Abstract
Cytosolic sulfotransferases catalyze the sulfonation of hormones, metabolites, and xenobiotics. Many of these proteins have been shown to form homo- and heterodimers. An unusually small dimer interface was previously identified by Petrotchenko et al. (FEBS Lett 490, 39-43, 2001) by crosslinking, protease digestion, and mass spectrometry, and verified by site-directed mutagenesis. Analysis of the crystal packing interfaces in all 28 available crystal structures consisting of 17 crystal forms shows that this interface occurs in all of them. With a small number of exceptions, the publicly available databases of biological assemblies contain either monomers or incorrect dimers. Even crystal structures of mouse SULT1E1, which is a monomer in solution, contain the common dimeric interface, although distorted and missing two important salt bridges.
Introduction
Sulfonation is the process of transferring sulfonate to organic molecules, including drugs, xenobiotics, hormones, and proteins. There are two broad classes of enzymes that catalyze the sulfonation reaction -- the membrane sulfotransferases and cytosolic transferases. A nomenclature for the cytosolic enzymes has been provided by Blanchard et al.1. Within this nomenclature, all such proteins are called “SULT” followed by a family identifier (numbers 1, 2, etc.), a subfamily identifier (letters A, B, etc.), an isoform identifier, for proteins encoded by different genes (numbers 1, 2, etc.), and in some cases a splice-form identifier (“_v1”, “_v2”, etc.). Families were defined by a minimum sequence identify of 45% and subfamilies by a minimum sequence identity of 60%. Many branches of the superfamily have been identified in plants and animals, not all of which exist in humans. The human genes comprise SULT1A1, SULT1A2, SULT1A3, SULT1A4, SULT1B1, SULT1C2, SULT1C3, SULT1C4, SULT1E1, SULT2A1, SULT2B1, and SULT4A1.1,2 The SULT3, SULT5, and SULT6 families do not exist in humans but are present in mice and other species. A total of 28 structures of 13 different gene products of this family have been determined by X-ray crystallography. The fold bears remote similarity to nucleotide kinases. Sulfatases are similar to alkaline phosphatases, indicating an evolutionary relationship between sulfonation and phosphorylation3.
Most of the cytosolic sulfotransferases exist as dimers, including both homodimers and heterodimers, although the physiological significance of the dimer is not known. Experimental data indicating dimer formation are available for human SULT1A14, rat SULT1A15,6, human SULT1A37, human SULT1E18, Guinea pig SULT1E19, hamster SULT2A110, human SULT2A111, rat SULT2A112, human SULT2A313, and C. elegans ST114. Petrotchenko et al.8 used a combination of cross-linking reagents, protease digestion, and mass spectrometry to identify peptides involved in the dimerization interface of human SULT1A3. The peptides were consistent with a small dimer interface that they observed in the X-ray structures of human SULT1A313 and human SULT1A115 then available. This symmetric interface consists of two anti-parallel extended backbone segments with four backbone-backbone hydrogen bonds between them, and two salt bridges formed by the first residue of the motif (Lys/Arg) from one protein and the last residue of the ten-residue motif (Glu) from its partner in the dimer and vice versa. Two residues in the center of the motif (Thr-Val at positions 5 and 6) make strong symmetric contacts between the monomers. Thus they dubbed the 10-residue region the KTVE motif (residues 1, 5, 6, 10), which are the predominant residues at these positions in cytosolic sulfotransferases. Dimeric human SULT1E1 contains the same residues at these positions, while monomeric mouse SULT1E1 contains the sequence PE in place of TV. Mutation of TV to PE in human SULT1E1 produced a monomer, while mutations of mouse PE to TV created a homodimer, establishing the validity of the motif as the dimerization interface.8
Recently, we performed an extensive study of protein crystals across protein families, and identified interfaces common to large numbers of different crystal forms in individual families16. Using a benchmark of known monomers and dimers/oligomers, we established that the observation of an interface across a small number of crystal forms of homologous but non-identical proteins (<90% identity) indicates that the interface is likely to have biological significance. In this paper, we show that the Petrotchenko interface is observed in all crystal structures of the cytosolic sulfotransferases, and that it is not present in the distantly related heparan sulfotransferases or retinol dehydratases. Strangely, even mouse SULT1E1 contains the Petrotchenko dimer in its crystal but with a distorted geometry such that the N/C-terminal salt bridges within the motif are not formed. We also show that the database annotations for the biological assemblies of these proteins are diverse and in nearly all cases do not correspond to the interface identified by Petrotchenko et al.
Methods
The program MolIDE, version 1.617,18, was used to search the Uniref100 database19 with PSI-BLAST20 for sequences related to human SULT1E1. The position-specific scoring matrices from this search were used to search the sequences of proteins of known structure in the Protein Data Bank for proteins related to cytosolic sulfotransferases.
Neighbors in the crystals of proteins in the asymmetric unit of the crystal structures were constructed using crystallographic symmetry operations. Crystal forms were differentiated from one another as described in our recent paper16. This differentiation was performed by comparing interfaces in each crystal with those in the others. Crystals that share most or all interfaces were classified as a single crystal form. Conversely those with different interfaces were considered separate crystal forms, even if they shared the same crystal symmetry space group. Once the different crystal forms were separated, the unique interfaces in each form were compared to those in the others to identify common interfaces among the crystal forms. Coordinate files for these dimers were then saved for visual inspection. The program NACCESS21 was used to calculated buried surface area in each dimer.
Information on the biological units or assemblies were obtained from three publicly available resources using the program ProtBuD22: the PDB itself23; the Protein Quaternary Server (PQS)24, and PISA25 and compared. In the case of the PDB, biological units were built using the symmetry operators contained in the XML versions of the PDB entries26. In two cases, these differed from the biological unit coordinates obtained from the RCSB Protein Data Bank website for unknown reasons.
Results
A PSI-BLAST search within the program MolIDE was used to identify cytosolic sulfotransferases in the PDB starting with the sequence of human SULT1E1. This resulted in a total of 41 PDB entries. 28 of these proteins are cytosolic sulfotransferases, and 13 are other proteins with similar folds including the heparan sulfotransferase, a bacterial sulfotransferase, StaL, and retinol dehydratase. An analysis of the 28 crystal structures of the cytosolic sulfotransferases is presented in Table I. The structures comprise 17 different crystal forms of 11 different proteins (excluding the splice variants) and are sorted in the table by their crystal forms. In two cases, different proteins crystallized into the same crystal forms.
Table I.
PDB entries of cytosolic sulfotransferases.
| PDB | Species | Gene | Space Group | CF# | ASU | PDB-BU | PQS-BU | PISA-BU | Surface area |
|---|---|---|---|---|---|---|---|---|---|
| 3bfx | Human | SULT1C2 | P 1 2 1 | 1 | D (455) | D (455) * | M | M | 387, 376 |
| 1j99 | Human | SULT2A1 | P 21 21 2 | 2 | M | M | M | M | 372 |
| 1ov4 | Human | SULT2A1 | P 21 21 2 | 2 | M | D (365) | M | M | 365 |
| 2d06 | Human | SULT1A1 | P 21 21 2 | 3 | D (373) | M | M | M | 373 |
| 2a3r | Human | SULT1A3 | P 21 21 2 | 3 | D (226) | M | M | M | 368 |
| 2ad1 | Human | SULT1C4 | P 32 2 1 | 4 | M | M | M | M | 375 |
| 1cjm | Human | SULT1A3 | P 32 2 1 | 4 | M | M | D (544) | M | 371 |
| 1aqu | Mouse | SULT1E1 | P 21 21 2 | 5 | D (705) | D (705) | M | D (705) | 317, 308 |
| 1aqy | Mouse | SULT1E1 | P 21 21 2 | 5 | D (729) | D (729) | T (729, 534, 413) | D (729) | 413, 534 |
| 1bo6 | Mouse | SULT1E1 | P 21 21 2 | 5 | D (757) | D (757) | D (757) | D (757) | 353, 343 |
| 1ls6 | Human | SULT1A1 | P 21 21 2 | 6 | M | M | M | M | 382 |
| 1z28 | Human | SULT1A1 | P 21 21 2 | 6 | M | M | M | M | 378 |
| 1z29 | Human | SULT1A2 | C 1 2 1 | 7 | M | M | M | M | 381 |
| 1zd1 | Human | SULT4A1 | C 1 2 1 | 8 | D (208.05) | M | M | M | 385 |
| 2q3m | Arabidopsis | FLAVONOL SULT | C 2 2 21 | 9 | M | M | D (1750) | D (1750) | 358 |
| 1q44 | Arabidopsis | FLAVONOL SULT | C 2 2 21 | 9 | M | M | D (1776) | D (1776) | 371 |
| 2h8k | Human | SULT1C3 VARIANT D | C 2 2 21 | 10 | D (326) | D (326) | M | M | 326 |
| 2gwh | Human | SULT1C4 | I 4 2 2 | 11 | D (379) | D (379) | M | M | 379 |
| 1g3m | Human | SULT1E1 | P 1 21 1 | 12 | D (373) | D (373) | M | M | 373 |
| 1hy3 | Human | SULT1E1 | P 1 21 1 | 12 | D (312) | D (312) | M | M | 312 |
| 3ckl | Human | SULT1B1 | P 1 21 1 | 13 | D (1103) | D (1103) * | M | D(1103) | 379 |
| 1efh | Human | SULT2A1 | P 21 21 21 | 14 | D (840) | D (840) | D (840) | D (840) | 364 |
| 2z5f | Human | SULT1B1 | P 21 21 21 | 15 | D (565) | D (565) | M | M | 387 |
| 1q1q | Human | SULT2B1 ISO A | P 41 21 2 | 16 | M | M | M | M | 357 |
| 1q1z | Human | SULT2B1 ISO B | P 41 21 2 | 16 | M | M | M | M | 364 |
| 1q20 | Human | SULT2B1 ISO B | P 41 21 2 | 16 | M | M | M | M | 368 |
| 1q22 | Human | SULT2B1 ISO B | P 41 21 2 | 16 | M | M | M | M | 363 |
| 2reo | Human | SULT1C3 | P 61 2 2 | 17 | M | M | M | M | 385 |
Gene names are as given in Uniprot (http://uniprot.org). CF# designates each unique crystal form in arbitrary order. ASU is the asymetric unit size (M=monomer; D=dimer; T=tetramer). PDB-BU, PQS-BU, and PISA-BU are the biological assemblies provided by the PDB, PQS, and PISA respectively. Surface areas in Å2 for the interfaces in the ASU and BUs are given for the dimers and tetramers. Surface area in the last column is the surface area of the Petrotchenko dimers, as shown in Figure 1. Oligomers that contain the Petrotchenko dimer in the ASU and BUs are shown in bold italic type.
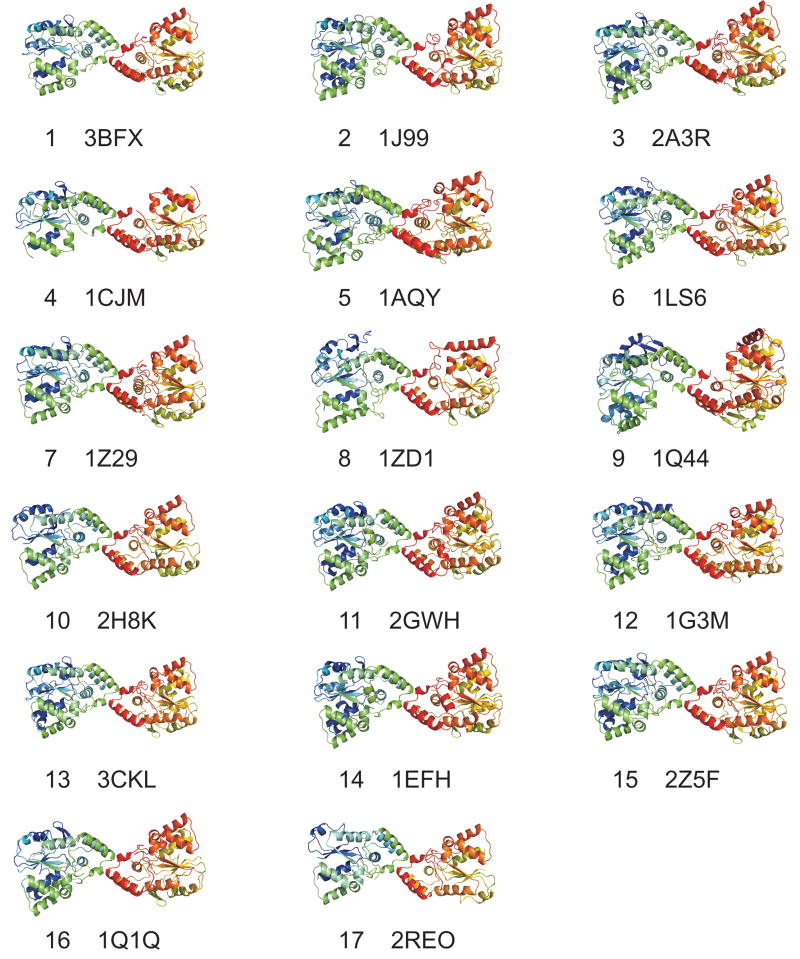
Using the method described in Xu et al.16, we compared the protein-protein interactions in these crystals to identify any common interfaces that they might contain. This method uses protein structure alignment to identify corresponding residues in homologous proteins, and a function Q which expresses the fraction of residue-residue interactions in one interface that are in common with another. Several small interfaces (200-400 Å2) were found in common between only two crystal forms each, and most of these were found only in identical sequences (data not shown). However, one interface was present in all 17 crystal forms of the cytosolic sulfotransferases. It was not present in the heparan sulfotransferase, StaL, or retinol dehydratase crystals. The common dimer structures for one member of each crystal form are shown in Figure 1. Their surface areas are given in the last column of Table I. These interfaces correspond to that identified by Petrotchenko et al. from crosslinking, protease digestion, and mass spectrometry on SULT1A3.
Figure 1.
Interfaces common among all crystals in the PDB of cytosolic sulfotransferases, numbered by crystal form as listed in Table I.
Table I also contains information on the asymmetric units and biological units provided by three publicly available sources: the PDB itself in their biological unit description in the XML version26 of PDB entries; the Protein Quaternary Server (PQS)24 from the EBI; and PISA25, also from the EBI. The PDB contains biological units as hypothesized by the authors of each structure. PQS is a mixture of manually annotated and automatically annotated biological assemblies. PISA assemblies are automatically determined on the basis of estimated chemical thermodynamic parameters of enthalpy and entropy. In this table, “M” stands for monomer, “D” for dimer, and “T” for tetramer. Those in bold italic type contain the common dimer found in all 17 crystal forms. The surface areas for all non-monomeric structures are also given in parentheses. The common dimer occurs in the asymmetric units of only 5 of 28 crystal structures. In four of these five, it is also in the PDB's biological unit, while it also appears in the PDB biological unit of one other entry, 1OV427. PQS has the common dimer in only one entry, 1AQY (mouse SULT1E1) as part of a larger tetramer. PISA does not identify the common interface in any of its biological assemblies for the cytosolic sulfotransferases.
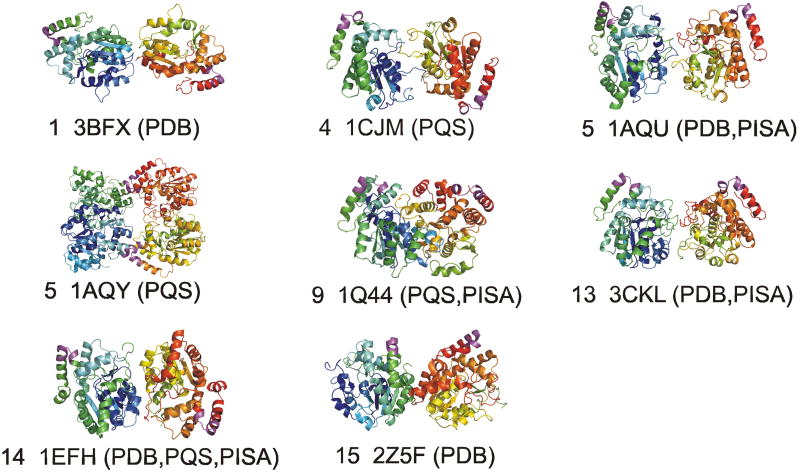
On the other hand, the PDB has seven incorrect dimers in its biological units and all of these are identical to the asymmetric unit dimers. This indicates that it is common for crystallographers to make the unwarranted assumption that the asymmetric unit assembly corresponds to the biological assembly. PQS has done this twice, while also creating three other dimers from monomeric asymmetric units. PISA has 5 non-biological dimers identical to the asymmetric units, and two constructed from asymmetric monomers. Incorrect dimers or tetramers for PDB, PQS, and PISA are shown in Figure 2. In each image, the biological interface is colored in magenta in each monomer. There is little similarity among the different dimers, despite high sequence identity among the proteins involved (in the range of 40-80%). Aloy and Russell showed that it is usually the case that proteins with greater than 30% sequence identity form oligomers in similar ways28. The dimers in Figure 1 affirm this conclusion, while the annotated dimers from the public databases shown in Figure 2 do not.
Figure 2.
Incorrect dimers in publicly available databases of biological assemblies. The crystal form from table I is given, along with the database names that contain the dimer shown. The dimer interface motif is shown in magenta. PQS has a tetramer for 1AQY that contains two copies of the Petrotchenko dimer (blue and yellow monomers, bottom; and green and red monomers, top).
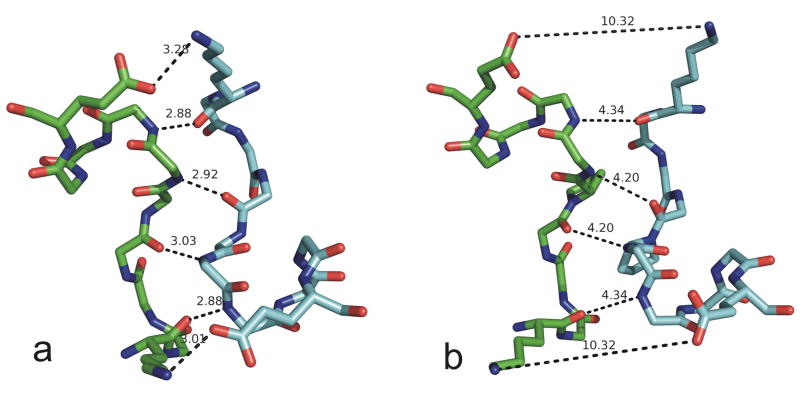
We examined the common interface in more detail. A close-up view of the dimer interface of human SULT1E1 from PDB entry 1G3M29 is shown in Figure 3a. The dimerization sequence motifs are given in Table II, along with the hydrogen bond distances found in most of the interfaces. In nearly all of the structures, there are six hydrogen bonds between the two monomers. In each case, the hydrogen bond exists in both directions, i.e. atom x of monomer A hydrogen bonds to atom y of monomer B, and atom x of monomer B hydrogen bonds to atom y of monomer A. The three symmetric hydrogen bonds are: 1) backbone N of residue 1 to backbone O of residue 7; 2) backbone N of residue 4 to backbone O of residue 6; and 3) side-chain Nζ of Lys 1 to side-chain Oε1 or Oε2 of Glu 10. These six hydrogen bonds are shown for human SULT1E1 in Figure 3a, labeled by their position in the motif (residues 1-10). In the flavonol SULT from Arabidopsis, the Lys is an Arg. In PDB entry 2H8K (unpublished), SULT1C3, crystal form 10, the Glu side chain is disordered. However, this is a low-resolution structure (3 Å resolution). Examination of the electron density via the Uppsala Electron Density Server30 shows significant density between the Glu CB and Lys NZ that is likely to be the salt bridge observed in the other structures.
Figure 3.
Dimer interfaces in: a) human SULT1E1 (PDB entry 1AQU) and b) mouse SULT1E1 (PDB entry 1G3M). Distances are given in Å for the hydrogen bonds between the monomers. These interactions are from top to bottom, left to right: 1) OE110-NZ1; 2) N7-O1; 3) N6-O4; 4) O4-N6; 5) O1-N7; 6) NZ1-OE110, where the residues are numbered from 1 to 10 in the motif as given in Table II.
Table II.
Hydrogen bond distances between monomers in biological dimers of cytosolic sulfotransferases.
| PDB | Species | Gene | CF# | Sequence | O1-N7 | O4-N6 | K1-E10 |
|---|---|---|---|---|---|---|---|
| 3bfx | Human | SULT1C2 | 1 | KNHFTVAQNE | 3.0 | 3.1 | 3.1 |
| 1j99 | Human | SULT2A1 | 2 | KNHFTVAQAE | 2.8 | 2.9 | 2.9 |
| 2a3r | Human | SULT1A3 | 3 | KTTFTVAQNE | 2.8 | 3.0 | 3,5 |
| 2d06 | Human | SULT1A1 | 3 | KTTFTVAQNE | 2.8 | 3.0 | 3.5 |
| 1cjm | Human | SULT1A3 | 4 | KTTFTVAQNE | 3.0 | 2.9 | 3.0 |
| 2ad1 | Human | SULT1C4 | 4 | KKHFTVAQNE | 2.9 | 2.9 | 3.0 |
| 1aqu | Mouse | SULT1E1 | 5 | KNHFPEALRE | 4.3 | 4.2 | 10.3 |
| 1ls6 | Human | SULT1A1 | 6 | KTTFTVAQNE | 3.0 | 3.2 | 3.1 |
| 1z29 | Human | SULT1A2 | 7 | KTTFTVAQNE | 2.7 | 2.8 | 3.0 |
| 1zd1 | Human | SULT4A1 | 8 | KDIFTVSMNE | 2.8 | 2.8 | 3.1 |
| 1q44 | Arabidopsis | FLAVONOL SULT | 9 | RDTLSESLAE | 3.0 | 3.2 | 4.0 (R1) |
| 2h8k | Human | SULT1C3 VARIANT D | 10 | KNYFTVAQNE | 3.1 | 3.4 | missing E10 |
| 2gwh | Human | SULT1C4 | 11 | KKHFTVAQNE | 2.9 | 2.9 | 2.9 |
| 1g3m | Human | SULT1E1 | 12 | KNHFTVALNE | 2.9 | 2.9 | 3.3 |
| 3ckl | Human | SULT1B1 | 13 | KNYFTVAQNE | 2.9 | 3.0 | 3.0 |
| 1efh | Human | SULT2A1 | 14 | KNHFTVAQAE | 2.7 | 2.9 | 3.2 |
| 2z5f | Human | SULT1B1 | 15 | KNYFTVAQNE | 2.9 | 2.8 | 2.7 |
| 1q1q | Human | SULT2B1 ISO A | 16 | KNHFTVAQSE | 2.9 | 3.2 | 3.3 |
| 2reo | Human | SULT1C3 | 17 | KNYFTVAQNE | 2.9 | 2.9 | 2.7 |
One structure that is quite different, however, is mouse SULT1E1. This interface from PDB entry 1AQU31 is shown in Figure 3b. The backbone hydrogen bonds are too long -- 4.2 Å and 4.3 Å, and the interface orientation is distorted enough such that the salt bridge on either end of the motif is not formed. The relevant atomic distances are 10.3 Å. This protein was found to be monomeric in gel filtration experiments by Petrotchenko et al. In particular the mouse SULT1E1 sequence contains the sequence PE at the center of the motif (positions 5 and 6) rather than TV in the other proteins. Petrotchenko found that mutating PE to TV in mouse SULT1E1 caused it to dimerize in solution, while mutating human TV to PE caused the human protein to be monomeric. It is surprising, to say the least, that this small interface still forms in the crystals of mouse SULT1E1, albeit with loss of the favorable interactions that stabilize this interface in the other proteins. This would seem to indicate that some proteins may form interfaces similar to others in the same family in crystals, but weakly enough that they may not be present under physiological conditions.
It is possible that the flavonol sulfotransferase from Arabidopsis thaliana is also a monomer, given that residue 6 of the dimerization motif is also Glu, as it is in mouse SULT1E1. The salt bridge hydrogen bond length is rather long at 4 Å.
Discussion
Cytosolic sulfotransferases are important metabolic and detoxifying enzymes in humans. Missense mutations or downregulation of some of these proteins are associated with susceptibility to various cancers, probably due to lowered ability to metabolize and eliminate xenobiotics32-34. We have investigated the puzzling small interface that was determined to be the dimer interface, and found it in all of the available crystal structures of cytosolic sulfotransferases. Despite publication of the paper by Petrotchenko et al in early 2001, this small interface is not annotated as the dimer interface in the publicly available databases with only a few exceptions. At total of 21 of 28 structures were deposited in the PDB after the 2001 paper appeared, and only 3 of these are annotated with the correct dimer in the PDB. The biological role of this interface, if any, is unknown, as the monomeric mouse SULT1E1 is presumably active8. The interface is 25-30 Å away from the active site in all of the crystal structures (not shown), which is roughly in the center of the protein, as determined from the location of the sulfation donor analogues and the substrates.
While experiments are often performed on proteins to determine the molecular weight of the fully formed assembly under roughly physiological conditions, there are few cases where experiments designed to determine the interfaces involved are performed. In many cases, the correct interface appears to be obvious from inspection of the crystal, and this is especially true if there are multiple crystal forms available, especially of non-identical proteins in the family. In the example given here, an analysis of all the available crystals would have indicated the correct physiologically relevant dimer, which could then be tested with further experiments. In the near future, we will provide an online tool for performing this analysis over the whole PDB, so that it will be available as new structures are determined.
Acknowledgments
This work was supported under NIH grant R01 GM73784.
References
- 1.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14(3):199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90(1):5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 3.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: a structural classification of proteins database. Nucleic Acids Res. 2000;28(1):257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falany CN, Vazquez ME, Heroux JA, Roth JA. Purification and characterization of human liver phenol-sulfating phenol sulfotransferase. Arch Biochem Biophys. 1990;278(2):312–318. doi: 10.1016/0003-9861(90)90265-z. [DOI] [PubMed] [Google Scholar]
- 5.Homma H, Kamakura M, Nakagome I, Matsui M. Purification of a rat liver phenol sulfotransferase (P-STG) with the aid of guanidine hydrochloride treatment. Chem Pharm Bull (Tokyo) 1991;39(12):3307–3312. doi: 10.1248/cpb.39.3307. [DOI] [PubMed] [Google Scholar]
- 6.Kiehlbauch CC, Lam YF, Ringer DP. Homodimeric and heterodimeric aryl sulfotransferases catalyze the sulfuric acid esterification of N-hydroxy-2-acetylaminofluorene. J Biol Chem. 1995;270(32):18941–18947. doi: 10.1074/jbc.270.32.18941. [DOI] [PubMed] [Google Scholar]
- 7.Dajani R, Sharp S, Graham S, Bethell SS, Cooke RM, Jamieson DJ, Coughtrie MW. Kinetic properties of human dopamine sulfotransferase (SULT1A3) expressed in prokaryotic and eukaryotic systems: comparison with the recombinant enzyme purified from Escherichia coli. Protein Expr Purif. 1999;16(1):11–18. doi: 10.1006/prep.1999.1030. [DOI] [PubMed] [Google Scholar]
- 8.Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M. The dimerization motif of cytosolic sulfotransferases. FEBS Lett. 2001;490(1-2):39–43. doi: 10.1016/s0014-5793(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 9.Hobkirk R, Glasier MA, Brown LY. Purification and some characteristics of an oestrogen sulfotransferase from guinea pig adrenal gland and its non-identity with adrenal pregnenolone sulfotransferase. Biochem J. 1990;268(3):759–764. doi: 10.1042/bj2680759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouthillier M, Bleau G, Chapdelaine A, Roberts KD. The purification of 3 beta-hydroxysteroid sulfotransferase of the hamster epididymis. J Steroid Biochem. 1985;22(6):733–738. doi: 10.1016/0022-4731(85)90279-1. [DOI] [PubMed] [Google Scholar]
- 11.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulfotransferase. Biochem J. 1989;260(3):641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogura K, Satsukawa M, Okuda H, Hiratsuka A, Watabe T. Major hydroxysteroid sulfotransferase STa in rat liver cytosol may consist of two microheterogeneous subunits. Chem Biol Interact. 1994;92(1-3):129–144. doi: 10.1016/0009-2797(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen LC, Petrotchenko EV, Negishi M. Crystal structure of SULT2A3, human hydroxysteroid sulfotransferase. FEBS Lett. 2000;475(1):61–64. doi: 10.1016/s0014-5793(00)01479-4. [DOI] [PubMed] [Google Scholar]
- 14.Hattori K, Inoue M, Inoue T, Arai H, Tamura HO. A novel sulfotransferase abundantly expressed in the dauer larvae of Caenorhabditis elegans. J Biochem. 2006;139(3):355–362. doi: 10.1093/jb/mvj041. [DOI] [PubMed] [Google Scholar]
- 15.Bidwell LM, McManus ME, Gaedigk A, Kakuta Y, Negishi M, Pedersen L, Martin JL. Crystal structure of human catecholamine sulfotransferase. J Mol Biol. 1999;293(3):521–530. doi: 10.1006/jmbi.1999.3153. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Canutescu AA, Wang G, Shapovalov M, Obradovic Z, Dunbrack RL., Jr Statistical analysis of interface similarity in crystals of homologous proteins. J Mol Biol. 2008;381(2):487–507. doi: 10.1016/j.jmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canutescu AA, Dunbrack RL., Jr MollDE: a homology modeling framework you can click with. Bioinformatics. 2005;21(12):2914–2916. doi: 10.1093/bioinformatics/bti438. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Canutescu AA, R L, Dunbrack J. SCWRL and MolIDE: Programs for protein side-chain prediction and homology modeling. Nature Protocols. 2008 doi: 10.1038/nprot.2008.184. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33(Database issue):D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of database programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbard SJ, Thornton JM. NACCESS. London: Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- 22.Xu Q, Canutescu A, Obradovic Z, Dunbrack RL., Jr ProtBuD: a database of biological unit structures of protein families and superfamilies. Bioinformatics. 2006;22(23):2876–2882. doi: 10.1093/bioinformatics/btl490. [DOI] [PubMed] [Google Scholar]
- 23.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23(9):358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.05.022. in press. [DOI] [PubMed] [Google Scholar]
- 26.Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21(7):988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 27.Chang HJ, Shi R, Rehse P, Lin SX. Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex. J Biol Chem. 2004;279(4):2689–2696. doi: 10.1074/jbc.M310446200. [DOI] [PubMed] [Google Scholar]
- 28.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332(5):989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Shevtsov S, Petrotchenko EV, Pedersen LC, Negishi M. Crystallographic analysis of a hydroxylated polychlorinated biphenyl (OH-PCB) bound to the catalytic estrogen binding site of human estrogen sulfotransferase. Environ Health Perspect. 2003;111(7):884–888. doi: 10.1289/ehp.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala Electron-Density Server. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 31.Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC. Crystal structure of estrogen sulfotransferase. Nat Struct Biol. 1997;4(11):904–908. doi: 10.1038/nsb1197-904. [DOI] [PubMed] [Google Scholar]
- 32.Pachouri SS, Sobti RC, Kaur P, Singh J, Gupta SK. Impact of polymorphism in sulfotransferase gene on the risk of lung cancer. Cancer Genet Cytogenet. 2006;171(1):39–43. doi: 10.1016/j.cancergencyto.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Dandara C, Li DP, Walther G, Parker MI. Gene-environment interaction: the role of SULT1A1 and CYP3A5 polymorphisms as risk modifiers for squamous cell carcinoma of the oesophagus. Carcinogenesis. 2006;27(4):791–797. doi: 10.1093/carcin/bgi257. [DOI] [PubMed] [Google Scholar]
- 34.Modena P, Lualdi E, Facchinetti F, Veltman J, Reid JF, Minardi S, Janssen I, Giangaspero F, Forni M, Finocchiaro G, Genitori L, Giordano F, Riccardi R, Schoenmakers EF, Massimino M, Sozzi G. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24(33):5223–5233. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]