Introduction
Knowledge-based statistical energy functions are widely used in protein structure modeling and prediction 1. They are usually constructed based on statistical analysis of predefined interacting units from a set of selected high-resolution structures. The interacting units can be either coarse-grained structural components, such as Cα atoms for representing a whole residue, or atomistic structural components as in all-atom representation. The energy function is potential of mean force, or free energy cost, required for generating the observed distribution of the interacting units in the real structures from a zero-interaction reference state. Thus, the choices of interacting units are crucial for the effectiveness of the energy functions. One of the key issues is the orientation dependence in the interaction between the units. This is because the chemical bond connectivity is often ignored in constructing statistical energy functions leading to mis- or under-representation of anisotropic orientation preference in molecular interactions.
In the literature, substantial efforts have been made to model anisotropic orientation preference 2–9. An early attempt employed a side-chain-specific local reference frame to construct distance- and orientation-dependent residue-based statistical potentials for proteins 10. In a subsequent work 4, it was shown that contacts between side-chains and main-chains are important, and a Cα-SC-Pep model was introduced to represent orientation dependence. In a more recent highly coarse-grained potential, called OPUS-Ca 8, orientation preference was introduced into a distance-dependent pairwise potential. In that case, the orientation dependence between two side-chains was described by the relative orientation between two Cα-Cβ vectors. It was found that inclusion of this effect improved the potential’s ability to recognize the native state and to improve Z-scores in decoy set tests. Orientation dependence for homodimeric 11 and heterodimeric 12 interactions among seven hydrophobic residues in water has also been included in an analytical modeling of potentials of mean force.
Although a certain degree of success in describing orientation dependence was achieved in the aforementioned work, there is still much room for improvement. Recently, a new type of potential, called OPUS-PSP, was developed to maximally capture the orientation dependence in side-chain interactions 13. OPUS-PSP is an orientation-dependent statistical all-atom potential derived from side-chain packing.
Here, we first briefly outline the general framework of OPUS-PSP, followed by the results of its performance on decoy set tests. Then, we will discuss a major application of OPUS-PSP on side-chain conformation modeling via a method called OPUS-Rota 14. Most importantly, based on the lessons learned from our own work and others, we will discuss issues and insights in the modeling of orientation dependence in molecular interactions.
Theoretical Framework of OPUS-PSP
OPUS-PSP is constructed from two major components: (a) a novel set of 19 rigid-body blocks that define the geometry of the interaction units, and (b) a knowledge-based energy function based on packing statistics of these blocks. In addition, a repulsive Lennard-Jones term is used to deter steric clashes. Coarse-graining and symmetry are also employed to improve the statistics.
Definitions of Rigid-body Blocks and Relative Orientation
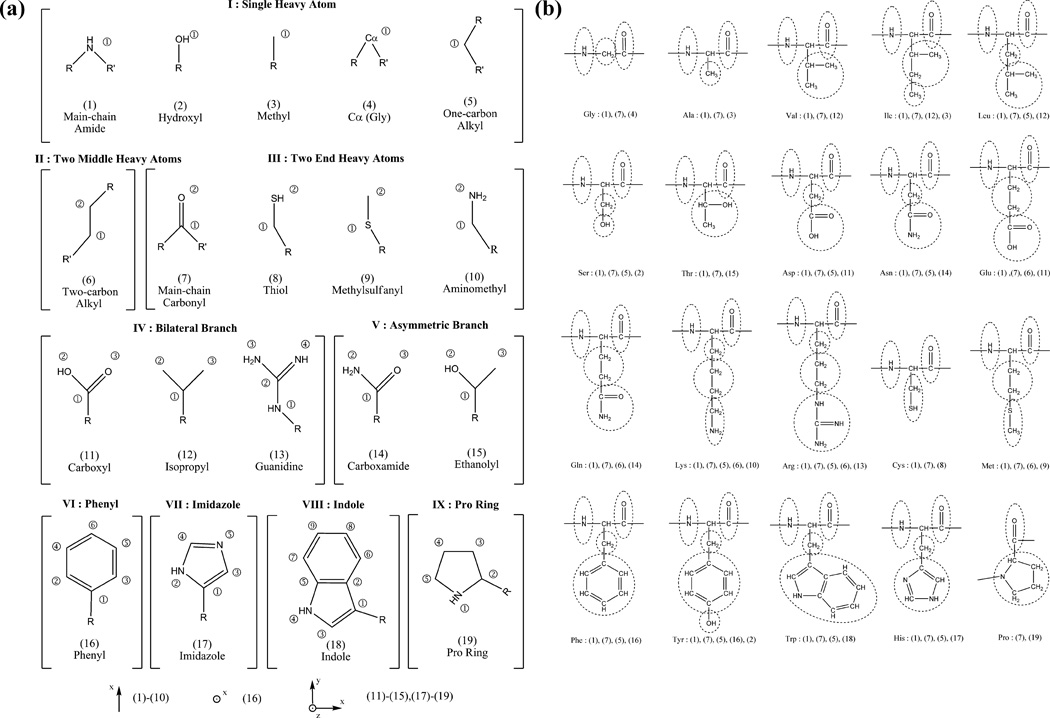
First, to form the basis set of interaction units, the chemical structures of 20 residues are decomposed into a set of 19 rigid-body blocks (shown in Fig.1a). Those blocks share three important characteristics: (a) all atoms in a block are chemically bonded and belong to the same residue, (b) each block is treated as a rigid body, (c) all non-hydrogen heavy atoms are assumed to be in the same plane. For the proline ring of block type 19, assumptions (b) and (c) are approximate, and we found that they are reasonable in constructing OPUS-PSP. Furthermore, the alpha carbon atoms of all residues except Pro and Gly are not included in the basis set. We do so by assuming that the heavily shielded alpha carbons have minimal influence on side-chain packing and our results support this assumption. In this representation, each residue contains more than one block, but each block appears only once in a single residue. Fig.1b shows the block compositions of the 20 residue types. For notational consistency, we shall denote residue types (20 total) with m and n, block types (19 total) with a and b, block indices with α and β, and atomic indices with i and j.
Figure 1.
Rigid-body blocks in OPUS-PSP. (a) Definition of 19 block types. Blocks are categorized into nine symmetry classes denoted by Roman numerals. Block classes I, II, III and VI are line shapes, and the others are plane shapes. R and R’ are not considered parts of the blocks but are shown to indicate connectivity only. The reference frames for line shapes and plane shapes are schematically shown alongside their corresponding block types at the bottom of the figure. (b) Block composition of residues. All blocks (block types denoted by numbers in parentheses and defined in Fig.1a) are circled for all amino acids. This figure is adopted from Fig.1 in reference 13.
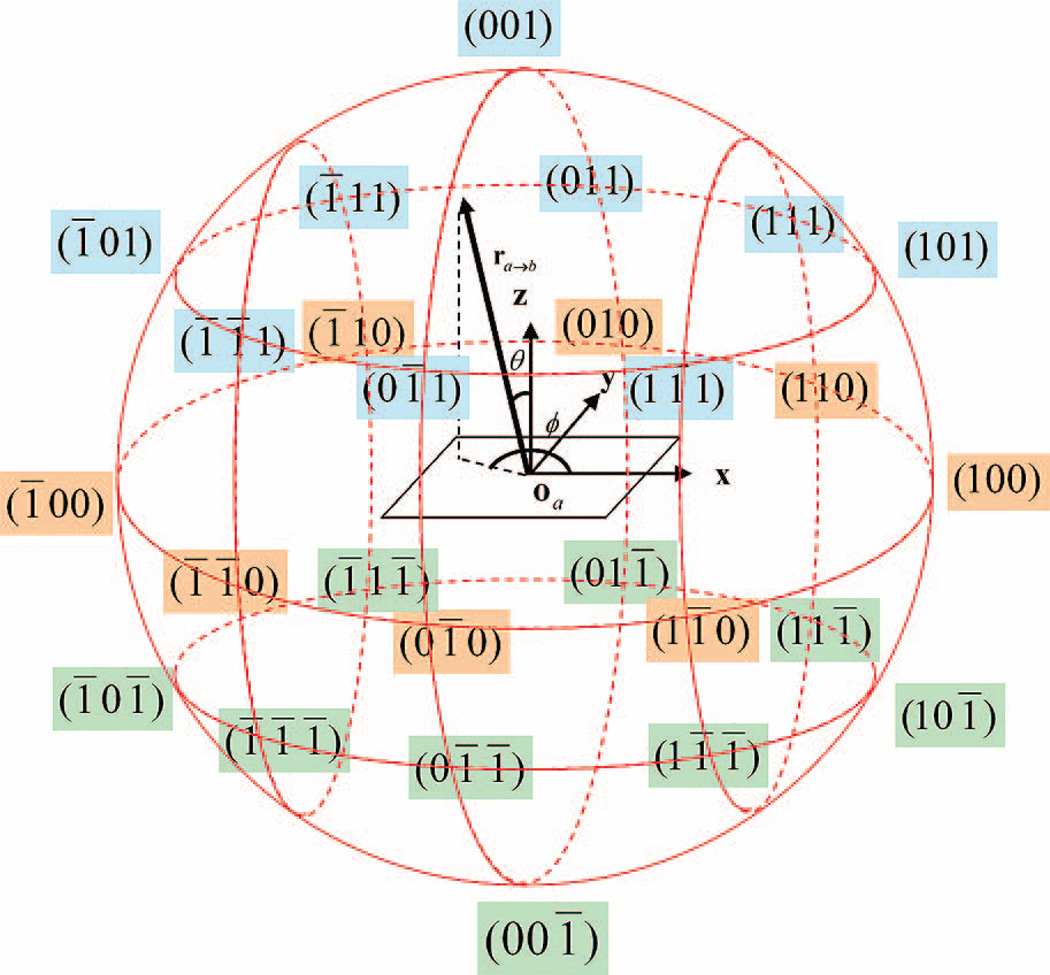
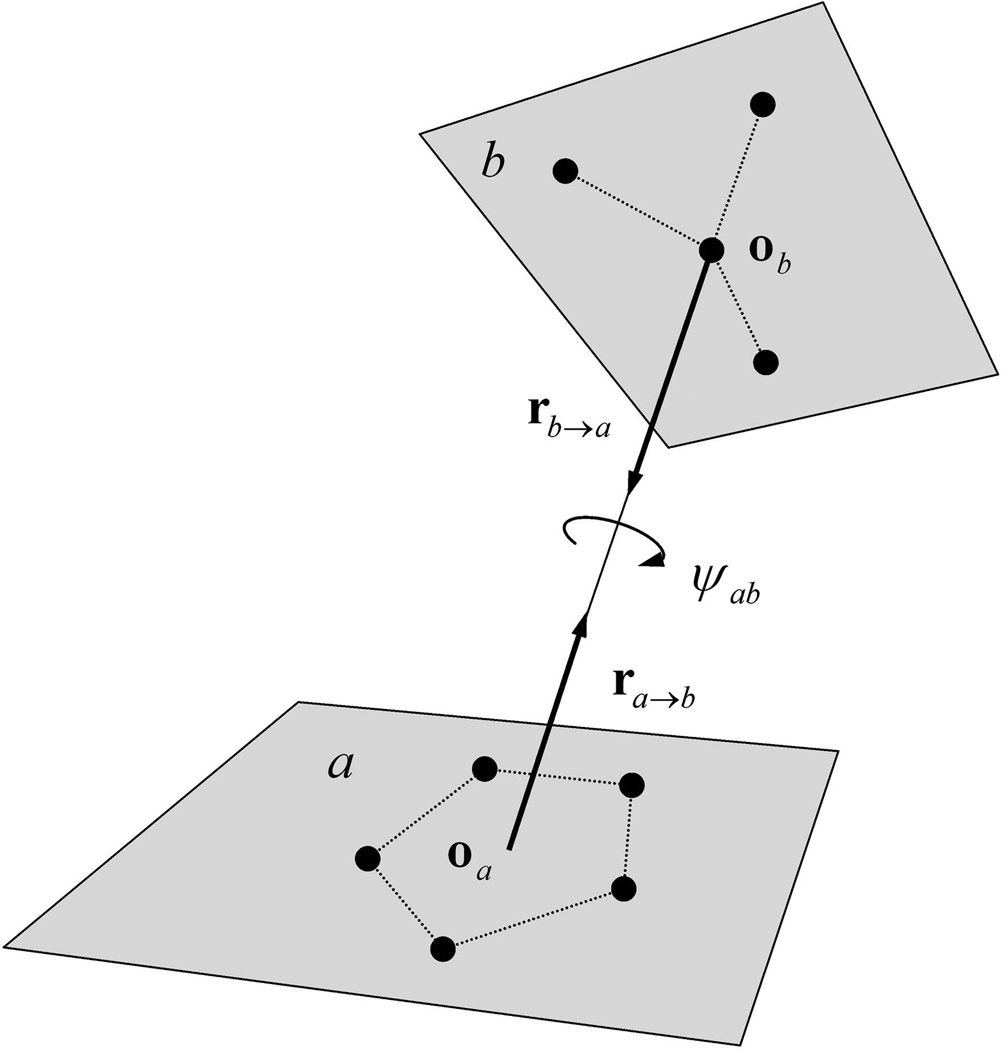
A special coordinate system is designed to define the relative orientation of a pair of blocks. As illustrated in Fig.2, the relative orientation of block types a and b is defined using three variables: two relative direction vectors ra→b and rb→a, and an inter-rotation angle ψab along the axis connecting the origins of the two blocks in their respective molecular reference frames. These coordinates describe the axial rotation around the line linking the origins of the two blocks and the pivot motion around the origin of each block, respectively. The relative orientation of a pair of blocks is completely defined by these three variables (computed in the laboratory reference frame), coupled with the molecular reference frame for each block.
Figure 2.
The definition of relative orientation of blocks in OPUS-PSP. If block types a and b are in contact, then ra→b and rb→a are the relative direction vectors and ψab is the inter-rotation angle along the axis connecting the origins oa and ob of the two blocks. This figure is adopted from Fig.2 in reference 13.
Energy Function
OPUS-PSP contains an orientation-dependent packing energy term Eorient and a repulsive energy term Erepul:
| (1) |
where wrepul is a weight parameter optimized against a small subset of decoy sets 13.
To calculate the first term, the total orientation-dependent packing energy Eorient , we first define the packing energy for a pair of blocks by,
| (2) |
Here, pobs is the probability of a particular orientation state for block types a and b in contact with respect to all observed contact states for any block pair extracted from the non-redundant structure database, and pref is the contact probability of all possible occurrences of that state without packing interactions (the reference state). The quantity Ωab = (ra→b,rb→a,ψab) designates the relative orientation of a and b, and kBT is the Boltzmann constant (set to unity). The value of Eorient is obtained by summing the packing energies of all pairs of blocks in contact (“block contact pairs”) between all pairs of non-consecutive residues:
| (3) |
Here, δ(α,β) is a delta function whose value is one when blocks α and β are in contact and zero otherwise, and B(α) = a maps block a to its block type a. The second term in Equ.3 is Eˆ(a,b) = n(a,b)E(Ωab,a,b), where n(a,b) is a weighting term for block size defined as the average number of pairs of heavy atoms in contact between block types a and b (we define an “atom contact pair” as two atoms whose pairwise distance is less than 5 Å). The weighting term is evaluated by random sampling in the manner of the reference state probability calculation. This is necessary because larger blocks contribute more atom contact pairs and therefore more energy. In calculating Eorient, the contribution is restricted to side-chain-side-chain and main-chain-side-chain interactions only. The main-chain-main-chain hydrogen bonding and other short-range interactions are not included.
The repulsive term Erepul is defined as:
| (4) |
where ELJ(i, j) is a repulsive (no attractive term) Lennard-Jones (LJ) potential for two atoms i and j. Like Eorient, the summation in the LJ term ignores interactions between pairs of main-chain atoms and between two atoms in the same residue. Note that Eorient and Erepul are typically orthogonal so over-counting is not an issue.
Coarse-graining of Orientation Bins and Symmetry
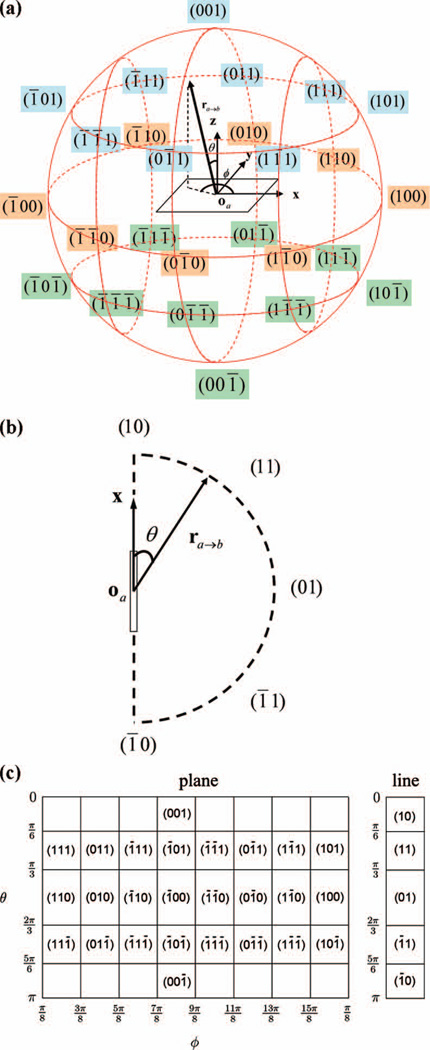
It is necessary to coarse-grain the orientation space and exploit the symmetry of the 19 blocks given the limited amount of non-homologous protein data available. As shown in Fig.1a, these blocks are classified into nine symmetry classes that belong to two basic groups: plane shapes (IV, V, VII-IX) and line shapes (I-III, VI). Note that VI is regarded as a line shape due to the six-fold axial symmetry of the phenyl ring.
For each plane-shaped block, the relative direction with respect to the molecular reference frame of the block is coarse-grained into 26 bins (illustrated in Fig.3a). For each line-shaped block, the cylindrical symmetry allows usage of five latitudinal bins (shown in Fig.3b). Fig.3c describes the θ and ϕ ranges of each relative direction bin. The inter-rotation angle is coarse-grained into four bins spanning π/2 radians each. In our study, we found that a choice of 26 directional bins is appropriate for plane-shaped blocks in order to balance the trade-off between the number of bins and the available structure data for statistical analysis.
Figure 3.
The definition of the relative direction bins for line-shaped and plane-shaped blocks in OPUS-PSP. (a) 26 relative direction bins for plane-shaped blocks (classes IV, V, VII-IX). Each bin is denoted by the index (nxnynz) and is derived from the spherical angles θ and ϕ of vector ra→b in the reference frame of block a. (b) 5 relative direction bins for line-shaped blocks (classes I-III, VI). Each bin is denoted by the index (nxny) and is derived from the angle θ between the primary axis (x-axis) and vector ra→b formed from the origin oa of block a to the origin ob of block b. (c) The direction bin indices plotted on a Mercator projection, for illustration only (a Mercator projection is a cylindrical map projection and the most common geographic map projection). The ranges for spherical angles θ and ϕ are indicated on the axes of the map. For plane shapes, the first or last row of the map represents a single bin at each of the poles rather than eight individual cells. The 5 bins for line shapes (on the right) areconsolidated from the 26 latitudinal bins of the plane shapes. This figure is adopted from Fig.3 in reference 13.
For two blocks in contact, the maximal number of bins is 26 × 4 × 26 = 2704. However, in practice, certain redundant bins are consolidated based on the intrinsic molecular symmetry of the blocks. This leads to a much smaller number of bins.
Performance of OPUS-PSP on Decoy Set Recognition
The performance of OPUS-PSP was examined in benchmark studies using the popular decoy set collections: Decoys ‘R’ Us 15, HR 16, Rosetta (and Rosetta2) 17,18, MOULDER 19, structal (http://dd.compbio.washington.edu/), and the decoy sets collected by Gilis 20, which we call the Gilis collection. The results are presented in Table 1. Out of all the benchmarks, only the MM-PBSA 21 and MJ_2005 potentials 7 outperformed OPUS-PSP on the structal decoy sets. These decoy sets contain decoys generated by comparative modeling of globins and immunoglobulins (60% of them have a Cα RMSD less than 2.5 Å from the native conformation). For the ig_structal and ig_structal_hires sets, OPUS-PSP can do better if main-chain interactions between pairs of block types {1,5,6,7} are also included in the total energy calculation.
Table 1.
OPUS-PSP performance on various decoy sets. (a) OPUS-PSP performance compared to other potentials. (b) OPUS-PSP performance on Decoys ‘R’ Us. This table is adopted from Table 1 in the original OPUS-PSP paper 13.
| (a) | ||
|---|---|---|
| Top 1/Total No.a | Mean Z | |
| Decoys ‘R’ Us 18,45–48 | ||
| OPUS-PSP | 31/34 | −5.37 |
| HPMF 49 | 29/32b | −4.18 |
| DOPE 39 | 28/32 | -- |
| MSE 50 | 21/23 | −5.78 |
| DFIRE 38 | 27/32 | −4.52 |
| MJ_2005 7 | 27/34 | −5.93 |
| DFIRE-SCM 51 | 23/32 | −4.36 |
| MM-PBSA 21 | 23/34 | −1.95 |
| DGR 52 | 21/25 | −5.25 |
| DWL 53 | 21/32 | −3.66 |
| TE13 54 | 14/25 | −3.53 |
| CALSP 55 | 15/25 | -- |
| Rosetta 6,18,56 | 14/32c | -- |
| MOULDER 19 | ||
| OPUS-PSP | 19/20 | −4.60 |
| DOPE | 19/20c | -- |
| Rosetta | 19/20c | -- |
| DFIRE | 19/20c | -- |
| DFIRE-SCM | 19/20c | -- |
| HR 16 | ||
| OPUS-PSP | 135/148 | −7.50 |
| HR 16 | 113/150 | -- |
| TE13 | 92/148d | -- |
| Rosetta (X-ray) 18 | ||
| OPUS-PSP | 37/41 | −6.56 |
| DFIRE | 31/41 | −3.91 |
| DFIRE-SCM | 33/41 | −4.90 |
| CALSP | 28/41 | −4.16 |
| Rosetta2 17,18 | ||
| OPUS-PSP | 23/41 | −2.71 |
| OPUS-PSP (X-ray) | 22/25 | −4.49 |
| DOPE | 11/41e | −1.50 |
| Rosetta 1+2 f (X-ray) 17,18 | ||
| OPUS-PSP | 34/35 | −6.76 |
| HPMF | 30/35 | −4.42 |
| hg_structal g | ||
| OPUS-PSP | 18/29 | −1.76 |
| MM-PBSA | 20/29 | −1.60 |
| MJ_2005 | 22/29 | −2.76 |
| ig_structal g | ||
| OPUS-PSP | 46/61h | −2.79 |
| MJ_2005 | 49/61 | −3.55 |
| ig_structal_hires g | ||
| OPUS-PSP | 19/20h | −3.03 |
| MJ_2005 | 19/20 | −4.31 |
| Gilis 20 | ||
| OPUS-PSP | 43/45 | −5.58 |
| (b) | ||||
|---|---|---|---|---|
| PDB code | Decoy set size | Rank | Z-score | |
| 4state_reduced | ||||
| 1 | 1ctf | 631 | 1 | −4.23 |
| 2 | 1r69 | 676 | 1 | −4.52 |
| 3 | 1sn3 | 661 | 1 | −5.35 |
| 4 | 2cro | 675 | 1 | −3.77 |
| 5 | 3icb | 654 | 1 | −2.72 |
| 6 | 4pti | 688 | 1 | −5.97 |
| 7 | 4rxn | 678 | 1 | −4.32 |
| fisa | ||||
| 8 | 1fc2 | 501 | 312 | 0.25 |
| 9 | 1hdd-C | 501 | 1 | −4.10 |
| 10 | 2cro | 501 | 1 | −5.05 |
| 11 | 4icb | 501 | 1 | −7.40 |
| fisa_casp3 | ||||
| 12 | 1bg8-A | 1201 | 1 | −6.01 |
| 13 | 1bl0 | 972 | 1 | −6.00 |
| 14 | 1eh2 | 2414 | 1 | −4.42 |
| 15 | 1jwe | 1408 | 1 | −7.95 |
| 16 | smd3 | 1201 | 1 | −6.73 |
| lattice_ssfit | ||||
| 17 | 1beo | 2001 | 1 | −9.58 |
| 18 | 1ctf | 2001 | 1 | −6.78 |
| 19 | 1dkt-A | 2001 | 1 | −6.75 |
| 20 | 1fca | 2001 | 1 | −6.13 |
| 21 | 1nkl | 2001 | 1 | −4.40 |
| 22 | 1pgb | 2001 | 1 | −7.79 |
| 23 | 1trl-A | 2001 | 1 | −4.81 |
| 24 | 4icb | 2001 | 1 | −5.95 |
| lmds | ||||
| 25 | 1b0n-B | 498 | 1 | −4.74 |
| 26 | 1bba | 501 | 501 | 3.66 |
| 27 | 1ctf | 498 | 1 | −8.99 |
| 28 | 1dtk | 216 | 1 | −6.07 |
| 29 | 1fc2 | 501 | 409 | 0.94 |
| 30 | 1igd | 501 | 1 | −7.77 |
| 31 | 1shf-A | 438 | 1 | −7.87 |
| 32 | 2cro | 501 | 1 | −7.17 |
| 33 | 2ovo | 348 | 1 | −5.87 |
| 34 | 4pti | 344 | 1 | −8.15 |
“Total No.” is the total number of decoy sets used for a specific decoy set collection, and this number may vary from study to study in the literature even for the same collection.
OPUS-PSP recognizes 30 of the 32 decoy sets used for HPMF
Results taken from 39
Results taken from 16
Results taken from 57
The total number of 35 is a subset of X-ray structures in the combined Rosetta and Rosetta2 collections
OPUSPSP includes main-chain interactions of block types { 1,5,6,7}.
OPUS-Rota: A Fast and Accurate Method for Side-chain Modeling
Side-chain conformation modeling is of the most severe bottlenecks in the high-accuracy refinement of computationally predicted structures. Aided by OPUS-PSP, OPUS-Rota 14 is a new method developed for such a purpose.
Rotamer libraries are most commonly and successfully used by side-chain modeling methods to reduce the space of conformations that must be sampled and there are many rotamer-based side-chain modeling method as summarized in the OPUS-Rota paper 14. In the rotamer approach, side-chain conformations are limited to a small set of most-likely positions (rotamers) taken from a rotamer library derived from X-ray structures.
Fast rotamer methods such as SCWRL 22 can quickly locate the global minimum by using a simple pair-wise energy function and dead-end elimination (DEE) 23,24. The accuracy of such methods is limited because the energy function used is over-simplified 25,26. Methods that use more accurate energy functions, such as NCN 27 and LGA 28, are significantly slower because of computationally expensive long-range and multi-body terms. High computational cost limits the application of these methods since the speed of execution in side-chain modeling is very important in the iterative process of structure prediction.
Brief Outline of the OPUS-Rota Algorithm
The total energy function used in OPUS-Rota has four terms:
| (5) |
Here Eorient is the side-chain packing potential OPUS-PSP 13, which is a short-range, pair-wise and coarse-grained all-atom potential that allows for fast and accurate energy evaluation during intensive sampling. The second term Evdw is a modified 6–12 Lennard-Jones (LJ) potential also used in OPUS-PSP, Erot is a term related to rotamer frequency, and Esolvation is a solvation energy term. The three weights worient = 0.15, wvdw =1.0 , and wsolvation = 0.1 are obtained by optimizing against a small set of high resolution structures.
The third rotamer frequency term Erot has the same form used in SCWRL 22. However, the contributions of bulky ring side-chains {Phe,Tyr,Trp,or His} are scaled up by a factor of three. The rotamer frequencies are taken from Dunbrack’s rotamer library 29.
Similar to what was used in the literature 30, the solvation energy Esolvation takes the form:
| (6) |
where Si is the solvent accessible surface area (SASA) of atom i, and Δσi is the atomic solvent parameter from Sharp et al.31. To rapidly calculate SASA, OPUS-Rota adopts the pair-wise approximation method of Zhang et al.32.
OPUS-Rota uses simulated annealing by heat bath Monte Carlo as a sampling method 33, which is able to rapidly identify near-native conformations when combined with neighbor list techniques and efficient energy updates. In OPUS-Rota, the move set for a given main-chain conformation is the collection of rotamer states from Dunbrack’s rotamer library 29, selected in order of highest to lowest probability until the cumulative probability reaches at least 99.5%. In this way, almost all possible rotamers can be sampled.
Performance of OPUS-Rota
The performance of OPUS-Rota was benchmarked with 65 high-resolution X-ray structures used in the literature 27,34. The analysis was carried out both for overall (all residues) and for core residues. Core residues are defined as residues with solvent accessible ratio below 17% (53.5% of residues are found to be core residues by this definition). The accuracy of χ1 is defined as the percentage of residues whose predicted χ1 dihedral is no more than 40° from the native value. The accuracy of χ1+2 is defined as the percentage of residues for which both χ1 and χ2 are in the 40° range.
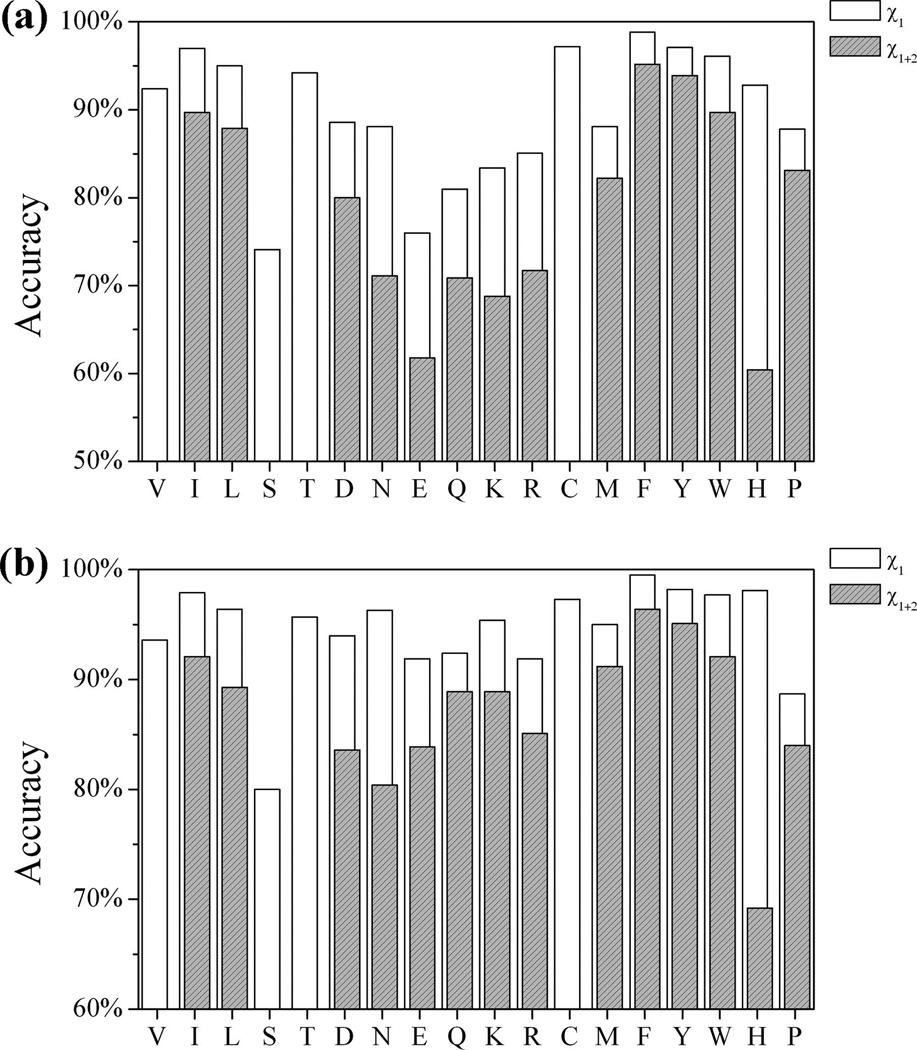
Fig.4 shows the accuracy of OPUS-Rota for each residue type. Serine has the lowest χ1 accuracy for all residues and for core residues. Polar and charged residues have lower χ1+2 accuracy, especially flexible surface residues. Hydrophobic and aromatic residues consistently have high accuracy except for His, which has high χ1 accuracy (overall ~93%) but low χ1+2 accuracy (overall ~60%, core ~70%). This is probably due to lack of knowledge of protonation states.
Figure 4.
The accuracy of OPUS-Rota for each residue type. (a) Overall χ1 and χ1+2 accuracies. (b) Core residue χ1 and χ1+2 accuracies (core residues are defined as the residues whose solvent accessible ratio is below a cutoff of 17%). This figure is adopted from Fig.2 in reference 14.
OPUS-Rota outperforms other related methods in terms of combined speed and accuracy. As shown in Table 2, on the 65-protein test set mentioned above, OPUS-Rota is much faster than all other methods except SCWRL 22, which is similar in speed. In addition, OPUS-Rota is much more accurate than SCRWL and comparably accurate with the rest. The computational efficiency of OPUS-Rota scales linearly with protein size.
Table 2.
The accuracy and speed of OPUS-Rota and several other side-chain modeling methods on the 65-protein test set. This table is adopted from Table 2 in the original OPUS-Rota paper 14.
| All residues | Core residuesa | Execution time | References | |||
|---|---|---|---|---|---|---|
| χ1(%) | χ1+2(%) | χ1(%) | χ1+2(%) | |||
| OPUS-Rota | 89.0 | 79.1 | 94.5 | 88.7 | 9.6 min c | |
| SCWRL | 83.6 | 70.3 | 88.8 | 79.2 | 2.2 min +5 hb,c | Ref. 22 |
| NCN | 89.3 | 77.5 | 94.1 | 87.4 | 24 h e | Ref. 27 |
| LGA | 88.5 | 74.1 | 93.7 | 84.6 | 14 h e | Ref. 28 |
| SPRUCE | 86.7 | 74.0 | 93.7 | 86.7 | 20 h d | Ref. 34 |
| Rosetta | 85.1 | 72.7 | 91.5 | 84.5 | 43.7 h c | Ref. 58 |
| SCAPorig f | 84.1 | 70.7 | 90.7 | 82.5 | 2.1 h c | Ref. 25 |
| SCAPmodi f | 83.1 | 70.1 | 91.4 | 84.0 | 24 h e | Ref. 27 |
Tests on OPUS-Rota, SCWRL, SPRUCE, Rosetta, and SCAPorig use the same definition of core residues (SPRUCE uses different solvent parameters and a different cutoff), while NCN, LGA and SCAPmodi define the core as having <20% accessible surface area in the native structure according to the method by Lee & Richards 59. All the definitions result in a similar portion of core residues ~53.5% 34.
SCWRL requires >5 hours for protein 1qlw, but only 2.2 minutes for the remaining 64 proteins.
Times for OPUS-Rota, SCWRL, Rosetta, and SCAPorig are for a single run on one Intel Xeon 2.8-GHz processor (by the software provided by the authors).
SPRUCE is run on one Intel Xeon 3.2-GHz processor 34.
Data for run times are from 27.
For real applications in structure prediction, both SCWRL and OPUS-Rota were also tested on the Wallner & Elofsson homology modeling benchmark set 35. It was found that OPUS-Rota performs consistently better than SCWRL when sequence identity is higher than 40% (see Fig.3 in reference 14). When sequence identity is lower than 40%, both methods have low accuracy, which is an expected result because the template structures are so far away from the target structures. This indicates that the quality of side-chain modeling heavily depends on the accuracy of the main-chain coordinates.
Discussion and Future Perspective
The most important feature of OPUS-PSP is its unique basis set of 19 rigid-body blocks that captures the essential elements of anisotropic orientation-dependent molecular interactions. OPUS-PSP is designed to maximally sense the change of relative orientation between two packed blocks, even when there is insignificant change in the packing distance. To the best of our knowledge, this is a feature that no other potential possesses.
OPUS-PSP is not a distance-dependent potential. The effect of packing distance between atoms is implicitly contained in its form. For example, if two blocks are in contact with native packing orientation, then the atomic contact criteria used in OPUS-PSP and the orientation parameters will restrict the distances between the atoms because of the fixed sizes of the blocks.
OPUS-PSP does not model solvation effects explicitly, but these effects are implicitly contained in its form as well; e.g., hydrophobic blocks will surely prefer to pack against each other. Although OPUS-PSP may be used in combination with other solvation models if necessary, it may be advantageous to avoid modeling explicit solvation effects in other cases. For example, in modeling membrane protein packing, OPUS-PSP may have an edge relative to other methods as the solvation dependence in this case may be very different from that of soluble proteins. Even though OPUS-PSP is constructed from a structure database of soluble proteins, the microenvironments of side-chain packing in membrane proteins should be similar to those of soluble proteins.
In constructing any statistical potential, the choice of reference state is very important 36,37. The Boltzmann expression in Equ. 2 is a general way of developing the potential, and the accuracy of the potential can be improved by proper modeling of either pobs or pref , or both. The significance of the choice of pref is evident in the development of the DFIRE 38 and DOPE 39 potentials. In OPUS-PSP, both pobs and pref are modeled very differently, in which case the statistics of pobs are generated based on the 19-block basis set, and those of pref are generated by self-avoided random sampling of blocks with different sizes 13. OPUS-PSP is also the first potential in which the geometry of interacting groups is explicitly considered in constructing the reference state.
OPUS-PSP is presently a discrete potential. In principle, it can be extended in two different ways. The first is to transform the discrete potential into a square-well potential and use it as a native contact potential between blocks. This is advantageous because the 19 blocks are expected to capture the essential elements of molecular interactions in an orientation-sensitive fashion. Such a contact potential can be combined with a funnel-like molecular mechanics potentials. In this way, OPUS-PSP may be used essentially as a bias to deepen the native state energy well without altering the long range interactions. Note, the contact potential is short-range in nature, i.e., only sensitive to native-like packing patterns between blocks. The second is to revise OPUS-PSP to be continuous so that derivatives can be obtained for molecular simulation 40. However, a substantial re-parameterization may be needed to achieve this.
A distinct feature of OPUS-PSP is that the interactions between pure main-chain atoms are excluded. However, many other studies showed that those interactions are important and highly correlated with the side-chain interactions 4,5,41,42. Thus, revising the block basis set and including main-chain atoms may be directions for future improvement.
OPUS-PSP is a pairwise potential that allows for very rapid computational evaluation. This feature is critically important for some applications such as the side-chain conformational modeling method OPUS-Rota 14. Along with its strong overall performance, OPUS-Rota performs particularly well in modeling aromatic side-chains due to several design features. First, the contributions of aromatic residues in the rotamer frequency term are enhanced. Second, the vdW potential is softened for aromatic side-chains, which enables the aromatic side-chains to find their preferred rotamer angles, especially inside the densely-packed protein core. Third, OPUS-PSP is inherently more sensitive to the orientation of the aromatic planes.
A major challenge in side-chain modeling is the issue of main-chain flexibility. The most successful methods, including OPUS-Rota, perform well when the main-chain is in its native conformation, yet the accuracy of side-chain placement decreases quickly once the main-chain deviates from its native state. There is of course a question of the significance of “native state” side-chain placement if the main-chain is not in its native state. Main-chain and side-chain states are tightly coupled; if one is not in its native state, neither will the other. Thus, the ultimate way to solve this problem is to refine the main-chain and side-chain simultaneously 43,44. There is another issue of causality between the main-chain and side-chain conformations. Most prediction methods try to position the main-chain first and then place the side-chains afterward. In reality, however, it is not unreasonable to assume that the main-chain conformation is dramatically influenced by side-chain packing. This is clear from the success of OPUS-PSP in decoy set recognition. OPUS-PSP does not explicitly account for pure main-chain interactions, yet it can consistently and accurately recognize the native state out of a large number of decoys. This result seems to imply that side-chain packing is crucial for native state formation, i.e., it is difficult to form a perfectly native protein backbone without having all the side-chains in place. This is also in line with the common observation that main-chain hydrogen bonding interactions are not specific, as any pair of residues can form hydrogen bonds, while only specific pairs of side-chains can be packed together favorably.
Acknowledgements
The author thanks financial support of grants from the National Institutes of Health (R01-GM067801), the National Science Foundation (MCB-0818353), and the Welch Foundation (Q-1512). Critical reading of manuscript by Athanasios D. Dousis and helpful discussion with Mingyang Lu are acknowledged.
References
- 1.Skolnick J. In quest of an empirical potential for protein structure prediction. Curr Opin Struct Biol. 2006;16:166–171. doi: 10.1016/j.sbi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Bahar I, Jernigan RL. Coordination geometry of nonbonded residues in globular proteins. Fold Des. 1996;1:357–370. doi: 10.1016/S1359-0278(96)00051-X. [DOI] [PubMed] [Google Scholar]
- 3.Liwo A, Oldziej S, Pincus MR, Wawak RJ, Rackovsky S, Scheraga HA. A united-residue force field for off-lattice protein-structure simulations .I. Functional forms and parameters of long-range side-chain interaction potentials from protein crystal data. Journal of Computational Chemistry. 1997;18:849–873. [Google Scholar]
- 4.Buchete NV, Straub JE, Thirumalai D. Orientational potentials extracted from protein structures improve native fold recognition. Protein Sci. 2004;13:862–874. doi: 10.1110/ps.03488704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee A, Bhimalapuram P, Bagchi B. Orientation-dependent potential of mean force for protein folding. Journal of Chemical Physics. 2005;123:014901. doi: 10.1063/1.1940058. [DOI] [PubMed] [Google Scholar]
- 6.Misura KM, Morozov AV, Baker D. Analysis of anisotropic side-chain packing in proteins and application to high-resolution structure prediction. J Mol Biol. 2004;342:651–664. doi: 10.1016/j.jmb.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 7.Miyazawa S, Jernigan RL. How effective for fold recognition is a potential of mean force that includes relative orientations between contacting residues in proteins? J Chem Phys. 2005;122:024901. doi: 10.1063/1.1824012. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Lu M, Chen M, Li J, Ma J OPUS-Ca. A Knowledge-based Potential Function Requiring Only Cα Positions. Prot. Sci. 2007;16:1449–1463. doi: 10.1110/ps.072796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchete NV, Straub JE, Thirumalai D. Dissecting contact potentials for proteins: Relative contributions of individual amino acids. Proteins-Structure Function and Bioinformatics. 2008;70:119–130. doi: 10.1002/prot.21538. [DOI] [PubMed] [Google Scholar]
- 10.Buchete N-V, Straub JE, Thirumalai D. Anisotropic coarse-grained statistical potentals improve the ability to identify native-like protein structures. J. Chem. Phys. 2003;118:7658–7671. [Google Scholar]
- 11.Makowski M, Sobolewski E, Czaplewski C, Liwo A, Oldziej S, No JH, Scheraga H. A. Simple physics-based analytical formulas for the potentials of mean force for the interaction of amino acid side chains in water. 3. Calculation and parameterization of the potentials of mean force of pairs of identical hydrophobic side chains. J Phys Chem B. 2007;111:2925–2931. doi: 10.1021/jp065918c. [DOI] [PubMed] [Google Scholar]
- 12.Makowski M, Sobolewski E, Czaplewski C, Oldziej S, Liwo A, Scheraga HA. Simple physics-based analytical formulas for the potentials of mean force for the interaction of amino acid side chains in water. IV. Pairs of different hydrophobic side chains. J Phys Chem B. 2008;112:11385–11395. doi: 10.1021/jp803896b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu M, Dousis A, Ma J OPUS-PSP. An Orientation-dependent Statistical All-atom Potential Derived from Side-chain Packing. J. Mol. Biol. 2008;376:288–301. doi: 10.1016/j.jmb.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu M, Dousis AD, Ma J. OPUS-Rota: a fast and accurate method for side-chain modeling. Protein Sci. 2008;17:1576–1585. doi: 10.1110/ps.035022.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samudrala R, Levitt M. Decoys 'R' Us: a database of incorrect conformations to improve protein structure prediction. Protein Sci. 2000;9:1399–1401. doi: 10.1110/ps.9.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajgaria R, McAllister SR, Floudas CA. A novel high resolution Calpha--Calpha distance dependent force field based on a high quality decoy set. Proteins. 2006;65:726–741. doi: 10.1002/prot.21149. [DOI] [PubMed] [Google Scholar]
- 17.Tsai J, Bonneau R, Morozov AV, Kuhlman B, Rohl CA, Baker D. An improved protein decoy set for testing energy functions for protein structure prediction. Proteins. 2003;53:76–87. doi: 10.1002/prot.10454. [DOI] [PubMed] [Google Scholar]
- 18.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 19.John B, Sali A. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003;31:3982–3992. doi: 10.1093/nar/gkg460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilis D. Protein decoy sets for evaluating energy functions. J Biomol Struct Dyn. 2004;21:725–736. doi: 10.1080/07391102.2004.10506963. [DOI] [PubMed] [Google Scholar]
- 21.Lee MC, Yang R, Duan Y. Comparison between Generalized-Born and Poisson-Boltzmann methods in physics-based scoring functions for protein structure prediction. J Mol Model. 2005;12:101–110. doi: 10.1007/s00894-005-0013-y. [DOI] [PubMed] [Google Scholar]
- 22.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein RF. Efficient rotamer elimination applied to protein side-chains and related spin glasses. Biophys J. 1994;66:1335–1340. doi: 10.1016/S0006-3495(94)80923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desmet J, Maeyer MD, Hazes B, Lasters I. The dead-end elimination theorem and its use in protein side-chain positioning. Nature. 2002;356–542:539. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- 25.Xiang Z, Honig B. Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol. 2001;311:421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- 26.Hartmann C, Antes I, Lengauer T IRECS. A new algorithm for the selection of most probable ensembles of side-chain conformations in protein models. Protein Sci. 2007;16:1294–1307. doi: 10.1110/ps.062658307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson RW, Dutton PL, Wand AJ. Improved side-chain prediction accuracy using an ab initio potential energy function and a very large rotamer library. Protein Sci. 2004;13:735–751. doi: 10.1110/ps.03250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang S, Grishin NV. Side-chain modeling with an optimized scoring function. Protein Sci. 2002;11:322–331. doi: 10.1110/ps.24902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunbrack RL, Jr, Karplus M. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J Mol Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg D, McLachlan AD. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 31.Sharp KA, Nicholls A, Friedman R, Honig B. Extracting hydrophobic free energies from experimental data: relationship to protein folding and theoretical models. Biochemistry. 1991;30:9686–9697. doi: 10.1021/bi00104a017. [DOI] [PubMed] [Google Scholar]
- 32.Zhang N, Zeng C, Wingreen NS. Fast accurate evaluation of protein solvent exposure. Proteins. 2004;57:565–576. doi: 10.1002/prot.20191. [DOI] [PubMed] [Google Scholar]
- 33.Newman MEJ, Barkema GT. Monte Carlo methods in statistical physics; Clarendon Press. Oxford, New York: Oxford University Press; 1999. [Google Scholar]
- 34.Jain T, Cerutti DS, McCammon JA. Configurational-bias sampling technique for predicting side-chain conformations in proteins. Protein Sci. 2006;15:2029–2039. doi: 10.1110/ps.062165906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallner B, Elofsson A. All are not equal: A benchmark of different homology modeling programs. Protein Science. 2005;14:1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Betancourt MR, Thirumalai D. Pair potentials for protein folding: Choice of reference states and sensitivity of predicted native states to variations in the interaction schemes. Protein Science. 1999;8:361–369. doi: 10.1110/ps.8.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen WW, Shakhnovich EI. Lessons from the design of a novel atomic potential for protein folding. Protein Sci. 2005;14:1741–1752. doi: 10.1110/ps.051440705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Summa CM, Levitt M. Near-native structure refinement using in vacuo energy minimization. Proc Natl Acad Sci U S A. 2007;104:3177–3182. doi: 10.1073/pnas.0611593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose GD, Fleming PJ, Banavar JR, Maritan A. A backbone-based theory of protein folding. Proc Natl Acad Sci U S A. 2006;103:16623–16633. doi: 10.1073/pnas.0606843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzgerald JE, Jha AK, Colubri A, Sosnick TR, Freed KF. Reduced C(beta) statistical potentials can outperform all-atom potentials in decoy identification. Protein Sci. 2007;16:2123–2139. doi: 10.1110/ps.072939707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgiev I, Donald BR. Dead-end elimination with backbone flexibility. Bioinformatics. 2007;23:I185–I194. doi: 10.1093/bioinformatics/btm197. [DOI] [PubMed] [Google Scholar]
- 44.Li G, Liu Z, Guo J, Xu Y. An algorithm for simultaneous backbone threading and side-chain packing. Algorithmica. 2008;51:435–450. [Google Scholar]
- 45.Park B, Levitt M. Energy functions that discriminate X-ray and near native folds from well-constructed decoys. J Mol Biol. 1996;258:367–392. doi: 10.1006/jmbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- 46.Samudrala R, Xia Y, Levitt M, Huang ES. A combined approach for ab initio construction of low resolution protein tertiary structures from sequence. Pac Symp Biocomput. 1999:505–516. doi: 10.1142/9789814447300_0050. [DOI] [PubMed] [Google Scholar]
- 47.Xia Y, Huang ES, Levitt M, Samudrala R. Ab initio construction of protein tertiary structures using a hierarchical approach. J Mol Biol. 2000;300:171–185. doi: 10.1006/jmbi.2000.3835. [DOI] [PubMed] [Google Scholar]
- 48.Keasar C, Levitt M. A novel approach to decoy set generation: designing a physical energy function having local minima with native structure characteristics. J Mol Biol. 2003;329:159–174. doi: 10.1016/S0022-2836(03)00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin MS, Fawzi NL, Head-Gordon T. Hydrophobic potential of mean force as a solvation function for protein structure prediction. Structure. 2007;15:727–740. doi: 10.1016/j.str.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 50.McConkey BJ, Sobolev V, Edelman M. Discrimination of native protein structures using atom-atom contact scoring. Proc Natl Acad Sci U S A. 2003;100:3215–3220. doi: 10.1073/pnas.0535768100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C, Liu S, Zhou H, Zhou Y. An accurate, residue-level, pair potential of mean force for folding and binding based on the distance-scaled, ideal-gas reference state. Protein Sci. 2004;13:400–411. doi: 10.1110/ps.03348304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dehouck Y, Gilis D, Rooman M. A new generation of statistical potentials for proteins. Biophys J. 2006;90:4010–4017. doi: 10.1529/biophysj.105.079434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong Q, Wang X, Lin L. Novel knowledge-based mean force potential at the profile level. BMC Bioinformatics. 2006;7:324. doi: 10.1186/1471-2105-7-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tobi D, Elber R. Distance-dependent, pair potential for protein folding: results from linear optimization. Proteins. 2000;41:40–46. [PubMed] [Google Scholar]
- 55.Zhang J, Chen R, Liang J. Empirical potential function for simplified protein models: combining contact and local sequence-structure descriptors. Proteins. 2006;63:949–960. doi: 10.1002/prot.20809. [DOI] [PubMed] [Google Scholar]
- 56.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 57.Colubri A, Jha AK, Shen MY, Sali A, Berry RS, Sosnick TR, Freed KF. Minimalist representations and the importance of nearest neighbor effects in protein folding simulations. J Mol Biol. 2006;363:835–857. doi: 10.1016/j.jmb.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 58.Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein Sci. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]