Introduction
Over the last 2 years, several reports have suggested that submicroscopic chromosomal deletions that disrupt the gene neurexin 1 (NRXN1) increase the risk of developing schizophrenia. In this article, we will review the evidence for this association.
NRXN1 encodes NRXN1, a synaptic neuronal adhesion molecule. NRXNs are found presynaptically and are believed to interact with postsynaptic neuroligins (NLGNs) in excitatory and inhibitory synapses in the brain. The structure and possible function of NRXNs and NLGNs have recently been comprehensively reviewed.1
Vertebrate NRXNs and NLGNs are the only adhesion molecules for which a specifically synaptic function has been demonstrated.1 Current evidence suggests that NRXNs and NLGNs act transsynaptically to mediate essential signaling between presynaptic and postsynaptic specializations. Evidence from cell culture experiments and the study of mouse knockouts suggest that these molecules are required for synapse function but not for synapse formation, that they influence transsynaptic activation of synaptic transmission; and that their dysfunction impairs the properties of synapses and disrupts neural networks without completely abolishing synaptic transmission. NRXNs and NLGNs probably function by binding to each other and by interacting with intracellular proteins (most notably those with PDZ domain), but the precise mechanisms involved remain unknown.1 Whatever the mechanism and their relationship to synaptic function, it is clear that mice lacking NRXNs or NLGNs show marked deficits in synaptic transmission.1
There are 5 NLGN genes and 3 NRXN genes in humans (NRXN1 [2p16.3], NRXN2 [11q13], NRXN3 [14q31]). The 3 NRXN genes each encode an α protein and a β protein from independent promoters. NRXNs have a striking molecular diversity: the messenger RNA can be processed by alternative splicing, giving potentially thousands of distinct protein isoforms.2
There is strong evidence for an involvement of NLGNs and NRXN1 in autism (reviewed by Kumar and Christian3). Several studies reported an increased rate of deletions or other genetic variants in NRXN1 in subjects suffering with autism.4–11 Additionally, deletions in this gene have been implicated in mental retardation and developmental delay.12,13 A number of studies have reported NRXN1 deletions in cases with schizophrenia, which we will review in more detail.
NRXN1 Deletions in Schizophrenia
The first suggestion that NRXN1 deletions might confer risk of schizophrenia came from a study by Kirov et al.14 They used array comparative genome hybridization (array-CGH) to screen the genomes of 93 cases of schizophrenia recruited in Bulgaria with approximately 35 000 probes for chromosomal copy number variants (CNVs) at a resolution of about 80 000 bp. They identified a deletion of NRXN1 in a female case, which was also present in her affected brother. The deletion spanned the promoter and the first exon of the gene. Neither sibling had signs of premorbid cognitive problems or dysmorphic features. Given that deletions of NRXN1 were not seen in 372 controls, and at that time there were reports of NRXN1 deletions and point mutations in autism4,5 and one deletion in a subject with mental retardation,12 the authors hypothesized that deletions in this gene might confer risk to schizophrenia as well as to other neurodevelopmental disorders. Intriguingly, these authors also found a de novo duplication of 1.4Mb of chromosome 15 that involved, among other genes, amyloid beta A4 precursor protein-binding (APBA2) gene encoding a protein known as Mint2. Mint2 is a neuronal adaptor protein that binds directly to NRXNs in a PDZ domain–mediated interaction.
Shortly after this initial report, Walsh et al,15 also using array-CGH, reported identical twins with early-onset schizophrenia who shared a 115-kb deletion in the gene (disrupting exons in the 3′ end) among a total of 233 schizophrenia cases and 268 controls.
Array technology has progressed tremendously since these small early reports, and it is now possible routinely to detect CNVs using microarrays with up to 2 million probes covering the whole genome. Several groups have found deletions of NRXN1 in patients with schizophrenia using the new technologies. Rujescu et al16 examined NRXN1 for CNVs in 2977 schizophrenia patients and 33 746 control subjects from 7 European countries (Iceland, Finland, Norway, Germany, the Netherlands, Italy, and the UK). Sixty-one NRXN1 deletions, including a de novo deletion, and 5 duplications were detected. Of these, 12 deletions and 2 duplications occurred in schizophrenia cases (0.47%) compared with 49 deletions and 3 duplications (0.15%) among the >10-fold larger control sample (P = .13, odds ratio [OR] = 1.73; 95% confidence interval [CI] = 0.81–3.50). There was no common breakpoint, and the CNVs varied in size from 18 to 420 kb. Because the penetrance of NRXN1 CNVs is likely to vary according to the level of functional impact on the gene, these authors next restricted analysis to CNVs that disrupt exons. Seven were found in cases (0.24%) and five in controls (0.015%). Compared with all CNVs spanning NRXN1, those affecting exons were more strongly associated with schizophrenia and had a higher OR (P = .0027, OR = 8.97, 95% CI = 1.8–51.9).
A second large collaborative study, the International Schizophrenia Consortium,17 found 4 deletions in 3391 cases and 3 in 3181 controls from 5 European countries (Sweden, Bulgaria, Ireland, Portugal, and the UK). If we only consider those deletions that disrupt exons, as suggested by Rujescu et al,16 the rate is 3 vs 1 or approximately 0.09% in cases and 0.03% in controls. No duplications were found.
Need et al18 reported 3 NRXN1 deletions in a sample of 1013 cases and 1084 controls of European ancestry and 60 cases and 64 controls of African American ancestry.
Kirov et al19 examined 471 patients and 2792 controls from the United Kingdom. They found one deletion in NRXN1 in a case and 3 in controls, giving a 2-fold nonsignificant increase in the rate in cases (0.2% vs 0.1%). All are likely to disrupt exons.
Finally, M. Ikeda et al. (M. Ikeda, MD, PhD, B. Aleksic, MD, PhD, G. Kirov, MRCPsych, PhD, Y. Kinoshita, MD, PhD, Y. Yamanouchi, MD, PhD, T. Kitajima, MD, PhD, K. Kawashima, MD, PhD, T. Okochi, MD, T. Kishi, MD, PhD, I. Zaharieva, M.J. Owen, FRCPsych, PhD, M.C. O'Donovan, FRCPsych, PhD, N. Ozaki, MD, PhD, N. Iwata, MD, PhD, unpublished data, 2009) tested 560 patients and 547 controls from Japan and found one deletion in a case and none in controls. The deletion was 161 kb long but does not disrupt an exon.
In the analysis described below, we do not include the 4 patients described in the study of Vrijenhoek et al20 as they were included in the study by Rujescu et al.16
Frequency of NRXN1 Deletions in Schizophrenia Across All Available Studies
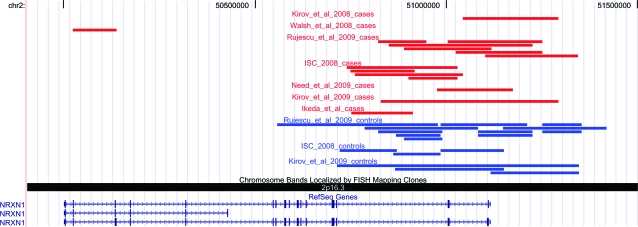
Given that deletions in NRXN1 are extremely rare and many of the studies are too small to provide statistically meaningful results, we reanalyzed all available data in order to find out whether there is convincing evidence for association. To make the results comparable across all studies, we included only deletions >100 kb, as all studies, apart from one,16 had used this selection criterion. We provide all the data in table 1 and visualize the exact breakpoints of each deletion on the background of the gene using UCSC tracks (http://wwwgenomeucscedu/) to enable readers to see the relationship between the deletions and exons (figure 1). In total, there were deletions in 17/8789 cases (0.19%) and in 17/42054 controls (0.04%), a highly significant excess (Fisher Exact test: P = .000013, OR = 4.78, 95% CI = 2.44–9.37). Reanalysis for deletions >100kb that only disrupt exons reveals a similar result (P = .000037, OR = 7.44, 95% CI = 3.22–17.18; table 1).
Table 1.
Data From All Available Studies on Deletions in Neurexin 1
| All Deletions >100 kb | Deletions >100 kb Disrupting Exons | ||||
| Study | Cases | Controls | Size (kb) | Cases | Controls |
| Kirov et al14 | 1/93 | 0/372 | 250 | 1/93 | 0/372 |
| Walsh et al15 | 1/233 | 0/268 | 115 | 1/233 | 0/268 |
| International Schizophrenia Consortium17 | 4/3391 | 3/3181 | 122–291 | 3/3391 | 1/3181 |
| Rujescu et al16 | 6/2977 | 11/33 746 | 100–419 | 5/2977 | 5/33 746 |
| Need et al18 | 3/1073 | 0/1148 | 200–418 | 3/1073 | 0/1148 |
| Kirov et al19 | 1/471 | 3/2792 | 231–631 | 1/471 | 3/2792 |
| Ikeda et al. (unpublished data, 2009, see text citation for all authors) | 1/560 | 0/547 | 161 | 0/560 | 0/547 |
| Totals | 17/8798 (0.19%) | 17/42054 (0.04%)a | 14/8798 (0.16%) | 9/42 054 (0.02%)b | |
Note: All deletions are filtered for >100kb in size
Fisher Exact test: P = .000013, odds ratio = 4.78, 95% confidence interval = 2.44–9.37.
Fisher Exact test: P = .0000037, odds ratio = 7.44, 95% confidence interval = 3.22–17.18.
Fig. 1.
Copy Number Variants in NRXN1 in All Studies, Filtered for >100 kb. The shorter form of the gene encodes for the NRXN1β form, while the longer one encodes for the NRXN1α form. Red bars are deletions in cases (n = 8798); blue are those in controls (n = 42 054).
Conclusions
The findings of studies to date provide strong evidence that deletions of NRXN1 confer a substantial increase in risk of schizophrenia. However, some degree of caution is still warranted. In the context of their rarity, there have still been relatively few subjects studied, and there are technical difficulties associated with detecting CNVs of variable size. Much of the available control data in our analysis came from the Icelandic sample reported by Rujescu et al.16 Given these controls are derived from a large population sample with a good phenotypic information, the rate of NRXN1 deletions observed is very likely to be an accurate representation in control populations. Moreover, it is also similar to the estimates from the majority of the other smaller studies. Nevertheless, the rarity of NRXN1 deletions means that the overall findings are quite strongly dependent upon that single study. There is therefore a need to study an even greater number of cases and controls in independent samples, using high-resolution platforms, which can detect smaller CNVs.
These findings at NRXN1 should be seen in the context of a number of recent demonstrations that risk of schizophrenia is increased by CNVs.21 In particular, there is very convincing evidence that large recurrent deletions affecting 1q21.1, 15q11.2, 15q13.3, and 22q11.2 confer risk.17,19 All 4 deletions contain multiple genes, and it might not be straightforward to understand which genes and pathways are implicated in risk of psychosis. In contrast, the deletions described in the present article only disrupt NRXN1. These findings could therefore represent a decisive step toward identifying a specific pathway implicated in the pathogenesis of schizophrenia.
Another key area of future research will be work aimed at understanding the nature of the pleiotropic effects that are associated with those CNVs that confer risk of schizophrenia. Interestingly, all the risk CNVs seem not only to associate with variable neurodevelopmental phenotypes, particularly autism, and variable degrees of cognitive dysfunction but can also occur in the absence of an apparent neurodevelopmental phenotype.22,23 The highly variable outcomes associated with these genomic abnormalities suggest that more detailed understanding of the mechanisms involved might point the way to new therapeutic opportunities.
References
- 1.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 3.Kumar RA, Christian SL. Genetics of autism spectrum disorders. Curr Neurol Neurosci Rep. 2009;9:188–197. doi: 10.1007/s11910-009-0029-2. review. [DOI] [PubMed] [Google Scholar]
- 4.Feng J, Schroer R, Yan J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Feng J, Schroer R, et al. Analysis of the neuroligin 4Y gene in patients with autism. Psychiatr Genet. 2008;18:204–207. doi: 10.1097/YPG.0b013e3282fb7fe6. [DOI] [PubMed] [Google Scholar]
- 10.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 11.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman JM, Baross A, Delaney AD, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahir FR, Baross A, Delaney AD, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 14.Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 15.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 16.Rujescu D, Ingason A, Cichon S, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Need AC, Ge D, Weale ME, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirov G, Grozeva D, Norton N, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen MJ, Williams HJ, O'Donovan MC. Schizophrenia genetics: advancing on two fronts. Curr Opin Genet Dev. 2009;19:266–270. doi: 10.1016/j.gde.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 22.O'Donovan MC, Kirov G, Owen MJ. Phenotypic variations on the theme of CNVs. Nat Genet. 2008;40:1392–1393. doi: 10.1038/ng1208-1392. [DOI] [PubMed] [Google Scholar]
- 23.van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome [published online ahead of print April 21, 2009] J Med Genet. doi: 10.1136/jmg.2008.063412. doi:10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]