrifugation (160,000g) to pellet F-actin and protein bound to F-actin. The pellet fractions were solubilized in SDS-sample buffer, the volume being equal to initial incubation volume. Equivalent amounts of pellet and supernatant fractions were analyzed by SDS-PAGE followed by staining using Novex colloidal blue (Invitrogen, Carlsbad, CA). To quantify F-actin binding, increasing amounts of F-actin were incubated with recombinant proteins as mentioned in the figure legends. The samples were then centrifuged and analyzed as mentioned above. The protein bands were quantified by densitometry using Quantity One analysis software (Biorad, Hercules, CA).
Observation of actin bundles
The following recombinant proteins were used in this assay: GST, GST(541–605)PRG, GST(541–605,Δ25)PRG, (541–605)PRG, monomeric (541–605)PRG-FKBP and dimeric (541–605)PRG-FKBP. In the case of (541–605)PRG and (541–605)PRG-FKBP, GST was removed by proteolytic cleavage. For inducible experiments, 25 µM (541–605)PRG-FKBP (lacking GST) was incubated without or with 25 µM AP20187 (Ariad Pharmaceuticals, Cambridge, MA) at room temperature for 15 min to prepare monomeric or dimeric (541–605)PRG-FKBP, respectively. Using these 25 µM stocks, monomeric and dimeric forms of (541–605)PRG-FKBP were diluted and used in the assay at 5 µM and 10 µM. In all other cases, 7 µM recombinant protein was used.
Fluorescence observation of actin bundles was performed as described previously (31). Briefly, 1.2 µM freshly polymerized F-actin was incubated alone or with the above described recombinant proteins in F-buffer (25 mM Hepes pH 7.5; 100 mM KCl, 0.2 mM CaCl2, 2 mM MgCl2, 2 mM EGTA, 0.2 mM ATP, 1 mM DTT) for 30 min on ice. Next, F-actin was stained with phalloidin conjugated to Alexa 594 at a dilution of 1:100 for 15 min on ice. The mixture was then applied to poly-L-lysine coated glass coverslips (BD Biosciences, Bedford, MA) and incubated for 20 min at room temperature. Adherent material was washed with F-buffer and observed using a 60× 1.4 NA oil immersion objective.
F-actin bundling assay
25 µM (541–605)PRG-FKBP was dimerized using AP20187, as described above. The 25 µM stock of dimerized (541–605)PRG-FKBP was appropriately diluted (0.5–16 µM) and incubated with 1 µM of F-actin in F-buffer at room temperature for 1 hour. This mixture was then subjected to a low speed centrifugation (10,000 g) for 30 min. Supernatant and pellet fractions were solubilized in SDS-sample buffer and analyzed as mentioned above.
Native gel electrophoresis
Native gel electrophoresis was performed in an 8% acrylamide gel prepared in 0.3 M Tris-HCl (pH 8.8). 25 µM of (541–605)PRG-FKBP was mixed with nondenaturing loading buffer (0.3 M Tris-HCl, pH 6.8, 50% glycerol, 0.5% bromophenol blue) and the samples were loaded and run at 4°C at 25 mA in 25 mM tris base, 200 mM glycine. The gel was stained using Novex colloidal blue.
F-actin polymerization assay
Pyrene-labeled actin (60% labeled) and Arp2/3 complex were obtained from Cytoskeleton, Inc (Denver, CO). Actin polymerization assay was performed as described previously (32). Briefly, pyrene labeled actin was incubated in buffer containing 5 mM Tris-HCl pH 7.5, 0.2 mM CaCl2 and 0.2 mM ATP for 1 h on ice. Following incubation residual F-actin was removed by centrifugation at 400,000g for 1 h. Polymerization reactions contained either labeled actin (1 µM) used alone or in combination with recombinant VCA domain (110 nM) of WAVE2 and Arp2/3 complex (30 nM) as negative and positive controls respectively. In addition labeled actin (1 µM) was also incubated with monomeric and dimeric forms of (541–605)PRG-FKBP (1 µM). The actin polymerization reaction was initiated in polymerization buffer containing 100 mM Tris-HCl pH 7.5, 10 mM MgCl2, 500 mM KCl, 10 mM EGTA and 0.2 mM ATP. Pyrene fluorescence was monitored continuously every 30 sec by a spectrofluorimeter (Perkin Elmer Luminesecence Spectrometer LS55) set at an excitation 365 nm and emission 407 nm.
RESULTS
Prediction of an actin-binding motif shared by PRG and frabin
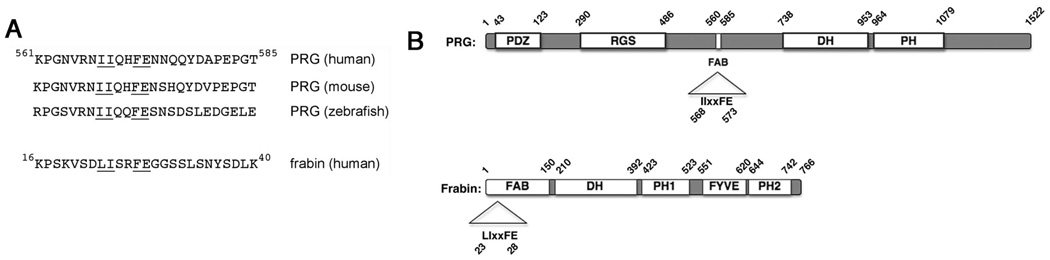
Our previous investigation suggested that a nine amino acid sequence, NIIQHFENN, consisting of amino acids 567–575, within the 561–585 actin-binding region is critical for the actin interaction of PRG (29). To examine the important elements of this sequence in more detail, here we have first compared the actin-binding region of human PRG to the similar region of the mouse and zebrafish PRG orthologs (Figure 1). Much of this region is highly conserved, even in zebrafish PRG, suggesting that actin-binding ability has been retained in PRG across species. Consistent with this, a recent report showed co-localization with F-actin of the zebrafish PRG when expressed in HEK293 cells (33).
Figure 1. Actin-binding motif of PRG and frabin.
(A) The amino acid sequence encompassing the predicted actin-binding motif in human PRG (amino acids 561–585) is shown along with sequences in similar locations in the mouse and zebrafish PRG orthologs. In addition, the predicted actin-binding motif (amino acids 16–40) of frabin is shown. The underlined amino acids indicate similarities among PRG orthologs and frabin, and the underlined amino acids represent a minimal actin-binding motif, L/IIxxFE, defined in this report. (B) The locations of the actin-binding motif in the overall structure of PRG and frabin are indicated.
In addition, in our previous study we speculated that a similar actin-binding motif is present in frabin, a RhoGEF for Cdc42. It has been shown that frabin binds to actin through its N-terminal 150 amino acids (34). Although the minimal sequence for actin binding was not defined in frabin, it was reported that mutation of leucine at position 23 to arginine abolished binding to actin (30). A comparison of the amino acid sequence surrounding L23 of frabin with the actin-binding region of PRG identifies some similarity, showing that two hydrophobic residues, leucine and/or isoleucine, an aromatic residue, phenylalanine, and an acidic residue, glutamic acid, are conserved among the PRG orthologs and human frabin (Figure 1). This sequence analysis thus suggests L/IIxxFE as a critical motif in the actin-binding domain of both PRG and frabin. The experiments described below confirm this prediction.
Subcellular localization of PDZ-RhoGEF
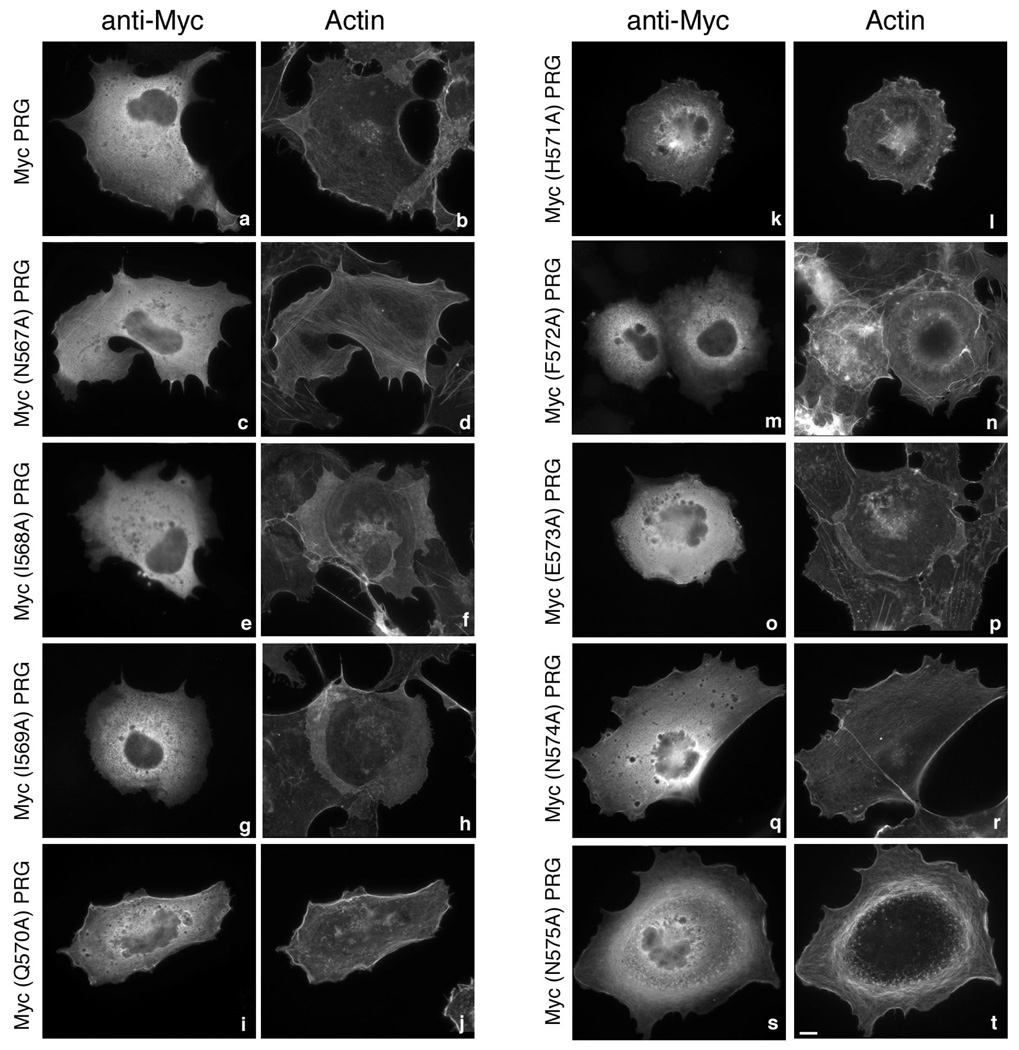
In PRG the stretch of nine amino acids, NIIQHFENN, at positions 567–575, play a crtitical role in localization of the protein to the peri-PM region in 293T and in COS-7 cells (29). To identify amino acids that are absolutely critical in binding to actin, we individually mutated each of these nine residues to alanine and examined PRG localization by immunofluorescence. PRG and its mutants containing an N-terminal Myc epitope tag, were expressed in COS-7 cells; all PRG mutants show similar expression levels (not shown). As observed earlier wild type (WT) PDZ-RhoGEF (Myc-PRG) displayed cytoplasmic as well as peri-PM localization (Figure 2a). In addition, Myc-PRG was also found to co-localize with cortical actin at the cell periphery (Figure 2b) as seen by staining for actin using fluorescently labeled phalloidin. Point mutants Myc(N567A)PRG (Figure 2, c and d), Myc(Q570A)PRG, Myc(H571A)PRG (Figure 2, i–l), Myc(N574A)PRG and Myc(N575A)PRG (Figure 2, q–t) displayed localization similar to Myc-PRG. These mutants were also observed to co-localize with cortical actin. On the other hand, mutants Myc(I568A)PRG, Myc(I569A)PRG (Figure 2, e–h), Myc(F572A)PRG and Myc(E573A)PRG (Figure 2, m–p) exhibited a complete loss of peri-PM localization and failed to co-localize with actin. These observations suggest that four amino acids, isoleucines at positions 568 and 569, phenylalanine at position 572 and glutamic acid at position 573 play a critical role in the peri-PM localization of PRG in COS-7 cells.
Figure 2. Subcellular localization of PRG and mutants.
COS-7 cells were transfected with 1 µg of an expression vector encoding Myc epitope–tagged PRG (a and b), Myc(N567A)PRG (c and d), Myc(I568A)PRG (e and f), Myc(I569A)PRG (g and h), Myc(Q570A)PRG (i and j), Myc(H571A)PRG (k and l), Myc(F572A)PRG (m and n), Myc(E573A)PRG (o and p), Myc(N574A)PRG (q and r) or Myc(N575A)PRG (s and t). 24 h post-transfection, cells were fixed and subjected to immunofluorescence microscopy as described in “Materials and Methods”. Expressed proteins were detected with an anti-Myc 9E10 antibody (a, c, e, g, I, k, m, o, q and s) followed by Alexa 594 conjugated to an anti-mouse antibody. Actin was visualized in the same cells by co-staining with Alexa 488 conjugated to phalloidin (b, d, f, h, j, l, n, p, r and t). Bar, 10 µm. Images shown are single cells representative of at least five separate experiments in which more than fifty cells were viewed in each experiment. Note that wild type PRG (a) and unaffected PRG mutants (c, i, k, q, s) show a modest co-localization with F-actin at the cell periphery, whereas I568A, I569A, F572A, and E573A mutants of PRG (e, g, m, o) reproducibly display a complete loss of staining at the cell periphery.
Subcellular localization of frabin
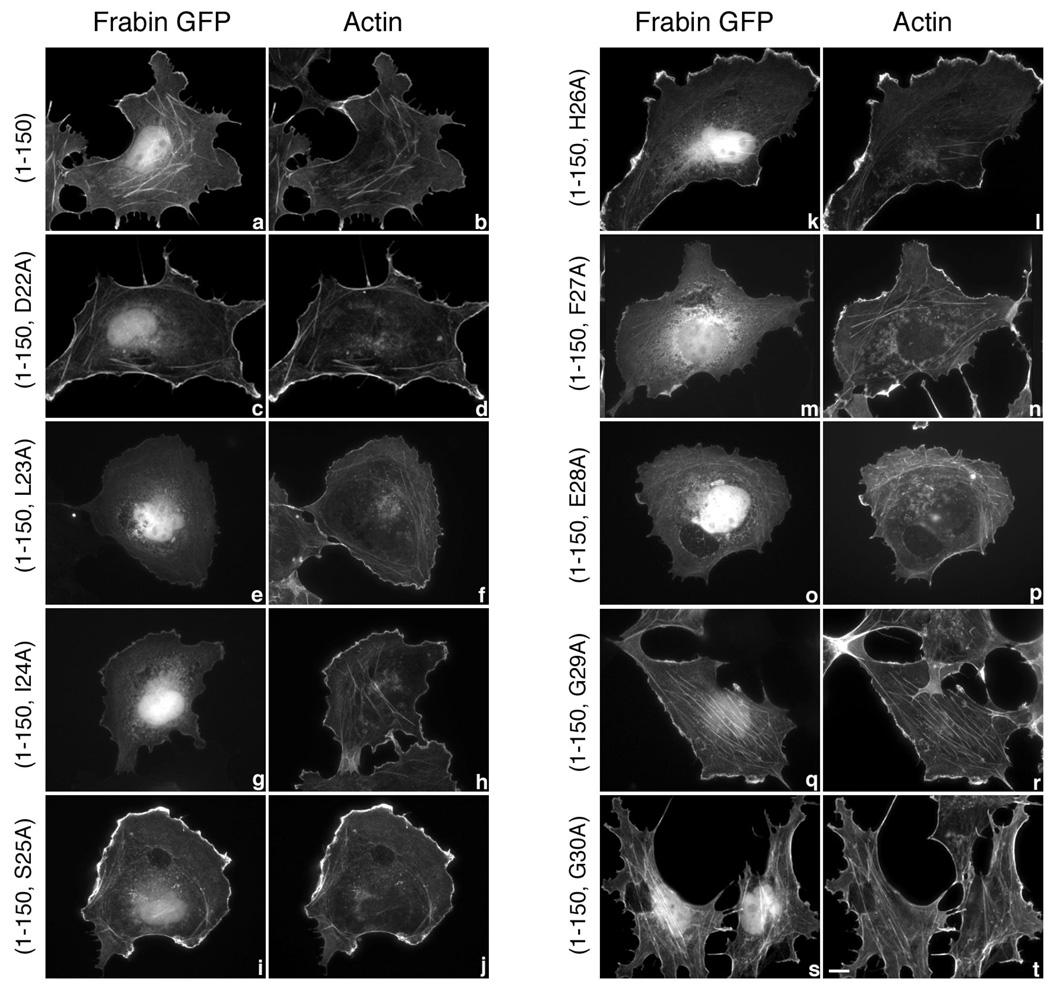
As with PRG, we examined the potential actin-binding motif of frabin by individually mutating to alanine each of the nine amino acids in the sequence DLISHFEGG at position 22–30. Mutations were introduced into an N-terminal 150 amino acid region of frabin fused to GFP that was previously shown to bind actin (30), and the subcellular distribution of the mutants was detected using fluorescence microscopy; all frabin proteins showed similar expression levels (not shown).
Frabin(1–150)GFP when transiently transfected in COS-7 cells showed a strong co-localization with actin both at the cell periphery and within the cell as seen by staining for actin using a fluorescently labeled phalloidin (Figure 3, a and b). Similarly, frabin(1–150, D22A)GFP (Figure 3, c and d), frabin(1–150, S25A)GFP, frabin(1–150, H26A)GFP (Figure 3, i–l), frabin(1–150, G29A)GFP and frabin(1–150, G30A)GFP (Figure 3, q–t) showed a strong co-localization with actin. Intriguingly, mutations that abolished co-localization with actin in similar positions of PRG, also abolished frabin’s co-localization with actin. Frabin(1–150, L23A)GFP, frabin(1–150, I24A)GFP (Figure 3, e–h), frabin(1–150, F27A)GFP and frabin(1–150, E28A)GFP (Figure 3, m–p) displayed little or no co-localization with actin, and the subcellular distribution of these point mutants was identical to that of GFP alone (data not shown). Thus, individual mutation of L23, I24, F27 and E28 caused a complete loss of co-localization of frabin with the actin cytoskeleton in COS-7 cells. PRG point mutants that resembled the frabin mutants also showed defective co-localization with actin cytoskeleton (Figure 2). Taken together, these observations indicate the importance of these four conserved residues in PRG and frabin localization to the actin cytoskeleton.
Figure 3. Subcellular localization of frabin mutants.
COS-7 cells were transiently transfected with 1 µg expression vectors encoding GFP-tagged frabin mutants, frabin(1–150) (a and b), frabin(1–150, D22A) (c and d), frabin(1–150, L23A) (e and f), frabin(1–150, I24A) (g and h), frabin(1–150, S25A) (i and j), frabin(1–150, H26A) (k and l), frabin(1–150, F27A) (m and n), frabin(1–150, E28A) (o and p), frabin(1–150, G29A) (q and r) and frabin(1–150, G30A) (s and t). Expressed proteins were visualized by GFP fluorescence (a, c, e, g, I, k, m, o, q and s), and actin was visualized (b, d, f, h, j, l, n, p, r and t) in the same cells by staining with Alexa 594 conjugated to phalloidin. Bar, 10 µm. Images shown are single cells representative of at least five separate experiments in which more than fifty cells were viewed in each experiment. Note that frabin(1–150) (a) and unaffected frabin(1–150) mutants (c, i, k, q, s) show an intense GFP signal at the cell periphery and at intracellular stress fibers that co-localizes with F-actin, whereas L23A, I24A, F27A, and E28A frabin(1–150) (e, g, m, o) reproducibly display a weak GFP signal at the cell periphery. The weak signal at the cell periphery observed with the four frabin mutants (e, g, m, o) is identical to the background signal of GFP alone.
A conserved motif in PRG and frabin is critical for co-immunoprecipitation with actin
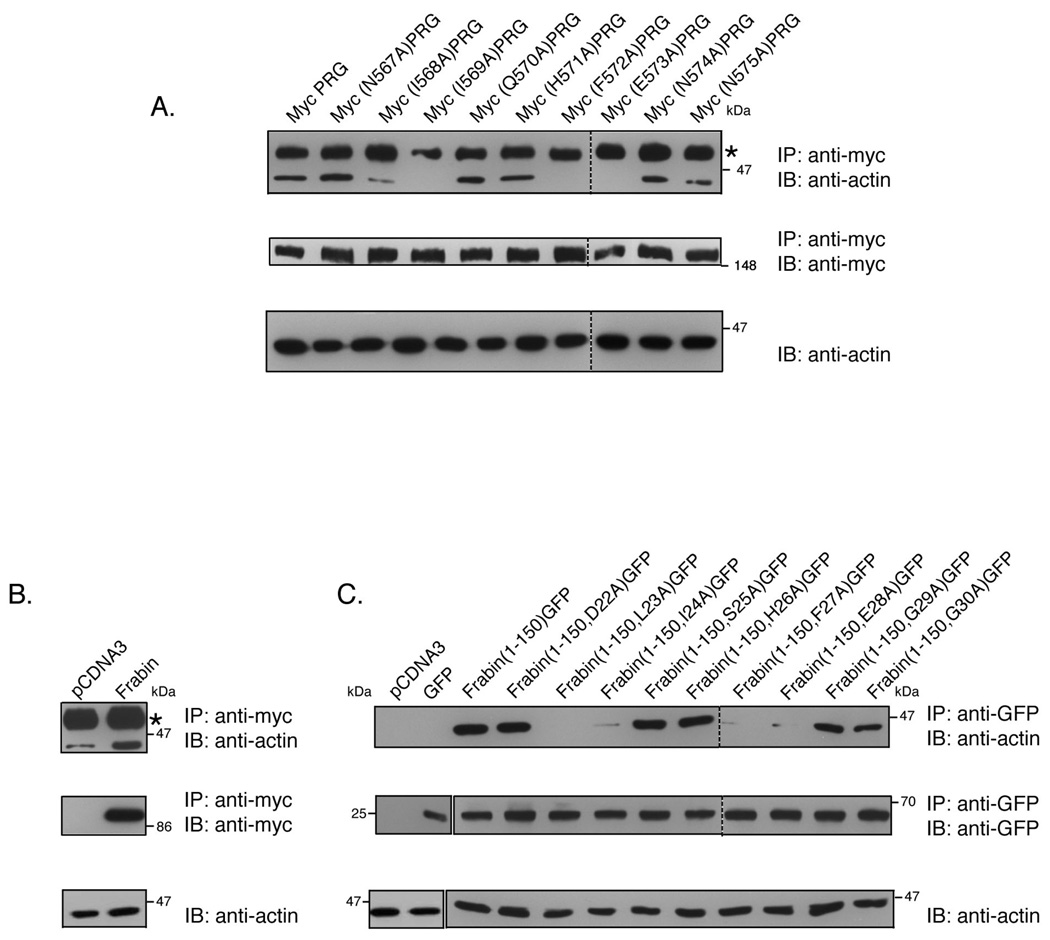
Next we performed co-immunoprecipitation assays to test the importance of these residues in the interaction of PRG and frabin with actin. COS-7 cells were transiently transfected with wild type and point mutants of PRG, frabin or frabin(1–150)GFP. Overexpressed proteins were immunoprecipitated, and immunoblots were probed with anti-actin antibody to determine co-immunoprecipitation.
As shown in Figure 4A (upper panel), actin co-immunoprecipitates with Myc PRG. Mutants Myc(N567A)PRG, Myc(Q570A)PRG, Myc(H571A)PRG, Myc(N574A)PRG and Myc(N575A)PRG that co-localize with actin were also able to efficiently co-immunoprecipitate with actin. However, Myc(I568A)PRG, Myc(I569A)PRG, Myc(F572A)PRG and Myc(E573A)PRG that failed to co-localize with actin, show little or no co-immunoprecipitation with actin.
Figure 4. PRG and frabin co-immunoprecipitate with actin.
(A) COS-7 cells were transiently transfected with 7 µg of an expression vector encoding PRG or the indicated mutants. PRG was immunoprecipitated from cell lysates with a mouse monoclonal anti-Myc antibody, and immunoprecipitates were analyzed by immunoblotting using anti-actin mAb (upper panel). Immunoprecipitation of the Myc-tagged PRG proteins was confirmed by immunoblotting of the immunoprecipitates using anti-Myc antibody (middle panel). Presence of actin in the lysates was detected by immunoblotting using anti-actin mAb (lower panel). (B) COS-7 cells were transfected with 7 µg empty vector or with expression plasmid for Myc-tagged full-length frabin. Cells were lysed and lysates were subjected to immunoprecipitation by monoclonal anti-Myc antibody. Immunoprecipitates were analyzed by immunoblot using anti-actin mAb (upper panel) and anti-Myc Ab (middle panel). Actin in the cell lysates was detected using anti-actin mAb (lower panel). (C) COS-7 cells were transiently transfected with 7 µg of empty vector, GFP alone or with the indicated GFP-tagged frabin mutants. Cell lysates were immunoprecipitated with polyclonal anti-GFP antibody and immunoprecipitates were probed for actin using anti-actin mAb (upper panel). Immunoprecipitation of GFP-tagged frabin constructs were confirmed by immunoblot of immunoprecipitates using anti-GFP antibody (middle panel), and presence of actin in the cell lysates was confirmed by anti-actin mAb (lower panel). Asterisks (*) indicate the antibody heavy chain that migrates slower than actin.
As expected, actin co-immunoprecipitated with full length frabin, whereas little or no co-immunoprecipitation of actin was observed with empty vector or GFP alone (Figure 4B and 4C). Frabin(1–150)GFP which strongly co-localizes with actin as observed by immunofluorescence (Figure 3), also efficiently co-immunoprecipitated with actin (Figure 4C); in addition, point mutants frabin(1–150, D22A)GFP, frabin(1–150, S25A)GFP, frabin(1–150, H26A)GFP, frabin(1–150, G29A)GFP and frabin(1–150, G30A)GFP, all of which colocalize with actin (Figure 3), strongly interact with actin as observed by co-immunoprecipitation. However, mutants frabin(1–150, L23A)GFP, frabin(1–150, I24A)GFP, frabin(1–150, F27A)GFP and frabin(1–150, E28A)GFP fail to co-immunoprecitate with actin suggesting the importance of these residues for interaction with actin. Thus, in both PRG and frabin identical residues play a role in interaction with actin.
PRG binds to F-Actin in vitro
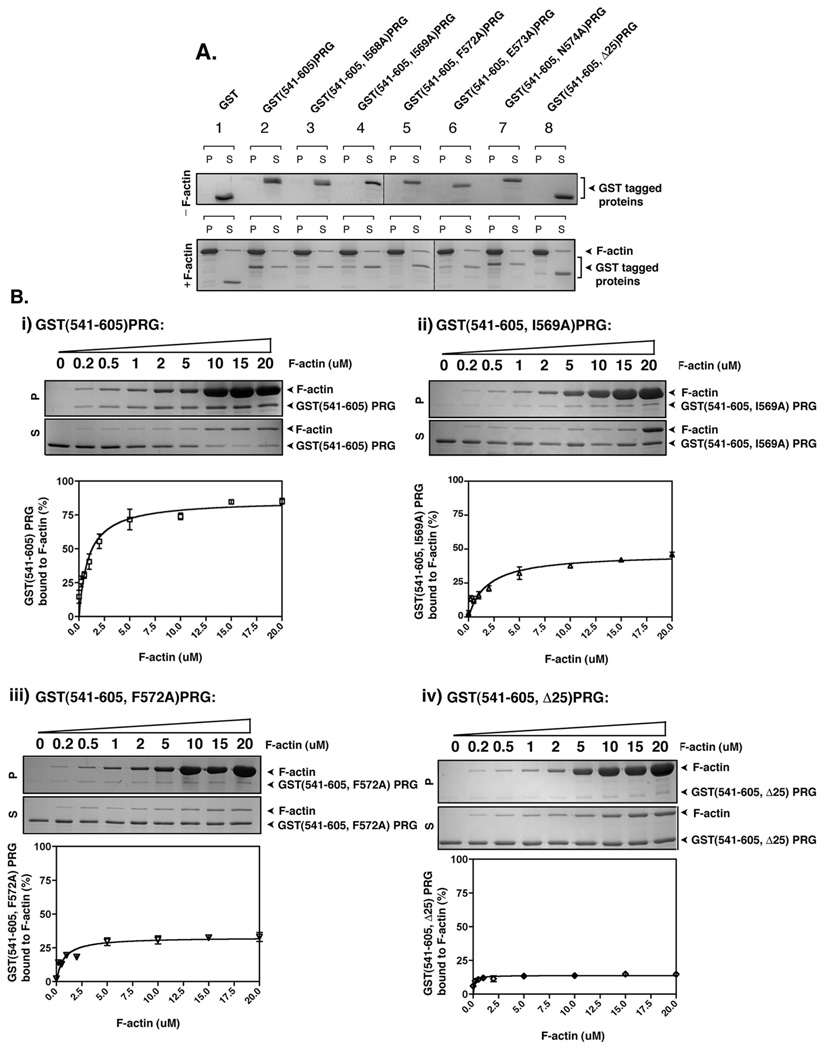
So far we have shown that PRG interacts with actin in cells. However, it has not been demonstrated whether PRG directly binds to actin through this 25 amino acid region or if instead binding to actin is mediated via other interacting proteins. To address this, we performed an actin co-sedimentation assay using purified proteins. GST-fused PRG comprising amino acids 541–605 with or without the 25 amino acid actin binding domain was used for this experiment. In addition, point mutants of the key residues that were observed earlier to be involved in interaction with actin (Figure 2 and Figure 4) were also made in the context of the PRG(541–605) GST fusion protein. Recombinant proteins (2 µM) were incubated with freshly polymerized F-actin (20 µM) and the ability of proteins to co-sediment with actin was examined (Figure 5). Co-sedimentation with F-actin in the pellet fraction (P) following a high speed centrifugation indicates binding to F-actin while separation into the soluble fraction (S) indicates no interaction with F-actin. As shown in Figure 5A, GST alone partitioned entirely in the soluble fraction both in the absence or in presence of F-actin (Figure 5A, lane 1). GST(541–605)PRG (Figure 5A, lane 2) was entirely in the soluble fraction without F-actin. However, in the presence of F-actin, GST(541–605)PRG partitioned to the F-actin bound pellet fraction, demonstrating for the first time direct binding to actin of PRG’s actin binding domain. Similarly, GST(541–605, N574A)PRG (Figure 5A, lane 7) also co-sedimented with F-actin in the pellet fraction. In contrast, GST(541–605, I568A)PRG (Figure 5A, lane 3) and GST(541–605, I569A)PRG (Figure 5A, lane 4) displayed impaired partitioning to the F-actin bound pellet fraction, and GST(541–605, F572A)PRG (Figure 5A, lane 5) and GST(541–605, E573A)PRG (Figure 5A, lane 6) displayed very little partitioning to the F-actin bound pellet fractions, suggesting the importance of these residues in F-actin binding in vitro. GST(541–605, Δ25)PRG (Figure 5A, lane 8), which lacks the actin binding domain, failed to cosediment with actin and was always in the soluble fraction regardless of the presence or absence of F-actin.
Figure 5. PRG binds to F-actin in vitro.
(A) 2 µg of the indicated recombinant proteins was incubated with or without 40 µg of freshly polymerized F-actin at room temperature for 30 min and then centrifuged at 160,000g for 90 min. Equal aliquots of resuspended pellet (P) and supernatant (S) were analyzed by SDS-PAGE and colloidal blue staining. (B) Quantification of F-actin binding activity of recombinant PRG mutants, GST(541–605)PRG (i), GST(541–605, I569A)PRG (ii), GST(541–605, F572A)PRG and (iv) GST(541–605, Δ25)PRG. 3 µM of the indicated mutants of PRG were incubated with variable concentrations F-actin (0.2 – 20 µM) and co-sedimented at 160,000g for 90 min. Equal aliquots of resuspended pellet (P) and supernatant (S) were analyzed by SDS-PAGE and colloidal blue staining (upper panels), and the bands were quantified by densitometry. The percentage of F-actin bound PRG mutants was calculated as the percentage recovered in the pellet (P) over total protein (S+P). The graphs represent the means ±S.D. of three independent experiments.
To examine more closely the binding of PRG to F-actin, we first tested dose dependent binding of GST(541–605)PRG with actin. A co-sedimentation assay was performed in which 3 µM of recombinant protein was incubated with variable concentrations of F-actin (0.2–20 µM). It was observed that GST(541–605)PRG co-sedimented with F-actin in a concentration dependent manner reaching saturation at approximately 5 µM of F-actin. At this concentration of F-actin approximately 75% of GST(541–605)PRG was bound to actin as determined by densitometric analysis (Figure 5B i). From the resulting binding curve, we also estimated that GST(541–605)PRG bound to F-actin with an apparent dissociation constant (Kd) of about 1 µM. However, in the case of GST(541–605, I568A)PRG, GST(541–605, I569A)PRG, GST(541–605, F572A)PRG and GST(541–605, E573A)PRG (Figures 5B ii and iii, and not shown) only about 25% of the protein bound to F-actin at 5 µM confirming that these mutants were defective in binding to actin. In the case of GST(541–605, Δ25)PRG (Figure 5B, iv) only 10% of the protein bound to actin at 5 µM clearly indicating the role of the 25 amino acid domain between amino acids 561–585 of PRG in binding to actin in vitro. Similar to GST(541–605)PRG, GST-frabin mutants were tested for binding to actin in vitro. While GST(1–150)frabin efficiently co-sedimented with F-actin as described previously (30), GST(1–150, L23A)frabin, GST(1–150, l24A)frabin, GST(1–150, F27A)frabin and GST(1–150, E28A)frabin displayed a defective binding to actin in vitro (data not shown). Thus, data obtained from co-sedimentation assays demonstrate the novel finding that PRG binds to F-actin in vitro, and that the L/IIxxFE sequence is critical for actin binding of both PRG and frabin.
Dimeric PRG actin-binding domain induces bundling of F-actin in vitro
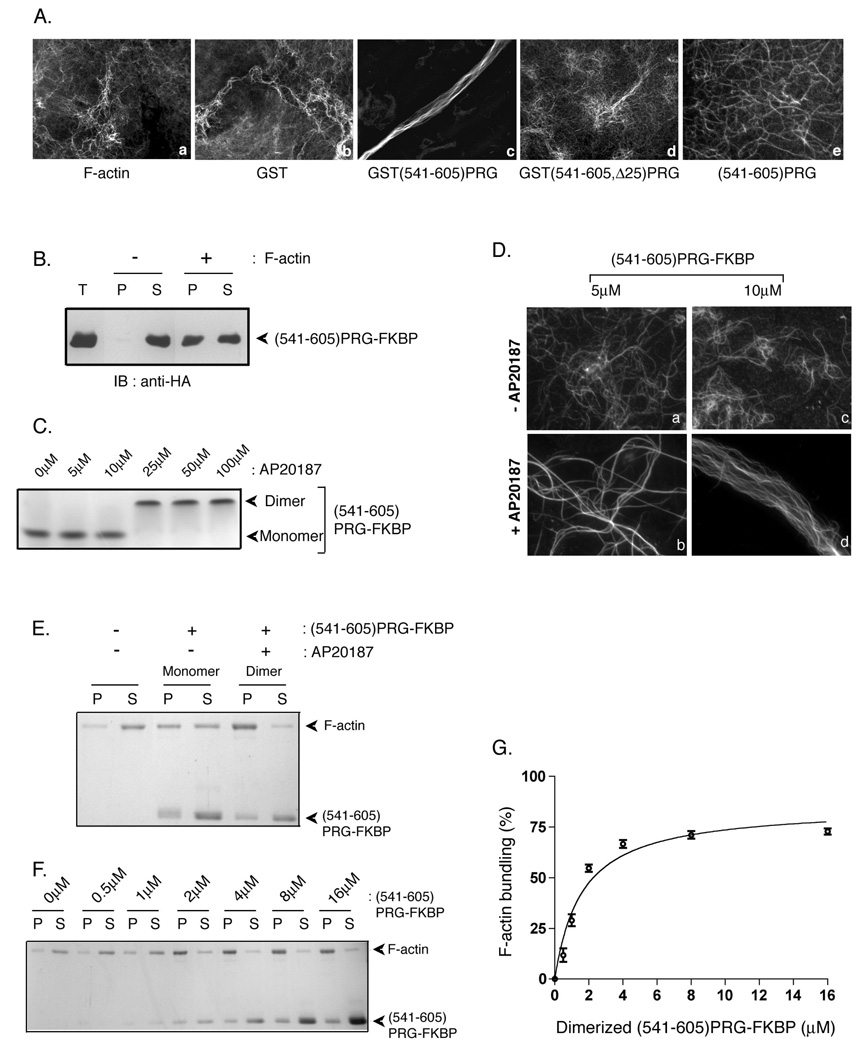
Next, we asked whether the PRG actin-binding domain could effect changes in F-actin structure. Interestingly, the PRG actin-binding domain exhibited an actin bundling activity. Freshly polymerized F-actin was incubated with recombinant GST-tagged proteins, actin was stained with fluorescently-labeled phalloidin, and actin filaments were visualized under fluorescence microscopy, using a previously described procedure (31). F-actin formed a meshwork of filaments when observed in the absence of added recombinant protein (Figure 6A, a) or in the presence of GST (Figure 6A, b). However, incubation with GST(541–605)PRG induced thick F-actin bundles (Figure 6A, c); actin bundles were not formed when F-actin was incubated with GST(541–605,Δ25)PRG (Figure 6A, d). Thus, these experiments indicate that GST(541–605)PRG directly promotes actin bundling.
Figure 6. PRG induces F-actin bundling in vitro.
(A) F-actin (1.2 µM) was incubated alone (a) or with 7 µM GST (b), GST(541–605)PRG (c), GST(541–605, Δ25)PRG (d), or (541–605)PRG in which GST was removed by proteolysis (e) for 30 min on ice. After staining with phalloidin, actin filaments and bundles were observed by fluorescence microscopy. (B) (541–605)PRG-FKBP (GST was removed by proteolysis) incubated with or without freshly polymerized actin was subjected to high speed co-sedimentation assay at 160,000g for 90 min. Equal aliquots of pellet and supernatant fractions along with total protein (T) were immunoblotted using anti-HA antibody to detect (541–605)PRG-FKBP. (C) 25 µM (541–605)PRG-FKBP was incubated with variable concentrations (5–100 µM) of AP20187 for 15 min at room temperature. Samples were subject to native gel electrophoresis followed by colloidal blue staining. 25 µM (541–605)PRG-FKBP was used to observe consistent colloidal blue staining. (D) (541–605)PRG-FKBP (5 µM and 10 µM) treated without (a and c) and with (b and d) AP20187 was incubated with 1.2 µM freshly polymerized F-actin. Following incubation, F-actin was stained with phalloidin and observed by fluorescence microscopy. (E) 2 µM monomeric or dimeric (541–605)PRG-FKBP was incubated with 1 µM freshly polymerized F-actin, and samples were centrifuged at 10,000g for 30 min. Equal amounts of pellet (P) and supernatant (S) fractions were resolved by SDS-PAGE and stained using colloidal blue. (F) To quantitate F-actin bundling activity of dimerized (541–605)PRG-FKBP, 0, 0.5, 1, 2, 4, 8 or 16 µM of dimerized (541–605)PRG-FKBP was incubated with 1 µM of F-actin. Samples were then centrifuged at 10,000g. amounts of pellet (P) and supernatant (S) fractions were run on SDS-PAGE and stained with colloidal blue. (G) F-actin in the 10,000g pellet fraction in (F) was quantitated using densitometry. The percentage of F-actin bundled using different amounts of dimerized (541–605)PRG-FKBP were calculated as the percentage recovered in the pellet (P) over total protein (S+P).The data represent the means ± S.D. of three independent experiments.
Actin bundling activity in a protein often requires either two independent F-actin binding sites or oligomerization of a protein containing a single F-actin binding site. Interestingly, frabin, which we show in this report to have a similar actin-binding motif as does PRG, has been reported to cause bundling of F-actin, although the mechanism is unclear (34). The 65 amino acid portion of PRG contained in GST(541–605)PRG likely contains only one actin-binding site due to its small size and our results that single mutations can abolish actin binding. GST is known to exist in solution as a dimer, and thus GST(541–605)PRG would exist as a dimer. Thus, we hypothesized that the dimerization of (541–605)PRG is necessary for actin bundling. This proposal is particularly relevant since a recent report indicated that PRG forms dimers in cells (22).
To test this, GST was removed following a proteolytic cleavage, and the resulting (541–605)PRG was incubated with F-actin. Interestingly, F-actin bundling was no longer observed when (541–605)PRG was incubated with F-actin (Figure 6A, e). To further test the possibility that dimers of the actin-binding domain of PRG induce F-actin bundling, we took advantage of an inducible dimerization system in which the rapamycin binding protein FKBP is fused to the protein of interest and dimerization is induced by a dimerizing rapamycin analog termed AP20187 (35). For this we made GST(541–605)PRG-FKBP. GST was cleaved off and the resultant (541–605)PRG-FKBP was first tested for its ability to cosediment with F-actin (Figure 6B). As expected, the protein was entirely in the soluble fraction in the absence of F-actin. In the presence of F-actin the protein co-sedimented with F-actin in the pellet fraction (Figure 6B) confirming the ability of (541–605)PRG-FKBP to bind F-actin. Next, (541–605)PRG-FKBP (25 µM) was treated with increasing concentrations of the dimerizer AP20187 (5–100 µM). A concentration dependent shift in the mobility of the protein in a native gel (Figure 6C) indicated the AP20187-dependent dimerization of (541–605)PRG-FKBP. (541–605)PRG-FKBP was then tested for its ability to induce bundling of F-actin. In the absence of AP20187, 5 µM and 10 µM monomeric (541–605)PRG-FKBP failed to cause bundling of F-actin (Figure 6D, upper panel). In contrast, (541–605)PRG-FKBP which was dimerized using AP20187 induced F-actin bundling (Figure 6D, lower panel). Incubation of 1.2 µM F-actin with 5 µM dimerized (541–605)PRG-FKBP caused thickening of F-actin (Figure 6D, b). Well organized F-actin bundles were observed (Figure 6D, d) when 1.2 µM F-actin was incubated with 10 µM of dimerized (541–605)PRG-FKBP, indicating a dose dependent ability to induce actin bundling.
To further study actin bundling by dimerized (541–605)PRG-FKBP, a low-speed centrifugation assay was performed (31). Briefly, 1 µM F-actin was incubated with either monomer or dimerized (541–605)PRG at room temperature for one hour. The protein –F-actin complex was then subjected to a low speed centrifugation (10,000g). Bundled F-actin is found in the pellet (P) fraction while unbundled F-actin remains in the soluble (S) fraction. In the absence of the dimerizer AP20187, (541–605)PRG promoted a partial shift of F-actin into the pellet fraction (Figure 6E). However, dimerization of (541–605)PRG-FKBP and incubation with F-actin resulted in a strong shift of F-actin into the pellet fraction, consistent with the ability of dimerized (541–605)PRG-FKBP to promote actin bundling. Moreover, dimerized (541–605)PRG displayed a concentration dependent actin bundling activity (Figure 6F and 6G). AP20187 alone, in the absence of (541–605)PRG-FKBP, did not cause bundling of actin (data not shown). From these results (Figure 6), we conclude that (541–605)PRG-FKBP causes F-actin bundling in vitro and this is regulated by the protein’s ability to dimerize.
Lastly, we examined the effect of recombinant (541–605)PRG on actin polymerization. Using a well-described pyrene-labeled actin assay (32, 36), neither monomeric nor dimeric (541–605)PRG was able to increase the rate of actin polymerization (Supplemental Figure S1). However, the positive control of the VCA domain of WAVE-2 plus Arp2/3 displayed the expected dramatic increase in actin polymerization. Thus, we conclude that actin binding domain of PRG alone does not affect polymerization of F-actin in vitro.
DISCUSSION
This report defines a novel actin-binding motif in PDZ-RhoGEF (PRG) and shows that this motif is responsible for directly binding F-actin in vitro and localizing PRG to the actin cytoskeleton in cells. In addition, we demonstrate that a similar actin-binding motif is found in the RhoGEF frabin. Extensive mutagenesis defines the consensus sequence for this actin-binding motif as L/IIxxFE. Lastly, we show that the PRG actin-binding region can promote the bundling of actin in vitro in a dimerization dependent manner.
Our previous work defined a 25 amino acid region at positions 561–585 in PRG that was necessary and sufficient for actin interaction and further indicated the importance of a nine amino acid stretch from 567–575 of sequence NIIQHFENN (29). Because this sequence did not appear to match any known actin-binding sequence modules (37), it was important to further define the critical amino acids in the actin-binding sequence of PRG. Each amino acid in the 567–575 sequence was individually substituted with alanine in full-length PRG. Individual mutation of isoleucine 568, isoleucine 569, phenylalanine 572, and glutamic acid 573 was sufficient to disrupt PRG’s co-localization and co-immunoprecipitation with actin (Figure 2 and Figure 4). Mutation of each of the other residues in the 567–575 sequence had no effect on localization or co-immunoprecipitation. Additionally, mutation of isoleucine 568, isoleucine 569, phenylalanine 572, and glutamic acid 573 in the context of GST(541–605)PRG inhibited direct binding to F-actin. Thus, these results define IIxxFE as the critical actin-binding motif in PRG.
Strikingly, visual inspection of the RhoGEF frabin revealed a sequence with identity to the actin-binding motif of PRG. Previously, actin binding was shown to exist in a fragment of frabin consisting of the N-terminal 1–150 amino acids. Although an actin-binding sequence within amino acids 1–150 was not defined, a mutation of leucine 23 to arginine disrupted actin binding and provided a clue that leucine 23 was likely part of an actin-binding sequence. The sequence surrounding leucine 23 in frabin is DLISHFEGG (amino acids 22–30), and thus we tested whether frabin utilized similar amino acids for actin binding as does PRG by individually mutating each residue in the 22–30 amino acid sequence. Consistent with the results for PRG, mutation of leucine 23, isoleucine 24, phenylalanine 27, and glutamic acid 28 abrogated the ability of frabin 1–150 to co-localize with actin in cells, co-immunoprecipitate with actin, and directly bind to actin (Figure 3 and Figure 4, and not shown). Mutation of each of the other residues in the 22–30 sequence did not disrupt frabin localization or interaction with actin. Thus, these experiments reveal the sequence LIxxFE as a critical actin-binding motif in frabin. Taken together, results presented here have defined for the first time the actin-binding motifs of PRG and frabin. Importantly, these results show that both PRG and frabin utilize almost identical amino acids for binding actin. Thus, we identify the sequence L/IIxxFE as a novel actin-binding motif.
Interestingly, this actin-binding motif may be more widespread. Recently, a 17 amino acid peptide from the yeast protein Abp140 was shown to possess actin-binding activity (38). Inspection of the peptide sequence, MGVADLIKKFESISKEE, reveals that it also contains the consensus motif LIxxFE. Although it was suggested that the Abp140 actin-binding sequence is not found in higher eukaryotes (38), our results suggest that mammalian proteins, such as PRG and frabin, utilize the same actin-binding motif and thus may bind actin similarly to the Abp140 peptide. However, it remains to be demonstrated that the LIxxFE amino acids play a critical role in Abp140 actin binding. It will be interesting to test whether PRG and frabin compete with the Abp140 peptide for actin binding. Furthermore, a motif search revealed that the L/IIxxFE sequence is found in numerous proteins. Many of these proteins have no known interaction with actin. A future challenge will be to determine in which proteins the presence of the L/IIxxFE motif represents a novel actin-binding function.
In addition to defining a novel actin-binding motif, our results indicate that the PRG actin-binding region, when dimerized, is able to induce the bundling of F-actin. Using two assays for in vitro F-actin bundling, fluorescence microscopy and low-speed centrifugation, it was demonstrated that the actin-bundling activity of the purified actin-binding region of PRG depended on dimerization (Figure 6). Specifically, GST is known to form dimers, and, consequently, removal of GST from GST(541–605)PRG abolished the actin-bundling activity of (541–605)PRG. In addition, we used an inducible homodimerization system to show that dimeric (541–605)PRG-FKBP induced actin bundling whereas monomeric (541–605)PRG-FKBP did not. Intriguingly, PRG, and the other RGS-RhoGEFs LARG and p115-RhoGEF, have been shown to exist as dimers, or possibly higher order oligomers, in cells (22, 23, 39, 40). Thus, we can speculate that PRG may affect the structure of the actin cytoskeleton in cells independent of or synergistic with its ability to activate Rho. Future studies will be needed to evaluate this proposal. Interestingly, frabin has been shown to also possess actin bundling activity (34). It has been suggested that frabin can directly reorganize a cell’s actin cytoskeleton independent of activation of its cognate small GTPase Cdc42, and this actin bundling activity of frabin may thus play a role in the generation of F-actin bundles in filopodia (34, 41). Other signaling proteins, including Ca2+/calmodulin-dependent kinase II (CaMKII) (42–44), have been shown recently to have unexpected roles in bundling or stabilizing F-actin independent of their well-characterized enzyme activity.
The precise functional significance of actin binding and/or bundling by PRG remains to be determined. Actin binding is unique for PRG among the RGS-RhoGEFs. LARG and p115RhoGEF do not contain the actin-binding motif, and they do not localize to the actin cytoskeleton. Interestingly, the IIxxFE motif is also found in zebrafish PRG (Figure 1), and indeed zebrafish PRG co-localizes with F-actin when expressed in HEK293 cells (33). Such conservation of the actin-binding motif suggests an evolutionary retained function for actin binding by PRG. Previous work showed that PRG mutants that fail to interact with actin displayed enhanced Rho-dependent signaling compared to wild type PRG upon expression in HEK293 or Neuro2A cells, suggesting that actin-binding may decrease PRG signaling function (29). However, the importance of actin binding by PRG remains to be addressed in a more physiological response; it will be interesting to define the role of the actin-binding domain in zebrafish PRG’s role in developmental processes involving ciliated epithelia. A fuller understanding of the receptor-dependent responses for which PRG is an integral component will be necessary to unravel the importance of actin binding by PRG.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Raja Bhattacharyya and Roshanak Irannejad for critically reading the manuscript; Dr. Yoshimi Takai for providing frabin cDNA; Dr. Natalia Zhukovskaya for help with actin fluorescence measurements and providing purified VCA domain of WAVE2; and Dr. Dong Soo Kang, Matthew Martz and Amrita Dawn for valuable experimental advice.
Abbreviations used
- GEF
guanine-nucleotide exchange factor
- RGS
regulator of G protein signaling
- PH
pleckstrin homology
- DH
Dbl homology
- PRG
PDZ-RhoGEF
Footnotes
This work was supported by NIH grant GM62884 (P.B.W.).
Supporting information available: A figure showing an actin polymerization assay is available as Supplemental Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 2.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 5.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers,lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 7.Buchsbaum RJ. Rho activation at a glance. J Cell Sci. 2007;120:1149–1152. doi: 10.1242/jcs.03428. [DOI] [PubMed] [Google Scholar]
- 8.Sternweis PC, Carter AM, Chen Z, Danesh SM, Hsiung YF, Singer WD. Regulation of rho Guanine nucleotide exchange factors by g proteins. Adv Protein Chem. 2007;74:189–228. doi: 10.1016/S0065-3233(07)74006-8. [DOI] [PubMed] [Google Scholar]
- 9.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 10.Hart MJ, Sharma S, elMasry N, Qiu RG, McCabe P, Polakis P, Bollag G. Identification of a novel guanine nucleotide exchange factor for the Rho GTPase. Journal of Biological Chemistry. 1996;271:25452–25458. doi: 10.1074/jbc.271.41.25452. [DOI] [PubMed] [Google Scholar]
- 11.Rumenapp U, Blomquist A, Schworer G, Schablowski H, Psoma A, Jakobs KH. Rho-specific binding and guanine nucleotide exchange catalysis by KIAA0380, a Db1 family member. FEBS Letters. 1999;459:313–318. doi: 10.1016/s0014-5793(99)01270-3. [DOI] [PubMed] [Google Scholar]
- 12.Togashi H, Nagata K, Takagishi M, Saitoh N, Inagaki M. Functions of a Rho-specific guanine nucleotide exchange factor in neurite retraction - Possible role of a proline-rich motif of KIAA0380 in localization. Journal of Biological Chemistry. 2000;275:29570–29578. doi: 10.1074/jbc.M003726200. [DOI] [PubMed] [Google Scholar]
- 13.Reuther GW, Lambert QT, Booden MA, Wennerberg K, Becknell B, Marcucci G, Sondek J, Caligiuri MA, Der CJ. Leukemia-associated Rho guanine nucleotide exchange factor, a Dbl family protein found mutated in leukemia, causes transformation by activation of RhoA. J Biol Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- 14.Taya S, Inagaki N, Sengiku H, Makino H, Iwamatsu A, Urakawa I, Nagao K, Kataoka S, Kaibuchi K. Direct interaction of insulin-like growth factor-1 receptor with leukemia-associated RhoGEF. J Cell Biol. 2001;155:809–820. doi: 10.1083/jcb.200106139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc Natl Acad Sci U S A. 2002;99:12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driessens MH, Olivo C, Nagata K, Inagaki M, Collard JG. B plexins activate Rho through PDZ-RhoGEF. FEBS Lett. 2002;529:168–172. doi: 10.1016/s0014-5793(02)03323-9. [DOI] [PubMed] [Google Scholar]
- 17.Hirotani M, Ohoka Y, Yamamoto T, Nirasawa H, Furuyama T, Kogo M, Matsuya T, Inagaki S. Interaction of plexin-B1 with PDZ domain-containing Rho guanine nucleotide exchange factors. Biochem Biophys Res Commun. 2002;297:32–37. doi: 10.1016/s0006-291x(02)02122-8. [DOI] [PubMed] [Google Scholar]
- 18.Yamada T, Ohoka Y, Kogo M, Inagaki S. Physical and functional interactions of the lysophosphatidic acid receptors with PDZ domain-containing Rho guanine nucleotide exchange factors (RhoGEFs) J Biol Chem. 2005;80:19358–19363. doi: 10.1074/jbc.M414561200. [DOI] [PubMed] [Google Scholar]
- 19.Longhurst DM, Watanabe M, Rothstein JD, Jackson M. Interaction of PDZRhoGEF with microtubule-associated protein 1 light chains: link between microtubules, actin cytoskeleton, and neuronal polarity. J Biol Chem. 2006;281:12030–12040. doi: 10.1074/jbc.M513756200. [DOI] [PubMed] [Google Scholar]
- 20.Chikumi H, Fukuhara S, Gutkind JS. Regulation of G protein-linked guanine nucleotide exchange factors for Rho, PDZ-RhoGEF, and LARG by tyrosine phosphorylation: evidence of a role for focal adhesion kinase. J Biol Chem. 2002;277:12463–12473. doi: 10.1074/jbc.M108504200. [DOI] [PubMed] [Google Scholar]
- 21.Iwanicki MP, Vomastek T, Tilghman RW, Martin KH, Banerjee J, Wedegaertner PB, Parsons JT. FAK, PDZ-RhoGEF and ROCKII cooperate to regulate adhesion movement and trailing-edge retraction in fibroblasts. J Cell Sci. 2008;121:895–905. doi: 10.1242/jcs.020941. [DOI] [PubMed] [Google Scholar]
- 22.Chikumi H, Barac A, Behbahani B, Gao Y, Teramoto H, Zheng Y, Gutkind JS. Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene. 2004;23:233–240. doi: 10.1038/sj.onc.1207012. [DOI] [PubMed] [Google Scholar]
- 23.Eisenhaure TM, Francis SA, Willison LD, Coughlin SR, Lerner DJ. The Rho guanine nucleotide exchange factor Lsc homo-oligomerizes and is negatively regulated through domains in its carboxyl terminus that are absent in novel splenic isoforms. J Biol Chem. 2003;278:30975–30984. doi: 10.1074/jbc.M303277200. [DOI] [PubMed] [Google Scholar]
- 24.Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 25.Wells CD, Liu MY, Jackson M, Gutowski S, Sternweis PM, Rothstein JD, Kozasa T, Sternweis PC. Mechanisms for reversible regulation between G13 and Rho exchange factors. J Biol Chem. 2002;277:1174–1181. doi: 10.1074/jbc.M105274200. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki N, Nakamura S, Mano H, Kozasa T. Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc Natl Acad Sci U S A. 2003;100:733–738. doi: 10.1073/pnas.0234057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart MJ, Roscoe W, Bollag G. Activation of Rho GEF activity by G alpha 13. Methods Enzymol. 2000;325:61–71. doi: 10.1016/s0076-6879(00)25431-1. [DOI] [PubMed] [Google Scholar]
- 28.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS. A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. Journal of Biological Chemistry. 1999;274:5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee J, Wedegaertner PB. Identification of a novel sequence in PDZ-RhoGEF that mediates interaction with the actin cytoskeleton. Mol Biol Cell. 2004;15:1760–1775. doi: 10.1091/mbc.E03-07-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda W, Nakanishi H, Tanaka Y, Tachibana K, Takai Y. Cooperation of Cdc42 small G protein-activating and actin filament-binding activities of frabin in microspike formation. Oncogene. 2001;20:3457–3463. doi: 10.1038/sj.onc.1204463. [DOI] [PubMed] [Google Scholar]
- 31.Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem. 2004;279:14929–14936. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, Schenck A, Bardoni B, Scott JD, Nairn AC, Greengard P. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 33.Panizzi JR, Jessen JR, Drummond IA, Solnica-Krezel L. New functions for a vertebrate Rho guanine nucleotide exchange factor in ciliated epithelia. Development. 2007;134:921–931. doi: 10.1242/dev.02776. [DOI] [PubMed] [Google Scholar]
- 34.Obaishi H, Nakanishi H, Mandai K, Satoh K, Satoh A, Takahashi K, Miyahara M, Nishioka H, Takaishi K, Takai Y. Frabin, a novel FGD1-related actin filament-binding protein capable of changing cell shape and activating c-Jun N-terminal kinase. J Biol Chem. 1998;273:18697–18700. doi: 10.1074/jbc.273.30.18697. [DOI] [PubMed] [Google Scholar]
- 35.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 36.Cooper JA, Pollard TD. Methods to measure actin polymerization. Methods Enzymol. 1982;85(Pt B):182–210. doi: 10.1016/0076-6879(82)85021-0. [DOI] [PubMed] [Google Scholar]
- 37.Maciver SK. 2004 http://www.bms.ed.ac.uk/research/others/smaciver/Encyclop/encycloABP.htm.
- 38.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabocka E, Wedegaertner PB. Disruption of oligomerization induces nucleocytoplasmic shuttling of leukemia-associated rho Guanine-nucleotide exchange factor. Mol Pharmacol. 2007;72:993–1002. doi: 10.1124/mol.107.035162. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, Strauch P, Rubtsov A, Donovan EE, Pelanda R, Torres RM. Lsc activity is controlled by oligomerization and regulates integrin adhesion. Mol Immunol. 2008;45:1825–1836. doi: 10.1016/j.molimm.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakanishi H, Takai Y. Frabin and other related Cdc42-specific guanine nucleotide exchange factors couple the actin cytoskeleton with the plasma membrane. J Cell Mol Med. 2008;12:1169–1176. doi: 10.1111/j.1582-4934.2008.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YC, Redmond L. CaMKIIbeta binding to stable F-actin in vivo regulates F-actin filament stability. Proc Natl Acad Sci U S A. 2008;105:15791–15796. doi: 10.1073/pnas.0804399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanabria H, Swulius MT, Kolodziej SJ, Liu J, Waxham MN. {beta}CaMKII regulates actin assembly and structure. J Biol Chem. 2009;284:9770–9780. doi: 10.1074/jbc.M809518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.