Abstract
Notch signaling is an evolutionarily conserved pathway essential for many cell fate specification events during metazoan development. We conducted a large-scale transposon-based screen in the developing Drosophila eye to identify genes involved in Notch signaling. We screened 10,447 transposon lines from the Exelixis collection for modifiers of cell fate alterations caused by overexpression of the Notch ligand Delta and identified 170 distinct modifier lines that may affect up to 274 genes. These include genes known to function in Notch signaling, as well as a large group of characterized and uncharacterized genes that have not been implicated in Notch pathway function. We further analyze a gene that we have named Amun and show that it encodes a protein that localizes to the nucleus and contains a putative DNA glycosylase domain. Genetic and molecular analyses of Amun show that altered levels of Amun function interfere with cell fate specification during eye and sensory organ development. Overexpression of Amun decreases expression of the proneural transcription factor Achaete, and sensory organ loss caused by Amun overexpression can be rescued by coexpression of Achaete. Taken together, our data suggest that Amun acts as a transcriptional regulator that can affect cell fate specification by controlling Achaete levels.
THE Notch pathway is a highly conserved signaling cascade that controls cell fate specification by inducing or inhibiting the adoption of cell fates in many contexts. Disruption of pathway function generally results in disruption of cell fate specification during development (Artavanis-Tsakonas et al. 1999; Portin 2002; Schweisguth 2004; Bray 2006; Nichols et al. 2007b). Despite a wealth of literature addressing many aspects of Delta–Notch signaling, numerous findings indicate that additional molecular components and mechanisms that affect the pathway remain to be discovered.
Drosophila Notch is a type I transmembrane receptor protein with two known ligands, Delta and Serrate, which are also type I transmembrane proteins (Fiuza and Arias 2007; D'Souza et al. 2008). Upon ligand binding, the Notch extracellular domain (NotchECD) enters the Delta-expressing cell via trans-endocytosis (Parks et al. 2000; Itoh et al. 2003; Nichols et al. 2007a). The transmembrane-bound Notch intracellular domain then undergoes two proteolytic cleavages. The first is thought to be mediated by an ADAM metalloprotease, Kuzbanian (Pan and Rubin 1997; Sotillos et al. 1997; Lieber et al. 2002), while the second is mediated by the γ-secretase complex, which includes Presenilin (Fortini 2002; Selkoe and Kopan 2003). These cleavages release the Notch intracellular domain into the cytoplasm, which subsequently translocates into the nucleus where it forms a transcriptional co-activation complex that includes Suppressor of Hairless and Mastermind (Mam). These complexes activate expression of Notch target genes [e.g., Enhancer of split-Complex, or E(spl)-C, genes] in a variety of contexts (Kopan 2002; Schweisguth 2004; Bray 2006). Regulation of Notch signaling occurs on several different levels (Schweisguth 2004; Bray 2006). For example, many proteins of the endocytic (e.g., Auxilin, Dynamin, Epsin, Numb, and Rab11) and ubiquitylation (e.g., Neuralized, Mindbomb1, and Deltex) machinery affect ligand and receptor localization as well as activation of ligand-dependent signaling and downregulation of the receptor (reviewed in Chitnis 2006; Le Borgne 2006; Nichols et al. 2007b; Brou 2009). Post-translational modification of proteins can also play a role in Notch regulation. For example, Notch itself can be modified by O-fucosyl transferase (a glycosyltransferase and chaperone), Fringe (a glycosyltransferase), and proteases such as Furin (Nichols et al. 2007b; Stanley 2007; Irvine 2008).
Drosophila wing, eye, and bristle development are excellent contexts in which to study mechanisms of Notch-mediated development and uncover additional components in the pathway. Notch can act in either an inductive or an inhibitory manner to promote the proper adoption of various cell fates during development of these tissues. In the developing eye, Notch signaling first promotes formation of a single R8 photoreceptor cell within a group of equivalent proneural cells and then prevents neighboring cells from adopting the R8 fate, thereby restricting the number of R8 photoreceptors to one per ommatidium (Baker and Yu 1997; Baker 2000; Lee et al. 2000). Notch signaling is subsequently required for differential specification of the R3/R4 photoreceptor fates (Del Alamo and Mlodzik 2006) and for induction of the R7 photoreceptor, cone cell, and primary pigment cell fates (Cooper and Bray 2000; Tomlinson and Struhl 2001; Tsuda et al. 2002; Carthew 2007; Nagaraj and Banerjee 2007).
In the bristle organ, development is initiated with the expression of the basic helix–loop–helix (bHLH) transcription factors, Achaete and Scute, within groups of developmentally equivalent proneural cells. Within these proneural groups, sensory organ precursors (SOPs) are singled out, and Delta–Notch signaling by SOPs represses Achaete expression in neighboring cells, preventing them from adopting the SOP fate. SOPs then divide into two cells, pIIa and pIIb. The daughters of pIIa form the shaft and socket of the mature bristle organ, while the granddaughters of pIIb (daughters of pIIIb) form the neuron and sheath cells. Notch signaling occurs between pIIa and pIIb and each successive pair of daughter cells to prevent adoption of inappropriate fates. For example, Notch signaling prevents the adoption of the pIIb fate in the presumptive pIIa cell and inhibits adoption of the neuronal fate in the presumptive sheath cell (Parks and Muskavitch 1993; Gho et al. 1996, 1999; Guo et al. 1996; Wang et al. 1997; Reddy and Rodrigues 1999; Le Borgne and Schweisguth 2003). Mis-regulation of Notch signaling during bristle organ development can lead to a variety of phenotypic defects, depending on the developmental stage during which disruption occurs. For example, reduction of Notch signaling during SOP specification results in the specification of multiple SOPs, leading to the development of multiple bristle organs, whereas reduction of Notch signaling during differentiation of the neuron and sheath cells leads to the specification of two neurons and no sheath cells (Hartenstein and Posakony 1990; Parks and Muskavitch 1993).
To identify additional functions that modulate Notch signaling during development, we designed a large-scale genetic screen using the Exelixis transposon collection housed in the Artavanis-Tsakonas laboratory at the Harvard Medical School (Artavanis-Tsakonas 2004). Our screen is based on the ability of genes to modify cell fate changes that result from Delta overexpression in the retina during development. We confirm the identification of 170 individual transposon insertions that potentially affect a total of 274 genes. We further characterize a phenotypic suppressor of Delta overexpression encoded by CG2446, a gene that we have named Amun (for an ancient Egyptian god also referred to as “the hidden one”). The Amun protein contains a DNA glycosylase domain, and Amun loss-of-function phenotypes include bristle and eye defects. Overexpression of Amun inhibits the formation of microchaeta sense organs by downregulating Achaete protein within microchaeta proneural equivalence groups. Our data therefore suggest that Amun is a nuclear factor that can regulate Achaete levels to control cell fate specification during sensory organ development.
MATERIALS AND METHODS
Fly stocks and culture:
The Exelixis transposon collection (Thibault et al. 2004) and all stocks from our laboratory were maintained using standard procedures. All crosses were performed at 25°, unless otherwise noted.
Drosophila strains used:
The following strains were used for the screen: the Exelixis collection housed in the laboratory of Spyros Artavanis-Tsakonas (Artavanis-Tsakonas 2004); GMR-Gal4 (Hay et al. 1994; Freeman 1996); 34B-Gal4 (Ingham and Fietz 1995); C96-Gal4 (Gustafson and Boulianne 1996), a gift from Barry Yedvobnick, Emory University (Atlanta); UAS-DeltaWT (on chromosome 2; Jacobsen et al. 1998); and UAS-DeltaΔICD (also known as DeltaD; Huppert et al. 1997). The following strains were used for the study of CG2446 (Amun): eyeless (ey)-Gal4 (Bose et al. 2006) (Bloomington Drosophila Stock Center); decapentaplegic (dpp)-Gal4/TM6B (Staehling-Hampton et al. 1994); pannier (pnr)-Gal4/TM3 (Heitzler et al. 1996), a gift from Gines Morata, Centro de Biología Molecular Severo Ochoa (Madrid); patched (ptc)-Gal4 (Speicher et al. 1994) (Bloomington Drosophila Stock Center); scabrous (sca)-Gal4/CyO (Mlodzik et al. 1990), a gift from Andrea Brand, University of Cambridge (Cambridge, UK); UAS-myr-mRFP/TM6B (Bloomington Drosophila Stock Center); stripeMD710(sr)-Gal4/TM6B (Calleja et al. 2002; Usui et al. 2004), a gift from Pat Simpson, University of Cambridge (Cambridge, UK); UAS-AmunRNAi and UAS-Dicer2 (Dietzl et al. 2007), obtained from the Vienna Drosophila RNAi Center; and P[lArB]A101.IF3 (neurA101-LacZ)/TM3 (Bellen et al. 1989), a gift of Hugo Bellen, Baylor College of Medicine (Houston).
Screen:
The screen was performed by assaying the effects of each of 10,447 transposon insertions from the Exelixis stock collection (supporting information, Table S1) (Artavanis-Tsakonas 2004) on the eye phenotype of GMR-Gal4 UAS-DeltaWT/+ (GMR>DeltaWT/+) flies. The collection is composed of four different transposon types: three piggyBac-based transposons (PB, RB, and WH) and one P-element-based transposon (XP). Two of the four transposons contain at least one upstream activating sequence (UAS) cassette (Brand and Perrimon 1993). The WH transposon contains a single terminal UAS cassette that can activate or interfere with (via antisense transcript production) the expression of neighboring genes. The XP transposon contains two terminal UAS cassettes that have the potential to alter the expression of genes located adjacent to either or both ends of the inserted transposon. The PB and RB vectors contain no UAS sequences; therefore, their effects on genes are limited to the site of insertion. Our primary screen consisted of crossing males carrying autosomal or viable X-linked insertions to GMR>DeltaWT-bearing virgin females and scoring the F1 progeny for changes in the rough-eye phenotype. Modifying transposons were categorized as enhancers or suppressors of weak, moderate, or strong intensity. Of 798 primary screen modifiers, 284 were retested with GMR>DeltaWT/+ to confirm modification. A negative secondary test was performed by crossing confirmed modifiers to flies carrying the GMR-Gal4 transgene alone to eliminate modifiers that affect eye development in the same manner as seen in the primary screen. Positive secondary analyses were performed to prioritize the candidate modifiers using phenotypes that result from expression of a dominant-negative Delta variant created by truncation of the Delta intracellular domain (DeltaΔICD; Huppert et al. 1997) in the developing wing vein [34B-Gal4 UAS-DeltaΔICD/+ (34B>DeltaΔICD)] or wing margin [UAS-DeltaΔICD/+; C96-Gal4/+ (C96>DeltaΔICD)]. We assessed enhancement and suppression of both of these phenotypes.
Annotation of hits:
All high-priority modifiers were annotated by aligning the relevant transposon-flanking sequence (Thibault et al. 2004) against the Drosophila melanogaster genome (FB2007_03 Dmel Release 5.4) using the FlyBase BLAST website (http://flybase.bio.indiana.edu/blast). A 10-kb genome browser snapshot was taken, and transposon-specific criteria were used to assess which genes were potentially affected by the transposon insertion. A gene was considered potentially disrupted if the transposon was inserted within the transcription unit or within 2 kb of the 5′-end or 1 kb of the 3′-end of the transcription unit. In addition, for UAS-containing transposons (XP and WH), a gene was considered a potential target for UAS-directed expression if it was within 5 kb of the transposon insertion site (and “downstream” of the UAS), unless there was a potential RNA polymerase II transcription stop site between the UAS and the gene in question. Genes identified as possible modifiers were placed into functional categories using previously published data when available and/or FlyBase (FB2007_03 Dmel Release 5.4) Gene Ontology terms.
Constructs and transgenic stocks:
A full-length cDNA clone of CG2446 (Amun) in the pOT2 vector (GH02702) was acquired from the Berkeley Drosophila Genome Project. A SalI–NotI fragment of Amun was created by using the forward primer (5′-GTCGACATGTCCAACGGCAAGGCG-3′) and reverse primer (5′-GTGCGGCCGCGATTCGCTGCGCAG-3′) (IDTDNA, http://www.idtdna.com), and the PCR fragment was purified and ligated into blunt-end TOPO (Invitrogen), restricted with SalI and NotI, and ligated into the Gateway vector pENTR1A (Invitrogen). Lambda recombinase (Invitrogen) was used to insert Amun into the following Gateway destination vectors (obtained from the Drosophila Genome Resource Center, Indiana University, Bloomington, IN): pTW (containing a UAS promoter) and pTWR [containing a UAS promoter and a monomeric Red Fluorescent Protein (mRFP) C-terminal tag]. w1118 transgenics containing these constructs were generated by Genetics Services (Cambridge, MA).
Molecular confirmation of the P{XP}d03329 insertion site:
Genomic DNA was prepared using a standard procedure (Parks et al. 2004). Inverse PCR was performed using a protocol adapted from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/pdfs/Exel_links/5__fly_iPCR_XP_pub.pdf). Briefly, genomic DNA was digested with Sau3AI and ligated to create circular DNA, and primer pairs 31A-31B and 51A-51B were used to amplify sequences flanking the transposon. PCR products were then sequenced, and BLAST was used to match the sequences obtained against the D. melanogaster genome. Two-sided PCR was also performed using a standard procedure (Parks et al. 2004). For amplification of the 5′-end of the P{XP}d03329 transposon insertion, 52B (forward) and d03329 reverse flank 5′-AGTCGCACACACAGAGACGTAGTT-3′ (reverse) primers were used. For amplification of the 3′-end of the transposon, d03329 forward flank 5′-ATGGGAATGACGAACGACGACGAA-3′ (forward) and XP-3SEQ (reverse) primers were used.
Immunohistochemistry:
Primary antibodies used (all mouse monoclonals) were the following: anti-Cut at 1:5 [Developmental Studies Hybridoma Bank (DSHB); Iowa University, Iowa City, IA]; anti-Achaete at 1:5 (DSHB and a gift from Teresa Orenic, University of Illinois, Chicago); and 22C10 at 1:100 (a gift from Seymour Benzer, California Institute of Technology, Pasadena, CA). Secondary antibodies used were Alexa488-conjugated goat anti-mouse at 1:500 (Molecular Probes, Eugene, OR) and horseradish-peroxidase-conjugated goat anti-mouse at 1:1000 (Jackson Immunoresearch, West Grove, PA). Peroxidase activity was visualized using 3, 3′-diaminobenzidine. β-Galactosidase activity was detected using Fe(CN)/X-gal staining solution (Hartenstein and Posakony 1990). Imaginal discs were stained as in Parks et al. (1997), except TPBS (0.3% Triton X-100, 0.02 M Na2HPO4, 0.14 M NaCl, pH 7.6) was used as the buffer for Achaete staining. Vybrant DyeCycle Green stain (Molecular Probes) was used to assess Amun∷RFP (Amun C-terminally-tagged with monomeric Red Fluorescent Protein) subcellular localization. For cone cell analysis, ∼15 retinas were dissected, and the number of cone cells in 20 ommatidia per retina was counted, providing an average of 300 ommatidia for each genotype tested. Student's t-test was used (two-tail distribution and two sample unequal variants; Microsoft Excel 2004) to compare the number of cone cells per ommatidium between genotypes. The SP5 Leica confocal microscope and Adobe Photoshop 7.0 were used to process images.
Gain-of-function clones:
For ectopic clonal expression, hs-Flp; Act5C>y+>Gal4/CyO virgins were crossed to UAS-Amun∷RFP/TM6B males. F1 first to second instar larvae were incubated at 37° for 1 hr to induce clones. Six hours after puparium formation (APF) nota were dissected and stained as in Parks et al. (1997) except TPBS was used as the buffer.
Phenotypic assessment of transgenic adults:
Adult wings and nota were submerged in mineral oil, and pictures were taken on a Zeiss Axioskop and Zeiss Stemi SV11 using the Zeiss AxioCam camera and Zeiss AxioCam Plug-In software, Version 1.0. Adult eye pictures were taken using the Leica MZ16 In-Focus system. All images were assembled using Adobe Photoshop 7.0.
Protein alignments:
Alignments were created in VectorNTI (Suite 7.1, for Mac OS X) using the AlignX program with the following protein sequences: D. melanogaster Amun (CG2446), NP_727552.1; Drosophila simulans GD15978, XP_002106704.1; Danio rerio Zgc:112496, AAH91543.1; Xenopus tropicalis LOC100145131, NP_001120112.1; Equus caballus LOC100066977, XP_001497177.1; Bos taurus LOC516108, XP_594248.3; and Monodelphis domestica LOC100020910, XP_001373236.1. Homo sapiens N-methylpurine-DNA glycosylase (MPG), NP_001041636.1; H. sapiens MutYH, NP_001041636.1; Mycobacterium tuberculosis ultraviolet N-glycosylase/AP lyase (Pdg), NP_338328.1; Bacillus subtilis DNA-3-methyladenine glycosylase (AlkA), YP_176647.1; D. melanogaster 8-oxoguanine DNA glycosylase (OGG1), NP_572499.2; H. sapiens 8-oxoguanine DNA glycosylase (OGG1), NP_002533.1; H. sapiens Nth1, NP_002519.1; and D. melanogaster Nth1, NP_610078.2.
RESULTS
Delta overexpression posterior to the morphogenetic furrow causes specific cell fate changes:
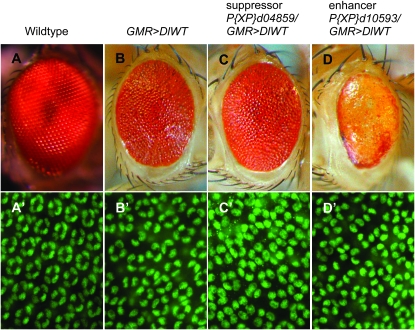
Notch signaling establishes spacing within the ommatidial array and regulates the specification of most, if not all, cell types within each ommatidium (see Introduction). To identify additional components of the Delta–Notch signaling pathway, we designed a genetic modifier screen based on overexpression of Delta in the developing retina under control of the GMR promoter, which drives gene expression posterior to the morphogenetic furrow (Hay et al. 1997). The GMR-Gal4 driver initiates expression in rows 4–6 of the developing eye disc (data not shown) when the photoreceptors R2, R3, R4, R5, and R8 are already present and photoreceptors R1 and R6 are joining the ommatidial precluster (Wolff and Ready 1993). GMR-driven wild-type Delta expression (GMR-Gal4 UAS-DeltaWT/+ or GMR>DlWT/+) leads to an adult eye that is glossy and reduced in size with irregular ommatidial spacing (Figure 1B). This phenotype is suitable for the identification of both suppressor and enhancer mutations and provides a suitable sensitized background for a genetic modifier screen.
Figure 1.—
Suppressors and enhancers of DeltaWT overexpression in the Drosophila eye. (A–D) Adult eyes. (A′–D′) Twenty-four-hour APF retinas stained with anti-Cut antibody to detect cone cells (green). (A and A′) A wild-type eye possesses an organized array of ommatidia (A); each ommatidium has four cone cells (A′). (B and B′) A GMR>DeltaWT/+ eye is small, glossy, and rough, with disorganized ommatidia (B) and an average of 3.34 cone cells/ommatidium (B′). (C and C′) A P{XP}d04859/GMR>DeltaWT eye. P{XP}d04859 mediates overexpression of Vha68-2. Adults have a larger, less glossy eye (C) with an average of 3.5 cone cells/ommatidium (C′); P < 0.005 compared with GMR>DlWT/+. (D and D′) A P{XP}d10593/GMR>DeltaWT eye. P{XP}d10593 mediates overexpression of Hr38. Adults have a smaller eye with loss of pigmentation and more disorganized ommatidia (D) with an average of 2.89 cone cells/ommatidium (D′); P < 0.005 compared with GMR>DlWT/+.
To understand how mis-regulation of Notch signaling by Delta overexpression leads to the observed adult eye phenotype, we examined the fates of two cell types induced by Notch signaling (see Introduction) after GMR-Gal4 expression is initiated: the R7 photoreceptor and the non-neuronal cone cells. We used the lacZ reporter XA12 to detect R7 photoreceptors (Van Vactor et al. 1991). In control third larval instar eye discs, we found a single XA12-positive cell per ommatidium, whereas in GMR>DlWT/+ eyes, we often detected multiple XA12-positive cells per ommatidium (see Figure S1). This indicates that overexpression of Delta results in the specification of excess R7 cells, suggesting an increase in Notch inductive signaling in this context. Wild-type ommatidia possess four cone cells, which strongly express Cut protein at 24 hr APF. In contrast to the increase in R7 cells, anti-Cut immunolabeling reveals a decrease in the number of cone cells per ommatidium in GMR>DlWT/+ retinas, as well as an overall disorganization of the ommatidial array (Figure 1B′). The average number of cone cells per ommatidium (n = 140) is 3.34 in the GMR>DlWT/+ eye, which is significantly less than the invariant number of four cone cells per ommatidium in wild-type eyes (Figure 1A′; see also Figure 3D). The decrease in cone cell numbers indicates a decrease in Notch inductive signaling in this context. Dominant-negative phenotypes resulting from overexpression of wild-type Delta have been observed previously in the notum (T. R. Parody, T. Zhong and M. A. T. Muskavitch, unpublished results) and in the wing pouch (De Celis and Bray 1997; Micchelli et al. 1997; Li and Baker 2004). The presence of Notch signaling gain-of-function and loss-of-function phenotypes in the GMR>DlWT/+ eye may be due to differential expression of GMR-Gal4 in different cell types or may reflect situations in which different cell types respond to ectopic Delta expression by Notch activation in some instances and by Notch inhibition in others. Taken together, our analyses indicate that overexpression of Delta posterior to the morphogenetic furrow causes specific cell fate changes for at least two cell types. These cell fate changes are consistent with either an increase in Notch signaling (R7 photoreceptor specification) or a decrease in Notch signaling (cone cell specification), depending on the developmental context assessed. The GMR>DlWT/+ genotype therefore provides an ideal genetic background for screening for modifiers of the effects of increased or decreased Notch signaling on the specification of distinct, well-characterized retinal cells.
Figure 3.—
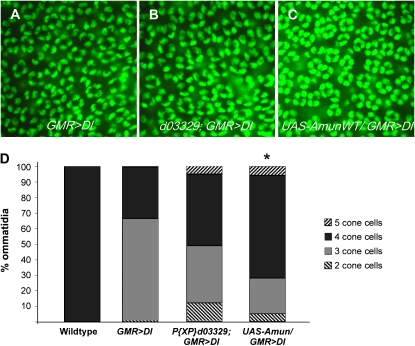
Overexpression of Amun suppresses the GMR>DeltaWT/+ cone cell phenotype. (A–C) Twenty-four-hour APF retinas stained with anti-Cut antibody (green) to detect cone cells. (A) A GMR>DeltaWT/+ retina exhibits a disorganized array of ommatidia with an average of 3.34 cone cells/ommatidium. (B) A P{XP}d03329/+;GMR>DeltaWT/+ retina exhibits suppression of the GMR>DeltaWT/+ cone cell phenotype, increasing the average to 3.5 cone cells/ommatidium. (C) A GMR>DeltaWT/+;UAS-AmunWT/+ retina exhibits greater suppression of the GMR>DeltaWT/+ cone cell phenotype, as compared to B, increasing the average to 3.73 cone cells/ommatidium (P < 0.005). (D) The proportional representation of ommatidia with two, three, four, or five cone cells. The percentage of ommatidia with four cone cells is greater for P{XP}d03329/+;GMR>DeltaWT/+ and GMR>DeltaWT/+;UAS-AmunWT/+ retinas than for GMR>DeltaWT/+ retinas. An asterisk denotes a statistically significant difference (P < 0.005) in average cone cells per ommatidium, as compared to GMR>DeltaWT/+.
A screen for suppressors and enhancers of Delta-dependent cell fate changes:
We performed a genetic modifier screen by assaying the effect of each of 10,447 transposon insertions on the GMR>DlWT/+ eye phenotype (see Table S1 and Table S3) from the Exelixis stock collection (Thibault et al. 2004). The transposons in this collection could act either through UAS-mediated overexpression of neighboring genes or by transposon-mediated reduction of gene function due to insertional gene disruption or antisense transcript synthesis (see materials and methods). Modifiers classified as “suppressors” yielded larger eyes with a more hexagonal appearance to the ommatidial array compared to GMR>DlWT/+ (Figure 1C). Modifiers classified as “enhancers” yielded a smaller, flatter eye with a “shinier” or “smoother” surface and/or loss of pigmentation compared to GMR>DlWT/+ (Figure 1D). We hypothesized that several classes of genes would be isolated as modifiers, including genes that directly regulate the Notch pathway (e.g., genes involved in processing, trafficking, and expression of Notch pathway members), genes that act in signaling pathways that interact with the Notch pathway (e.g., Ras/EGFR signaling), and genes that function in eye development independently of Notch signaling.
A total of 798 transposons modified the GMR>DlWT/+ phenotype in our primary screen (Table S1). Among these primary hits, 66% of the modifiers were UAS-containing XP transposons (see materials and methods), although XPs make up only 21% of the transposons screened (Table 1). The prevalence of XPs recovered suggests that the GMR>DlWT/+ phenotype is more easily modified by expression of neighboring genes via one of the UAS elements present in the XP transposon than by transposons that lack UAS elements and are more likely to disrupt genes by creating hypomorphic and/or null insertion alleles. Of the 798 modifying transposons, we chose to further analyze 284 transposons, most of which were classified as strong or moderate modifiers. Among these 284 primary hits, 260 transposons passed retesting against GMR>DlWT/+ (91% confirmation rate), and these were subsequently crossed to GMR-Gal4 to eliminate Delta-independent modifiers. This resulted in 170 “confirmed” modifiers, including 92 suppressors, 62 enhancers, 9 enhancers/suppressors (which exhibit aspects of both enhancement and suppression), and 7 modifiers that were lethal in combination with GMR>DlWT/+ (Table 1, Table S2, and Table S3). If we assume a similar confirmation rate for all 798 primary hits, we would predict our final hit rate as 6.9%. Our hit rate is comparable to the hit rate of 3.94% obtained by Kankel et al. (2007) in a screen of 15,500 lines from the same collection for phenotypic modification of reduced Notch signaling during wing-margin development using a C96-Gal4 UAS-MamDN (a dominant-negative Mam variant) genetic background.
TABLE 1.
Screen statistics of 170 confirmed modifying transposons
| Transposon type | Total in collection | Total screened | Passed retest and negative 2° testa | Enhancers | Suppressors | Lethal | Enhancers/Suppressors |
|---|---|---|---|---|---|---|---|
| PB | 3,548 | 2,421 | 7 | 2 | 4 | 1 | 0 |
| RB | 3,288 | 2,228 | 3 | 0 | 3 | 0 | 0 |
| WH | 5,637 | 3,632 | 8 | 0 | 7 | 0 | 1 |
| XP | 3,715 | 2,166 | 152 | 60 | 78 | 6 | 8 |
| Total | 16,188 | 10,447 | 170 | 62 | 92 | 7 | 9 |
A total of 10,447 transposons were screened, and 284 of the 798 primary hits were retested against GMR>DeltaWT/+. Of these, 260 passed retesting and were subsequently crossed to GMR-Gal4 (negative 2° test). A final set of 170 lines passed the negative secondary test. Enhancers/suppressors yield phenotypic characteristics associated with enhancement and suppression.
Of 798 primary hits, 284 were retested.
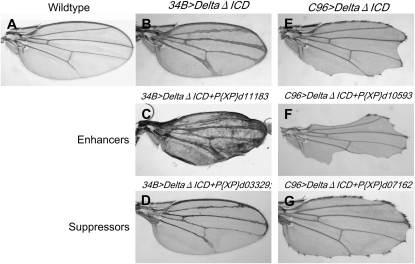
All 170 confirmed modifiers were crossed into two additional genetic backgrounds to assess their ability to modify Delta-dependent phenotypes affecting the wing vein and the wing margin. Expression of DeltaΔICD (Huppert et al. 1997) under the control of the 34B-Gal4 driver (Ingham and Fietz 1995) (34B>DeltaΔICD) causes the development of thickened wing veins (Figure 2B). DeltaΔICD expression under control of the C96-Gal4 driver (Gustafson and Boulianne 1996) results in notches in the wing margin (Figure 2E). Both phenotypes reflect reduced Notch signaling. Among the 170 confirmed modifiers, 20 enhanced and 11 suppressed the 34B>DeltaΔICD wing vein phenotype (see Figure 2, C and D, for examples; Table S2), while 33 enhanced and 25 suppressed the C96>DeltaΔICD wing-notching phenotype (see Figure 2, F and G, for examples; Table S2). Taken together, our secondary screen data indicate that the GMR>DlWT-based primary screen enriched for phenotypic modifiers of Notch-signaling-associated developmental defects.
Figure 2.—
Secondary tests for confirmed modifiers of the GMR>DeltaWT/+ phenotype. (A) A wild-type adult wing. (B) Adult 34B-Gal4 UAS-DeltaΔICD/+ (34B>DeltaΔICD) wings display thickened wing veins, consistent with reduced Notch signaling. (C) Adult 34B>DeltaΔICD/+;P{XP}d11183/+ wings exhibit enhancement of the 34B>DeltaΔICD wing-vein-thickening phenotype. P{XP}d11183 disrupts karst. (D) Adult P{XP}d03329/+;34B>DeltaΔICD/+ wings exhibit suppression of the 34B>DeltaΔICD wing vein-thickening phenotype. P{XP}d03329 mediates overexpression of Amun and CG1837. (E) Adult UAS-DeltaΔICD/+;C96-Gal4/+ (C96>DeltaΔICD) wings display notches along the wing margin, typical of reduced Notch signaling. (F) Adult P{XP}d10593/UAS-DeltaΔICD;C96-Gal4/+ wings exhibit enhancement of the C96>DeltaΔICD wing notching phenotype. P{XP}d10593 mediates overexpression of Hr38. (G) Adult UAS-DeltaΔICD/+;C96-Gal4/P{XP}d07162 wings exhibit suppression of the C96>DeltaΔICD wing notching phenotype. P{XP}d07162 disrupts Cysteine string protein.
Identification of loci that are dominant modifiers of Delta overexpression:
To understand the functional relevance of the recovered modifiers, we sorted genes potentially affected by these transposons into seven functional categories (cell–cell communication, cell metabolism, cytoskeletal/trafficking, gene regulation, lipid metabolism, protein metabolism, and transport) on the basis of previous published studies or on gene ontology terms associated with each gene in FlyBase. Our criteria for identifying affected genes varied by transposon type and are described in materials and methods. On the basis of these criteria, 152 modifying transposons had potential effects on 274 genes (195 genes with known or putative function and 79 genes of unknown function), 16 transposons resided in regions with no annotated genes, and two transposons have no sequence data available. We note that a few of the transposons have not been definitively placed at a unique site within the genome, and although most of the transposon insertion sites are accurate (Kankel et al. 2007), molecular characterization of all transposons would be required to positively confirm their assigned insertional positions. The distribution among functional categories of gene(s) potentially affected by these modifying transposons is summarized in Table 2.
TABLE 2.
Functional classification of 274 candidate genes potentially disrupted by 170 GMR>DeltaWT-modifying transposons
| Functional category | Total no. of genes |
|---|---|
| Cell–cell communication | 25 |
| Cell metabolism | 40 |
| Cytoskeletal/trafficking | 20 |
| Gene regulation | 62 |
| Lipid metabolism | 7 |
| Protein metabolism | 27 |
| Transporters | 14 |
| Novel/unable to assign | 79 |
| No identified CG in region | 16 |
| Total genes | 274 |
Members of a set of 274 candidate genes predicted to be affected by the 170 modifying transposons were placed within seven functional categories or classified as unknown. Since a few genes were recovered more than once, the number of transposon lines for each category may be higher than the number of genes represented by each category.
The 30 transposon insertions associated with cell–cell communication proteins include genes encoding known Notch pathway members (e.g., numb, kuzbanian; Bray 2006), as well as genes that have been recovered previously from Notch-based screens, such as patched, Ras85D, and puckered (Rottgen et al. 1998; Muller et al. 2005; Mahoney et al. 2006). We also identified genes encoding a number of cell–cell communication proteins not previously implicated in Notch signaling, such as the phosphatase Gilgamesh, the two immunoglobulin superfamily members Fasciclin 2 and ImpL2, and the two hormone-receptor-like genes Hr38 and Hr39. Overexpression of Hr38 by P{XP}d10593 enhances the GMR>DlWT/+ adult phenotype (Figure 1D), as well as the cone cell phenotype (Figure 1D′) (average of 2.89 cone cells/ommatidium; P < 0.005). Overexpression of Hr38 also enhances the wing notching phenotype of C96>DeltaΔICD (Figure 2F) and is lethal in combination with 34B>DeltaΔICD. These data suggest that overexpression of Hr38 reduces net Notch signaling in all of these contexts. Hr38 has been shown to mediate an ecdysteroid signaling pathway that is distinct from that involving the classical ecdysone receptor (EcR) (Baker et al. 2003). Ecdysteroids are hormones found only in arthropods that induce signals required for postembryonic development (Kozlova and Thummel 2000). The recovery of Hr38 as a Notch antagonist from our screen is not surprising considering that Notch signaling and EcR-mediated ecdysone signaling have recently been shown to act antagonistically in oogenesis during the switch from endoreplication (whole-genome amplification without cell division) to amplification (amplification of specific genes only, without cell division) (Sun et al. 2008). These results, considered in light of the emergence of Hr38 and Hr39 from our screen, suggest that multiple hormones and hormone receptors may work along with Notch signaling to specify cell fates in a variety of developmental contexts.
We recovered 22 transposons affecting genes that are likely to play roles in cytoskeletal regulation and/or intracellular trafficking. Ligand and receptor endocytosis, NotchECD trans-endocytosis, and intracellular trafficking are core regulatory elements in the activation and regulation of Notch signaling (see Introduction). Genes identified in this class include those encoding myosin-related proteins (jaguar/Myosin VI, myosin heavy chain, and myosin binding subunit) and actin-binding proteins (fimbrin, formin3, and diaphanous), peanut (a septin), short stop (a cytoskeletal protein), Cysteine string protein (a putative chaperone), and karst (βHeavy-spectrin). We also recovered Vha68-2, a subunit of the v-ATPase proton pump complex. v-ATPases are known to function in vesicle trafficking, membrane fusion, and acidification of organelles (Dow 1999). Interestingly, P{XP}d04859, which overexpresses Vha68-2, suppresses the GMR>DlWT/+ adult rough-eye phenotype (Figure 1C), the cone cell phenotype (average of 3.5/ommatidium cone cells; P < 0.005) (Figure 1C′), and the C96>DeltaΔICD wing-margin-notching phenotype (data not shown), suggesting that overexpression of Vha68-2 may increase Notch pathway activity in more than one context. Vha68-2 could play important roles during Notch signaling; for example, it may be required to maintain the pH necessary for dissociation of internalized Delta/NotchECD complexes and/or for proper intracellular trafficking of Notch and Delta proteins.
Our largest class of modifiers (excluding those affecting unknown genes) fall into the gene expression and transcriptional regulation (“gene regulation”) category. This group contains 67 independent transposon insertions affecting genes including pipsqueak, longitudinals lacking (lola), lilliputian, split ends (spen), tramtrack, TATA binding protein, and Suppressor of Triplolethal. Many of these genes are known to have roles during eye development (Neufeld et al. 1998; Wittwer et al. 2001; Voas and Rebay 2004). Some, like lola and spen, have been implicated previously in Notch signaling and are thought to antagonize the Notch pathway (Ferres-Marco et al. 2006; Doroquez et al. 2007). Others, such as lilliputian and tramtrack, are known to have interactions with other pathways, such as the Ras and ecdysteroid pathways, that are known to influence Notch signaling (see above) (Sundaram 2005; Hasson and Paroush 2006). We also identified genes encoding many largely uncharacterized proteins including CG9650, a zinc-finger-containing putative transcription factor that has been implicated in axon guidance in the embryo (McGovern et al. 2003). The modifying transposon, P{XP}d03295, overexpresses the entire protein and enhances the GMR>DlWT/+ eye, resulting in an eye devoid of pigment containing necrotic regions, indicative of cell death (data not shown). In contrast, analysis of cone cell development reveals that P{XP}d03295 significantly increases the number of cone cells per ommatidium in the GMR>DlWT/+ background (average of 3.87 cone cells/ommatidium; P < 0.005) (data not shown). This increase of cone cell number suggests that overexpression of CG9650 increases net Notch signaling during cone cell specification. The resulting necrotic eye, however, implies that CG9650 has additional roles during Drosophila eye development and/or in cell viability.
In summary, our screen has identified numerous known components or mediators of Notch signaling, as well as genes not previously associated with the pathway, many of which can be linked to specific signaling functions on the basis of their known roles in processes like intracellular trafficking or transcriptional regulation. We anticipate that this modifier collection will provide a rich resource for further investigations of the molecular mechanisms of Notch signaling.
P{XP}d03329 suppresses the GMR>DlWT/+ phenotype via overexpression of CG2446:
We chose to characterize one suppressor, P{XP}d03329, in more detail because of the previous implication of an adjacent gene in Notch signaling. P{XP}d03329 is located between two open reading frames, CG1837 and CG2446, and could potentially mediate overexpression of either gene. While CG1837 is an uncharacterized gene, CG2446 has been associated previously with Notch signaling in the adult eye and bristle organ (Abdelilah-Seyfried et al. 2001; Muller et al. 2005). EP(X)1503, a UAS-containing transposon insertion (Rorth 1996) upstream of the CG2446 open reading frame, has been identified as a modifier in several Drosophila screens (Abdelilah-Seyfried et al. 2001; Bourbon et al. 2002; Brody et al. 2002; Muller et al. 2005; Zhu et al. 2005). In one screen, EP(X)1503 expressed under control of the sca-Gal4 driver modified Notch and Hairless (a Notch pathway inhibitor; Schweisguth and Lecourtois 1998) loss-of-function phenotypes affecting adult bristles (Abdelilah-Seyfried et al. 2001). Overexpression of CG2446 in both genetic backgrounds resulted in various bristle phenotypes, including shaft-to-socket transformations and development of supernumerary macrochaetae. In a second screen, EP(X)1503 was recovered as a suppressor in a Hairless gain-of-function screen designed to identify modifiers of a small rough-eye phenotype caused by GMR-driven Hairless (Muller et al. 2005). We find that P{XP}d03329 not only mildly suppresses the GMR>DlWT/+ cone cell phenotype (see below), but also suppresses the 34B>DeltaΔICD wing vein-thickening phenotype (Figure 2D) and enhances the C96>DeltaΔICD wing notching phenotype (data not shown). Previous genetic interaction data for EP(X)1503, combined with our initial data regarding P{XP}d03329, suggest that CG2446 may function in eye and bristle development in conjunction with Notch signaling.
UAS-Amun suppresses the GMR>DeltaWT/+ eye phenotype in a manner similar to P{XP}d03329:
To verify the genomic insertion position of P{XP}d03329, we used inverse PCR and two-sided PCR to map the insertion to the X chromosome at coordinate 11,610,026 (FB2006_01 Dmel Release 5.1), which is ∼200 bp displaced from the insertion site previously annotated by Exelixis (Thibault et al. 2004). To confirm that overexpression of Amun causes the suppression of the GMR>DlWT/+ eye phenotype, we generated a transgene placing a full-length Amun open reading frame under the control of a UAS regulatory cassette and generated transgenic lines. Co-expression of UAS-DeltaWT and UAS-Amun under GMR-Gal4 control results in suppression of the GMR>DlWT/+ adult rough-eye phenotype to an extent similar to that seen with P{XP}d03329 (data not shown). We analyzed cone cells of GMR>DlWT/+;UAS-AmunWT/+ 24-hr APF retinas and found significant suppression of cone cell loss (an average of 3.73 cone cells/ommatidium; P < 0.005) (Figure 3, C and D). We also observed mild suppression when P{XP}d03329 was crossed to GMR>DlWT (Figure 3, B and D). Since loss of cone cells results from decreased Notch signaling, these results suggest that Amun function potentiates Notch signaling required for cone cell induction in the developing Drosophila eye. This interpretation is further corroborated by the observation that overexpression of Amun mediated by EP(X)1503 suppresses inhibition of Notch signaling that results from Hairless overexpression in the eye (Muller et al. 2005).
Amun is a nuclear protein and has a conserved DNA glycosylase domain:
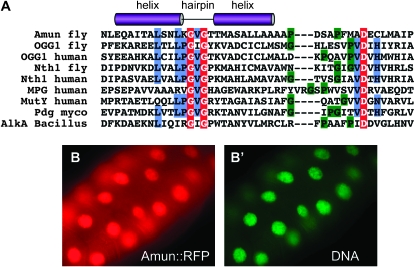
Amun is located cytogenetically at 10D6-10D7 and encodes a 550-amino-acid (aa) protein with a predicted molecular weight of ∼58.4 kDa. The entire protein sequence is highly conserved among drosophilid species (see Figure S2). The amino-terminal region of the protein also exhibits significant conservation among a set of putative orthologs from vertebrate and invertebrate species. In contrast, the carboxyl-terminal region of the protein exhibits no sequence similarity in animals beyond the drosophilids (see Figure S2), and it contains a predicted coiled-coil domain between aa 448 and aa 481 (Figure S2, region shaded in gray). Coiled-coil domains are thought to mediate protein–protein interactions and are found in proteins with diverse biological functions such as vesicle trafficking, cell signaling, and transcriptional regulation (Yu 2002). The amino-terminal region of Amun includes a putative DNA glycosylase domain (aa 116–151) that is highly conserved across phyla (Figure 4A). DNA glycosylases initiate an evolutionarily conserved base excision repair pathway by excising mismatched or altered bases that result from processes including oxidation, deamination, alkylation, and methylation (Dizdaroglu 2005). In addition, DNA glycosylases have been shown to act as transcriptional regulators (Choi et al. 2002; Cortazar et al. 2007).
Figure 4.—
Amun contains a putative DNA glycosylase domain and localizes to the nucleus. (A) Comparison of the Amun HhH DNA glycosylase domain to other known DNA glycosylases. The sequences used were the following: MPG, H. sapiens; MutY, H. sapiens; Pdg, M. tuberculosis; AlkA, B. subtilis; OGG1, D. melanogaster; Ogg1, H. sapiens; Nth1, D. melanogaster; Nth1, H. sapiens. There is little sequence similarity among these proteins; however, they share a conserved DNA-binding motif that consists of two α-helices (purple cylinders denote the approximate locations of these helices) connected by a hairpin loop with the consensus sequence LPG(V/I)G followed by a glycine/proline-rich region (green highlight) and a catalytically active aspartic acid residue (D, red highlight). The conserved H/N residue (blue highlight), following the catalytic D residue, differentiates between monofunctional (N) and bifunctional (H) glycosylases. Blue-highlighted L, P, and V residues are part of the consensus HhH domain. Red-highlighted white G residues are completely conserved in these DNA glycosylases. (B and B′) A dpp-Gal4/UAS-Amun∷RFP third larval instar salivary gland. Amun∷RFP (red) localizes to nuclei (B), as indicated by the DNA dye Vybrant Green (B′).
Protein sequences of DNA glycosylases vary significantly in size and possess little to no sequence conservation within their amino and carboxyl termini. However, many share a DNA glycosylase domain consisting of a leucine–proline–glycine–valine/isoleucine–glycine “hairpin loop” sequence flanked by two helices (α-helix–hairpin loop–α-helix, or HhH, domain) followed by a glycine/proline-rich region and a conserved, catalytically active aspartic acid, which donates an electron during DNA base excision (Figure 4A) (Krokan et al. 1997; Scharer and Jiricny 2001). Since the conserved sequence of Amun possesses a DNA glycosylase domain, we asked whether the protein localizes to the nucleus, where a DNA glycosylase/transcriptional regulator would be predicted to function. We engineered an RFP-tagged version of Amun and expressed it transgenically in Drosophila tissues, including salivary glands (Figure 4B); the notum (see Figure 8B); wing, eye, and leg imaginal discs; and S2 cells (data not shown). We find that overexpressed Amun∷RFP localizes to the nucleus in all tissues examined (we note that an antibody against the endogenous protein will be required to confirm that endogenous Amun is also nuclear), consistent with possible functions as a DNA glycosylase and/or transcriptional regulator.
Figure 8.—
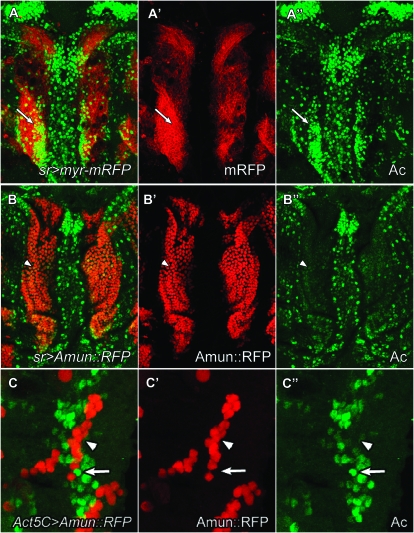
Amun overexpression downregulates Achaete levels in a cell-autonomous manner. (A and B) Nota from 9-hr APF pupae (27°) stained with anti-Achaete antibody (green). (A, A′, and A″) A sr-Gal/UAS-myr-mRFP notum. RFP (red) reflects expression of mRFP within the sr domain. Achaete-positive cells (arrows) mark the microchaeta proneural groups during this developmental stage. (B, B′, and B″) A sr-Gal4/UAS-Amun∷RFP notum shows a severe reduction or absence of Achaete expression (arrowheads) within the sr expression domain where Amun∷RFP is overexpressed. (C, C′, and C″) A 6- to 7-hr APF notum stained with anti-Achaete antibody (green in C and C″). Amun∷RFP is overexpressed randomly throughout the notal disc in Act5C-Gal4 clones (red). Achaete protein levels are downregulated in cells in which Amun∷RFP is overexpressed (arrowheads). Achaete-positive cells can be found directly adjacent to Amun∷RFP-positive cells (arrows) in clones in all regions of the notum.
Amun overexpression and reduction of Amun expression by RNA interference cause bristle defects:
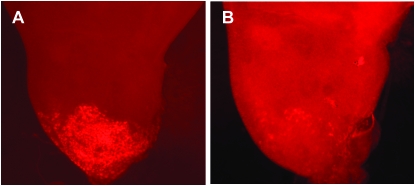
To understand the role of Amun during development, we examined the effects of loss of Amun function, as well as Amun overexpression, in different tissues during development. To examine the loss-of-function phenotypes of Amun, we used UAS-AmunRNAi transgenic flies obtained from the Vienna Drosophila RNAi Center (Dietzl et al. 2007). To show that this RNA interference (RNAi) strain can effectively reduce Amun protein expression, we co-expressed Amun∷RFP and AmunRNAi in the notum under the control of pnr-Gal4 (see below) and examined discs for the presence of RFP (see Figure S4 for a diagram of the pnr-Gal4 expression domain). In the absence of AmunRNAi, robust RFP accumulation is seen within the pnr expression domain (Figure 5A). In contrast, when AmunRNAi is co-expressed, a severe reduction in RFP is observed within the pnr expression domain (Figure 5B). Therefore, AmunRNAi effectively blocks Amun overexpression. Importantly, phenotypes induced by AmunRNAi can be rescued by overexpression of Amun∷RFP (see below), suggesting that Amun-RNAi also has effects on endogenous Amun levels.
Figure 5.—
AmunRNAi effectively reduces Amun∷RFP protein expression. Third larval instar wing/notal imaginal discs. (A) A pnr-Gal4 UAS-Amun∷RFP/+ disc (pnr>Amun∷RFP). Overexpression of Amun∷RFP can be detected via RFP expression (red) in the pnr expression domain. (B) A UAS-AmunRNAi/+; pnr>Amun∷RFP/+ disc. AmunRNAi successfully reduces Amun∷RFP expression, as seen by the substantial reduction of Amun∷RFP signal within the pnr expression domain.
Ubiquitous overexpression of Amun or AmunRNAi using the Act5C-Gal4 driver results in lethality between the first and second larval instar, suggesting that Amun is an essential gene necessary for aspects of embryonic and/or early larval development (data not shown). This is consistent with previous reports that Amun is an essential gene expressed during embryonic and larval development (Bourbon et al. 2002; Brody et al. 2002). When AmunRNAi is expressed under the control of ey-Gal4 (a transgene that drives expression in the early antennal-eye imaginal disc; Bose et al. 2006), we observe a reduction in eye size (see Figure S3) consistent with a role for Amun in eye development (see above). Reduction of Amun levels under control of the sca-Gal4 or ptc-Gal4 driver results in multiple bristle defects including missing, supernumerary, and misplaced macrochaetae, as well as probable shaft-to-socket transformations (Figure 6, E and F, respectively). Reduction of Amun under control of sr-Gal4 (a transgene that drives expression in a subset of microchaeta rows in the medial and lateral notum; Calleja et al. 2002) and pnr-Gal4 (a P-element insertion in the pnr gene that drives expression in the 10 medial microchaeta stripes; Heitzler et al. 1996) results in disorganized and smaller microchaetae (Figure 6, G and H, respectively). Importantly, as shown in Figure 6I, co-overexpression of Amun∷RFP with AmunRNAi rescues the loss-of-function phenotypes shown in Figure 6H, suggesting that the AmunRNAi phenotype results from specific reduction of endogenous Amun function.
Figure 6.—
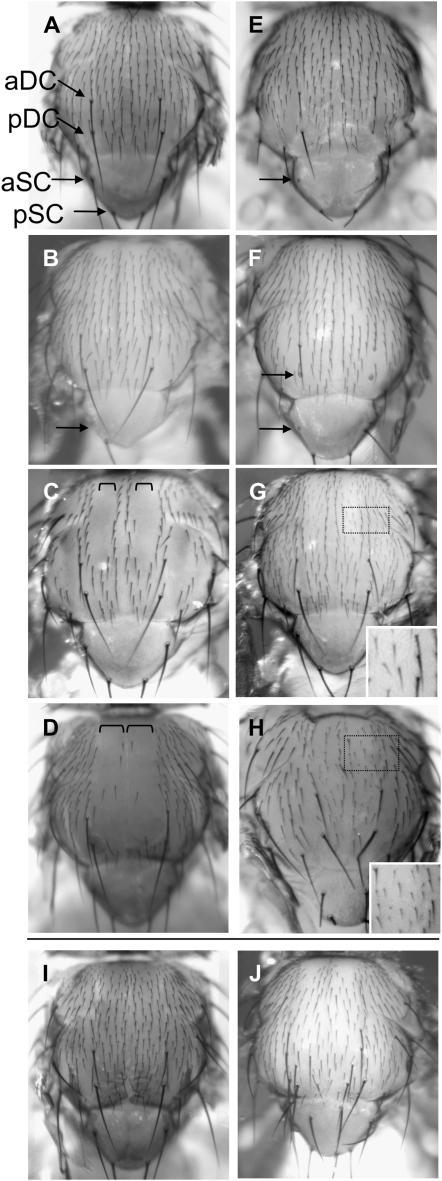
Amun loss-of-function, gain-of-function, and rescue experiments demonstrate a function for Amun during sensory organ development. (A) A wild-type adult notum. There are 10 organized rows of notal microchaetae including and between the two rows containing the dorsocentral macrochaetae (aDC and pDC). (B–D) Amun∷RFP overexpression phenotypes (assessed following growth at 27°). (E–H) Loss-of-function phenotypes that result from UAS-AmunRNAi expression (27° unless otherwise noted). (B) A ptc-Gal4/+;UAS-Amun∷RFP/+ notum results in loss of aSC (arrow) and pSC macrochaetae. (C) A sr-Gal4/UAS-Amun∷RFP notum exhibits severe loss of microchaetae in stripes 2 and 3 (highlighted with square brackets). (D) A pnr-Gal4 UAS-Amun∷RFP/+ notum exhibits severe loss of microchaetae across the central notum from stripe 1 to stripe 4 (highlighted with square brackets). (E) A UAS-AmunRNAi/+;UAS-Dicer2/ sca-Gal4 notum exhibits loss of aDC (data not shown), aSC (arrow), and pSC macrochaeta shafts (data not shown). (F) A UAS-AmunRNAi/+;UAS-Dicer2/ptc-Gal4 notum exhibits loss of aDC, pDC (arrow), and aSC (arrow) macrochaeta shafts, as well as misplacement of aSC macrochaetae. (G) A UAS-AmunRNAi/+;UAS-Dicer2/+;sr-Gal4/+ notum exhibits smaller microchaetae within the sr expression domain. Inset shows a magnified view of the medial notum. (H) A UAS-AmunRNAi/+;UAS-Dicer2/+;pnr-Gal4/+ notum (25°) displays smaller microchaetae and misplaced aDC macrochaetae. Inset shows a magnified view of the medial notum. (I) A UAS-AmunRNAi/+;pnr-Gal4 UAS-Amun∷RFP/+ notum. Co-expression of Amun∷RFP and AmunRNAi rescues the Amun∷RFP-induced gain-of-function phenotype (microchaeta loss) shown in D and the AmunRNAi-induced loss-of-function phenotype (smaller microchaetae) shown in H. (J) A UAS-Achaete/+;pnr-Gal4 UAS-Amun∷RFP/+ notum. Overexpression of Achaete partially rescues the Amun∷RFP overexpression phenotype (microchaeta loss) shown in D, supporting the hypothesis that loss of microchaetae occurs as a result of reductions in Achaete function. We also note the presence of ectopic macrochaetae, which may be due to Achaete overexpression. aDC, anterior dorsocentral; pDC, posterior dorsocentral; aSC, anterior scutellar; pSC, posterior scutellar.
Overexpression of Amun∷RFP under sca-Gal4 control results in a combination of missing, extra, and misplaced macrochaetae (data not shown), whereas overexpression under ptc-Gal4 control results in missing macrochaetae (Figure 6B). This range of bristle phenotypes is consistent with previous results obtained by overexpressing Amun in EP(X)1503 flies under sca-Gal4 control (see above) (Abdelilah-Seyfried et al. 2001). The most severe phenotype that we observed is the complete loss of microchaetae when Amun∷RFP is expressed using sr-Gal4 (Figure 6C) or pnr-Gal4 (Figure 6D). These data indicate that both reduced and increased Amun expression levels lead to cell fate specification and/or morphogenetic defects in the Drosophila eye and/or notum.
Loss of microchaetae is due to loss of bristle sensory organ precursor cells:
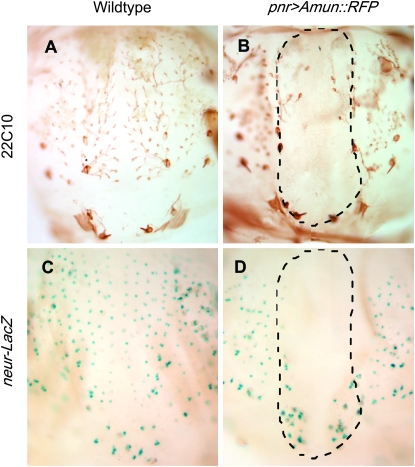
Loss of bristles following Amun overexpression could result from loss of SOPs or could reflect the loss of socket and shaft cells that would result from the adoption of the pIIb cell fate by the pIIa cell (resulting in multiple neurons and/or sheath cells in each organ). To determine whether Amun plays a role in the latter decision, we used MAb22C10 to stain neuron and shaft cells in the developing notum. In control nota at 31 hr APF, we detected a regular array of microchaeta neurons (Figure 7A). In contrast, there were few or no neurons or shaft cells discernible within the pnr expression domain following overexpression of Amun (Figure 7B), suggesting that Amun acts upstream of pIIa/pIIb specification during the development of the bristle organ. We then asked whether Amun plays a role in SOP specification. To test this possibility, we assayed SOP specification in the presence and absence of pnr-driven Amun∷RFP (pnr>Amun∷RFP) using neurA101, a lacZ insertion in the neuralized gene (Bellen et al. 1989), to mark SOP cells. Analysis of 15-hr APF nota stained for β-galactosidase activity reveals a regular array of microchaeta SOPs in control nota (Figure 7C) and the absence of SOPs within the pnr expression domain in pnr>Amun∷RFP nota (Figure 7D). This suggests that overexpression of Amun in the notum interferes with either the specification of SOPs or the formation of the proneural clusters within which SOPs are specified.
Figure 7.—
Amun-induced loss of notal microchaetae is due to the loss of bristle organs and sensory organ precursors in the developing notum. (A and B) Nota from 31-hr APF pupae (27°) stained with MAb22C10 to detect neurons and shaft cells. (A) A wild-type notum has organized rows of microchaeta neurons and shaft cells. (B) A pnr-Gal4 UAS-Amun∷RFP/+ notum lacks staining for neurons within the pnr expression domain (outlined by dashes), indicating that the absence of external shafts is not due to the transformation of shaft/socket cells into neurons. (C and D) Nota from 15-hr APF pupae (27°) stained for β-galactosidase activity. The neur A101 lacZ insertion in the neuralized gene is used to mark SOPs. (C) A neur A101 notum exhibits wild-type rows of microchaete SOPs. (D) A pnr-Gal4 UAS-Amun∷RFP/neurA101 notum lacks SOPs within the pnr expression domain (outlined by dashes).
Ectopic Amun expression downregulates the proneural transcription factor Achaete:
To determine whether failure of SOP specification in regions of elevated Amun expression is due to the absence of SOP proneural groups, we used Achaete immunolabeling to detect microchaeta proneural equivalence groups in the developing notum. Achaete is a bHLH transcription factor required for SOP specification within proneural equivalence groups (see Introduction). We compared the pattern of Achaete expression in sr>Amun∷RFP nota with that in control nota using sr>mRFP (a myristylated monomeric RFP; Andersen et al. 2005). Achaete-positive cells are detected in regions of control nota expressing mRFP at 9 hr APF (Figure 8A). In contrast, Achaete protein levels are severely reduced in the sr expression domain (microchaeta rows 2 and 3) following overexpression of Amun∷RFP (Figure 8B). Similar results are obtained with expression of Amun∷RFP under control of pnr-Gal4 (data not shown). These results suggest that the loss of microchaetae shown in Figure 6, C and D, is due to the absence of proneural groups and that overexpression of Amun downregulates levels of Achaete expression.
We investigated whether the downregulation of Achaete by Amun overexpression is cell autonomous or nonautonomous by overexpressing Amun∷RFP randomly throughout the disc in gain-of-function clones (Glittenberg et al. 2006) under control of the Act5C-Gal4 driver. Adults developing from larvae with gain-of-function clones exhibit numerous small patches of notal microchaeta loss (data not shown). Immunohistochemical analysis of clones in pupae reveals reductions in Achaete levels in cells that express Amun∷RFP within microchaeta proneural groups. At clone borders, strong Achaete staining is frequently detected in wild-type cells directly adjacent to Amun∷RFP-positive cells, which generally lack Achaete expression (Figure 8C). These observations indicate that overexpression of Amun exerts cell-autonomous effects on Achaete protein levels. This effect could reflect direct action of Amun on Achaete levels or an indirect action of Amun via other factors that regulate Achaete levels in the notum (see discussion).
We then asked whether or not loss of Achaete is responsible for the bristle-loss phenotype observed when Amun is overexpressed. Indeed, we find that overexpression of Achaete and Amun∷RFP within the pnr domain results in a significant rescue of the pnr>Amun∷RFP microchaeta-loss phenotype (Figure 6J). This result demonstrates that the primary cause of microchaeta loss induced by Amun overexpression is the loss of Achaete expression.
In contrast to Amun overexpression, reduction of Amun function using sr-Gal4>AmunRNAi causes smaller and disorganized microchaetae, but does not cause any apparent changes in microchaeta number, as might be expected if loss of Amun function affected Achaete levels. Indeed, we could not detect any overt changes in notal Achaete immunolabeling following knockdown of Amun using sr-Gal4 (data not shown). There are several possible explanations for this observation. First, it is possible that Amun does affect Achaete expression levels, but the change in level is not easily recognized with immunolabeling. Second, it is possible that expression of endogenous Achaete levels can be maintained by very low levels of Amun function and that knockdown mediated by AmunRNAi is not sufficient to cause a discernible effect on Achaete expression.
These observations suggest that while Amun can clearly affect Achaete expression and SOP specification, Amun probably also plays roles in other aspects of bristle development. Because Achaete is not known to be involved in bristle development beyond SOP formation (Bertrand et al. 2002), we suggest that Amun can act through other factors to control sensory organ development following SOP specification.
DISCUSSION
Drosophila continues to play a leading role in the discovery of genes and mechanisms implicated in developmental processes mediated by, or associated with, the Notch signaling pathway. We present the results of a transposon screen for the effects of loss-of-function and gain-of-function mutations in a genetic background sensitized for Delta-mediated cell fate changes. In addition, we characterize Amun, a nuclear protein identified as a suppressor in our screen. We show that Amun suppresses a dominant-negative effect of Delta overexpression on cone cell induction in the eye, suggesting that Amun can positively regulate Notch signaling in this context. Alternatively, Amun may function in a parallel or intersecting pathway to affect cone cell development. We also provide evidence that Amun can function early during the cellular patterning underlying mechanosensory bristle development by downregulating the expression of the proneural transcription factor Achaete (see below). The identification and initial characterization of Amun function reflect the potential of the ensemble of 170 transposon insertions identified in our screen for discovery of additional factors that affect Notch signaling mediated development.
A screen for Notch-mediated development:
The Exelixis collection covers ∼50% of Drosophila genes (Thibault et al. 2004) and contains many new alleles for genes that may prove to be involved in the Delta–Notch signaling pathway (e.g., Kankel et al. 2007 and this work) or other developmental pathways. The collection has also been screened by Kankel et al. (2007) in a search for modifiers of a Notch loss-of-function signaling phenotype in the wing margin using C96-driven MamDN. Among the 170 modifiers that we identified, 29 lines were also recovered by Kankel et al. (2007) (see Table S2) and 141 lines were recovered only in our screen. Among the putative genes recovered in both screens are several known Notch pathway members and genes that have been previously recovered from Notch-based screens (e.g., numb, wingless, puckered, and Ras85D). In addition, several genes that had not been implicated previously in Notch signaling were identified in both screens, supporting roles for their encoded products during Notch-mediated development. These genes include peanut (a septin), Oatp30B (an ion channel), Indy (a transporter), and Hr38 (a hormone receptor). Of potentially equal interest are the 11 transposon lines that modified phenotypes in both of the secondary tests in this work (Table S2). Genes potentially disrupted by these transposons include karst (βHeavy-spectrin), bifocal (a cytoskeletal regulator), diaphanous (an actin-binding protein), and caudal (a transcriptional regulator). Further characterization of these genes, as well as other genes recovered in our screen, will help provide a deeper understanding of the mechanisms that govern the Notch signaling pathway.
The function of Amun during Drosophila sensory organ development:
A number of our results suggest that Amun is required for cell fate determination during Notch-mediated bristle organ development. Reduction of Amun function and Amun protein overexpression in the developing notum, using several Gal4 drivers including pnr, ptc, sca, and sr, generate defects during microchaeta and macrochaeta development. Substantial loss of microchaetae is observed in the nota of adults that express Amun under pnr-Gal4 or sr-Gal4 control during development. Immunohistochemical analysis of developing nota and the Achaete expression rescue experiments demonstrates that this loss of microchaetae is due to loss of the bHLH transcription factor Achaete. The expression patterns of the proneural proteins Achaete and Scute are best characterized for the dorsocentral macrochaetae, for which cis-regulatory elements control the expression of these genes in specific patterns to establish proneural clusters (reviewed in Modolell and Campuzano 1998 and Calleja et al. 2002). These enhancer elements are thought to be activated directly by members of several signaling pathways, including Decapentaplegic and Wingless, as well as by other factors including Pannier (Pnr), Daughterless (Da), Chip, and members of the Iroquois complex (Araucan and Caupolican) (reviewed in Bertrand et al. 2002). The expression of achaete/scute is antagonized by several factors, including U-shaped and dCtBP, both of which bind Pnr to form a transcriptional corepressor complex (Cubadda et al. 1997; Sato and Saigo 2000; Biryukova and Heitzler 2008; Stern et al. 2009); Extramacrochaetae (Emc), which forms a heterodimer with Da to inactivate it (Ellis et al. 1990; van Doren et al. 1991); and the E(spl)-C proteins, which are downstream targets of Notch signaling (de Celis et al. 1996). In microchaeta proneural groups, Achaete is also known to be repressed by Hairy (Ohsako et al. 1994; van Doren et al. 1994), as well as by Notch signaling (Parks et al. 1997). We demonstrate here that the effect of Amun overexpression on Achaete levels is cell autonomous, suggesting that the action of Amun on achaete expression could be direct. However, while it is tempting to speculate that Amun regulates Achaete levels by directly binding to cis-regulatory elements that affect achaete expression, we cannot rule out the possibilities that Amun functions by repressing an activator of achaete (e.g., Da or Chip), by activating a repressor of achaete (e.g., Emc, Hairy, or the Notch pathway), or by destabilizing achaete mRNA or protein.
Reductions in Amun function by RNA interference result in small and disorganized microchaetae. In contrast to the Amun overexpression phenotype, the small microchaeta phenotype is not easily attributable to changes in Achaete expression, given that Achaete has no known roles in bristle development subsequent to SOP specification. It has been shown that bristle shaft size can be correlated with several processes. First, both the shaft and socket cells undergo endoreplication to form polyploid nuclei that are required to form the elongated shaft structure. The degree of endoreplication has been correlated with shaft size (Edgar and Orr-Weaver 2001; Weng et al. 2003). Second, shaft length can be affected by mutations in genes that affect actin bundle formation necessary for proper elongation of the shaft (Tilney et al. 2000). Third, there is a period of rapid protein synthesis during sensory bristle development that enables the shaft and socket cells to generate the high levels of protein required for the development of the socket and shaft structures (Lambertsson 1998; Marygold et al. 2007). Genes necessary for this process include small bristles [which exports mRNA from the nucleus into the cytoplasm (Korey et al. 2001)] and the Minute loci [genes encoding ribosomal proteins (Lambertsson 1998; Saeboe-Larssen et al. 1998; Marygold et al. 2007)], which can affect bristle shaft length. Preliminary data suggest that Amun is unlikely to affect endoreplication. We measured nuclei of microchaetae that develop in regions of the notum expressing sr-driven AmunRNAi and found no consistent effects on nuclear size as compared to the nuclei of cells of microchaetae in regions devoid of AmunRNAi (data not shown). We therefore favor the notion that Amun may be required for transcriptional regulation of specific genes involved in growth and elongation of the shaft or for the elevated levels of mRNA and protein synthesis required for shaft development.
Amun as a DNA glycosylase and transcriptional regulator:
Our finding that Amun can affect Achaete expression levels, together with our identification of Amun as a nuclear protein with a putative DNA glycosylase domain, are consistent with the hypothesis that Amun functions as a transcriptional regulator. While DNA glycosylases are best known for repair of damaged and mismatched bases, recent work indicates that they also play roles in transcriptional regulation. The mammalian DNA glycosylase thymine DNA glycosylase (TDG) acts as a transcriptional co-activator, when bound to CREB-binding protein (CBP) and p300 (Tini et al. 2002), to enhance CBP-activated transcription in cell culture (Cortazar et al. 2007). It also acts as a transcriptional corepressor when bound to thyroid transcription factor-1 (TTF1) to repress TTF1-activated transcription in cell culture (Cortazar et al. 2007; Kovtun and McMurray 2007). The Arabidopsis DNA glycosylase DEMETER is required to activate expression of the maternal MEDEA allele, an imprinted maternal gene essential for viability (Choi et al. 2002). In light of these studies, the nuclear localization of Amun is suggestive of a function for Amun as a transcriptional regulator.
In summary, we demonstrate that Amun is a nuclear protein essential for organismal viability and proper cell fate specification during metamorphosis of Drosophila tissues, including the eye and mechanosensory organs. We suggest that Amun affects at least two distinct processes during bristle organ development because of the distinct loss-of-function and gain-of-function bristle phenotypes associated with Amun. One pathway is critical for regulation of Achaete protein levels, and the other pathway affects sensory organ bristle shaft size. Because the sequence of Amun contains a putative DNA glycosylase domain, we reason that Amun may act as a transcriptional regulator, as previously demonstrated for other DNA glycosylases (e.g., TDG and DEMETER). Further characterization of Amun is necessary to identify distinct transcriptional targets and pathways on which it may act and to decipher its potential function as a DNA glycosylase during Drosophila development.
Acknowledgments
We thank S. Artavanis-Tsakonas for allowing us to screen the Exelixis collection and M. Kankel, D. Dimlich, and G. Doughty for help collecting males. We thank K. Klueg for generating the GMR-Gal4 UAS-DeltaWT stock. We thank T. Orenic, P. Simpson, H. Bellen, B. Yedvobnick, A. Brand, G. Morata, the Developmental Studies Hybridoma Bank, the Bloomington Drosophila Stock Center, the Drosophila Genome Resource Center, and the Vienna Drosophila RNAi Center for stocks and reagents. We are grateful to P. R. Hiesinger for discussion and critical reading of the manuscript and technical assistance. We also thank J. Rosenberg for technical assistance. This work was funded by National Institutes of Health grant GM33291 and the DeLuca Professorship awarded to M.A.T.M.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.099986/DC1.
References
- Abdelilah-Seyfried, S., Y. M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2001. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 157 455–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, R., Y. Li, M. Resseguie and J. E. Brenman, 2005. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J. Neurosci. 25 8878–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., 2004. Accessing the Exelixis collection. Nat. Genet. 36 207. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M. D. Rand and R. J. Lake, 1999. Notch signalling: cell fate control and signal integration in development. Science 284 770–775. [DOI] [PubMed] [Google Scholar]
- Baker, K. D., L. M. Shewchuk, T. Kozlova, M. Makishima, A. Hassell et al., 2003. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell 113 731–742. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., 2000. Notch signaling in the nervous system: pieces still missing from the puzzle. BioEssays 22 264–273. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., and S. Y. Yu, 1997. Proneural function of neurogenic genes in the developing Drosophila eye. Curr. Biol. 7 122–132. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., C. J. O'Kane, C. Wilson, U. Grossniklaus, R. K. Pearson et al., 1989. P-element mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3 1288–1300. [DOI] [PubMed] [Google Scholar]
- Bertrand, N., D. S. Castro and F. Guillemot, 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3 517–530. [DOI] [PubMed] [Google Scholar]
- Biryukova, I., and P. Heitzler, 2008. Drosophila C-terminal binding protein dCtBP is required for sensory organ prepattern and sharpens proneural transcriptional activity of the GATA factor Pnr. Dev. Biol. 323 64–75. [DOI] [PubMed] [Google Scholar]
- Bose, A., B. Kahali, S. Zhang, J. M. Lin, R. Allada et al., 2006. Drosophila CK2 regulates lateral-inhibition during eye and bristle development. Mech. Dev. 123 649–664. [DOI] [PubMed] [Google Scholar]
- Bourbon, H. M., G. Gonzy-Treboul, F. Peronnet, M. F. Alin, C. Ardourel et al., 2002. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 110 71–83. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Bray, S. J., 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7 678–689. [DOI] [PubMed] [Google Scholar]
- Brody, T., C. Stivers, J. Nagle and W. F. Odenwald, 2002. Identification of novel Drosophila neural precursor genes using a differential embryonic head cDNA screen. Mech. Dev. 113 41–59. [DOI] [PubMed] [Google Scholar]
- Brou, C., 2009. Intracellular trafficking of Notch receptors and ligands. Exp. Cell Res. 315 1549–1555. [DOI] [PubMed] [Google Scholar]
- Calleja, M., O. Renaud, K. Usui, D. Pistillo, G. Morata et al., 2002. How to pattern an epithelium: lessons from achaete-scute regulation on the notum of Drosophila. Gene 292 1–12. [DOI] [PubMed] [Google Scholar]
- Carthew, R. W., 2007. Pattern formation in the Drosophila eye. Curr. Opin. Genet. Dev. 17 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis, A., 2006. Why is Delta endocytosis required for effective activation of Notch? Dev. Dyn. 235 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y., M. Gehring, L. Johnson, M. Hannon, J. J. Harada et al., 2002. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110 33–42. [DOI] [PubMed] [Google Scholar]
- Cooper, M. T., and S. J. Bray, 2000. R7 photoreceptor specification requires Notch activity. Curr. Biol. 10 1507–1510. [DOI] [PubMed] [Google Scholar]
- Cortazar, D., C. Kunz, Y. Saito, R. Steinacher and P. Schar, 2007. The enigmatic thymine DNA glycosylase. DNA Repair (Amst.) 6 489–504. [DOI] [PubMed] [Google Scholar]
- Cubadda, Y., P. Heitzler, R. P. Ray, M. Bourouis, P. Ramain et al., 1997. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 11 3083–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis, J. F., and S. Bray, 1997. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124 3241–3251. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., J. de Celis, P. Ligoxygakis, A. Preiss, C. Delidakis et al., 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122 2719–2728. [DOI] [PubMed] [Google Scholar]
- del Alamo, D., and M. Mlodzik, 2006. Frizzled/PCP-dependent asymmetric neuralized expression determines R3/R4 fates in the Drosophila eye. Dev. Cell 11 887–894. [DOI] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu, M., 2005. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res. 591 45–59. [DOI] [PubMed] [Google Scholar]
- Doroquez, D. B., T. L. Orr-Weaver and I. Rebay, 2007. Split ends antagonizes the Notch and potentiates the EGFR signaling pathways during Drosophila eye development. Mech. Dev. 124 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, J. A., 1999. The multifunctional Drosophila melanogaster V-ATPase is encoded by a multigene family. J. Bioenerg. Biomembr. 31 75–83. [DOI] [PubMed] [Google Scholar]
- D'Souza, B., A. Miyamoto and G. Weinmaster, 2008. The many facets of Notch ligands. Oncogene 27 5148–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B. A., and T. L. Orr-Weaver, 2001. Endoreplication cell cycles: more for less. Cell 105 297–306. [DOI] [PubMed] [Google Scholar]
- Ellis, H. M., D. R. Spann and J. W. Posakony, 1990. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell 61 27–38. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco, D., I. Gutierrez-Garcia, D. M. Vallejo, J. Bolivar, F. J. Gutierrez-Avino et al., 2006. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439 430–436. [DOI] [PubMed] [Google Scholar]
- Fiuza, U. M., and A. M. Arias, 2007. Cell and molecular biology of Notch. J. Endocrinol. 194 459–474. [DOI] [PubMed] [Google Scholar]
- Fortini, M. E., 2002. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat. Rev. Mol. Cell Biol. 3 673–684. [DOI] [PubMed] [Google Scholar]
- Freeman, M., 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87 651–660. [DOI] [PubMed] [Google Scholar]
- Gho, M., Y. Bellaiche and F. Schweisguth, 1999. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development 126 3573–3584. [DOI] [PubMed] [Google Scholar]
- Gho, M., M. Lecourtois, G. Géraud, J. W. Posakony and F. Schweisguth, 1996. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 122 1673–1682. [DOI] [PubMed] [Google Scholar]
- Glittenberg, M., C. Pitsouli, C. Garvey, C. Delidakis and S. Bray, 2006. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 25 4697–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M., L. Y. Jan and Y. N. Jan, 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17 27–41. [DOI] [PubMed] [Google Scholar]
- Gustafson, K., and G. L. Boulianne, 1996. Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome 39 174–182. [DOI] [PubMed] [Google Scholar]
- Hartenstein, V., and J. W. Posakony, 1990. A dual function of the Notch gene in Drosophila sensillum development. Dev. Biol. 142 13–30. [DOI] [PubMed] [Google Scholar]
- Hasson, P., and Z. Paroush, 2006. Crosstalk between the EGFR and other signalling pathways at the level of the global transcriptional corepressor Groucho/TLE. Br. J. Cancer 94 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. M. Rubin, 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., R. Maile and G. M. Rubin, 1997. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 94 5195–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler, P., M. Haenlin, P. Ramain, M. Calleja and P. Simpson, 1996. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics 143 1271–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert, S. S., T. L. Jacobsen and M. A. T. Muskavitch, 1997. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development 124 3283–3291. [DOI] [PubMed] [Google Scholar]
- Ingham, P. W., and M. J. Fietz, 1995. Quantitative effects of hedgehog and decapentaplegic activity on the patterning of the Drosophila wing. Curr. Biol. 5 432–440. [DOI] [PubMed] [Google Scholar]
- Irvine, K. D., 2008. A notch sweeter. Cell 132 177–179. [DOI] [PubMed] [Google Scholar]
- Itoh, M., C. H. Kim, G. Palardy, T. Oda, Y. J. Jiang et al., 2003. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4 67–82. [DOI] [PubMed] [Google Scholar]
- Jacobsen, T. L., K. Brennan, A. M. arias and M. A. T. Muskavitch, 1998. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development 125 4531–4540. [DOI] [PubMed] [Google Scholar]
- Kankel, M. W., G. D. Hurlbut, G. Upadhyay, V. Yajnik, B. Yedvobnick et al., 2007. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics 177 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan, R., 2002. Notch: a membrane-bound transcription factor. J. Cell Sci. 115 1095–1097. [DOI] [PubMed] [Google Scholar]
- Korey, C. A., G. Wilkie, I. Davis and D. Van Vactor, 2001. small bristles is required for the morphogenesis of multiple tissues during Drosophila development. Genetics 159 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, I. V., and C. T. McMurray, 2007. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair (Amst.) 6 517–529. [DOI] [PubMed] [Google Scholar]
- Kozlova, T., and C. S. Thummel, 2000. Steroid regulation of postembryonic development and reproduction in Drosophila. Trends Endocrinol. Metab. 11 276–280. [DOI] [PubMed] [Google Scholar]
- Krokan, H. E., R. Standal and G. Slupphaug, 1997. DNA glycosylases in the base excision repair of DNA. Biochem. J. 325(Pt. 1): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson, A., 1998. The minute genes in Drosophila and their molecular functions. Adv. Genet. 38 69–134. [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., 2006. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18 213–222. [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., and F. Schweisguth, 2003. Unequal segregation of neuralized biases Notch activation during asymmetric cell division. Dev. Cell 5 139–148. [DOI] [PubMed] [Google Scholar]
- Lee, E. C., S. Y. Yu and N. E. Baker, 2000. The scabrous protein can act as an extracellular antagonist of notch signaling in the Drosophila wing. Curr. Biol. 10 931–934. [DOI] [PubMed] [Google Scholar]
- Li, Y., and N. E. Baker, 2004. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev. Biol. 4 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber, T., S. Kidd and M. W. Young, 2002. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, M. B., A. L. Parks, D. A. Ruddy, S. Y. Tiong, H. Esengil et al., 2006. Presenilin-based genetic screens in Drosophila melanogaster identify novel Notch pathway modifiers. Genetics 172 2309–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold, S. J., J. Roote, G. Reuter, A. Lambertsson, M. Ashburner et al., 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8 R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern, V. L., C. A. Pacak, S. T. Sewell, M. L. Turski and M. A. Seeger, 2003. A targeted gain of function screen in the embryonic CNS of Drosophila. Mech. Dev. 120 1193–1207. [DOI] [PubMed] [Google Scholar]
- Micchelli, C. A., E. J. Rulifson and S. S. Blair, 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124 1485–1495. [DOI] [PubMed] [Google Scholar]
- Mlodzik, M., N. E. Baker and G. M. Rubin, 1990. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 4 1848–1861. [DOI] [PubMed] [Google Scholar]
- Modolell, J., and S. Campuzano, 1998. The achaete-scute complex as an integrating device. Int. J. Dev. Biol. 42 275–282. [PubMed] [Google Scholar]
- Muller, D., S. J. Kugler, A. Preiss, D. Maier and A. C. Nagel, 2005. Genetic modifier screens on Hairless gain-of-function phenotypes reveal genes involved in cell differentiation, cell growth and apoptosis in Drosophila melanogaster. Genetics 171 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj, R., and U. Banerjee, 2007. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development 134 825–831. [DOI] [PubMed] [Google Scholar]
- Neufeld, T. P., A. H. Tang and G. M. Rubin, 1998. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, J. T., A. Miyamoto, S. L. Olsen, B. D'Souza, C. Yao et al., 2007. a DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, J. T., A. Miyamoto and G. Weinmaster, 2007. b Notch signaling—constantly on the move. Traffic 8 959–969. [DOI] [PubMed] [Google Scholar]
- Ohsako, S., J. Hyer, G. Panganiban, I. Oliver and M. Caudy, 1994. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 8 2743–2755. [DOI] [PubMed] [Google Scholar]
- Pan, D., and G. M. Rubin, 1997. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90 271–280. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., and M. A. T. Muskavitch, 1993. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev. Biol. 157 484–496. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., S. S. Huppert and M. A. T. Muskavitch, 1997. The dynamics of neurogenic signalling underlying bristle development in Drosophila melanogaster. Mech. Dev. 63 61–74. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. M. Klueg, J. R. Stout and M. A. T. Muskavitch, 2000. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127 1373–1385. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36 288–292. [DOI] [PubMed] [Google Scholar]
- Portin, P., 2002. General outlines of the molecular genetics of the Notch signalling pathway in Drosophila melanogaster: a review. Hereditas 136 89–96. [DOI] [PubMed] [Google Scholar]
- Reddy, G. V., and V. Rodrigues, 1999. Sibling cell fate in the Drosophila adult external sense organ lineage is specified by prospero function, which is regulated by Numb and Notch. Development 126 2083–2092. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottgen, G., T. Wagner and U. Hinz, 1998. A genetic screen for elements of the network that regulates neurogenesis in Drosophila. Mol. Gen. Genet. 257 442–451. [DOI] [PubMed] [Google Scholar]
- Saeboe-Larssen, S., M. Lyamouri, J. Merriam, M. P. Oksvold and A. Lambertsson, 1998. Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics 148 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., and K. Saigo, 2000. Involvement of pannier and u-shaped in regulation of decapentaplegic-dependent wingless expression in developing Drosophila notum. Mech. Dev. 93 127–138. [DOI] [PubMed] [Google Scholar]
- Scharer, O. D., and J. Jiricny, 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays 23 270–281. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F., 2004. Notch signaling activity. Curr. Biol. 14 R129–R138. [PubMed] [Google Scholar]
- Schweisguth, F., and M. Lecourtois, 1998. The activity of Drosophila Hairless is required in pupae but not in embryos to inhibit Notch signal transduction. Dev. Genes Evol. 208 19–27. [DOI] [PubMed] [Google Scholar]
- Selkoe, D., and R. Kopan, 2003. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 26 565–597. [DOI] [PubMed] [Google Scholar]
- Sotillos, S., F. Roch and S. Campuzano, 1997. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124 4769–4779. [DOI] [PubMed] [Google Scholar]
- Speicher, S. A., U. Thomas, U. Hinz and E. Knust, 1994. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development 120 535–544. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton, K., P. D. Jackson, M. J. Clark, A. H. Brand and F. M. Hoffmann, 1994. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 5 585–593. [PubMed] [Google Scholar]
- Stanley, P., 2007. Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 17 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, M. D., H. Aihara, G. A. Roccaro, L. Cheung, H. Zhang et al., 2009. CtBP is required for proper development of peripheral nervous system in Drosophila. Mech. Dev. 126 68–79. [DOI] [PubMed] [Google Scholar]
- Sun, J., L. Smith, A. Armento and W. M. Deng, 2008. Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram, M. V., 2005. The love-hate relationship between Ras and Notch. Genes Dev. 19 1825–1839. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36 283–287. [DOI] [PubMed] [Google Scholar]
- Tilney, L. G., P. S. Connelly, K. A. Vranich, M. K. Shaw and G. M. Guild, 2000. Actin filaments and microtubules play different roles during bristle elongation in Drosophila. J. Cell Sci. 113(Pt. 7): 1255–1265. [DOI] [PubMed] [Google Scholar]
- Tini, M., A. Benecke, S. J. Um, J. Torchia, R. M. Evans et al., 2002. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9 265–277. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., and G. Struhl, 2001. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol. Cell 7 487–495. [DOI] [PubMed] [Google Scholar]
- Tsuda, L., R. Nagaraj, S. L. Zipursky and U. Banerjee, 2002. An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell 110 625–637. [DOI] [PubMed] [Google Scholar]
- Usui, K., D. Pistillo and P. Simpson, 2004. Mutual exclusion of sensory bristles and tendons on the notum of dipteran flies. Curr. Biol. 14 1047–1055. [DOI] [PubMed] [Google Scholar]
- van Doren, M., H. M. Ellis and J. W. Posakony, 1991. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development 113 245–255. [DOI] [PubMed] [Google Scholar]
- van Doren, M., A. M. Bailey, J. Esnayra, K. Ede and J. A. Posakony, 1994. Negative regulation of proneural gene activity: hairy is direct transcriptional repressor of achaete. Genes Dev. 8 2729–2742. [DOI] [PubMed] [Google Scholar]
- Van Vactor, D. L. J., R. L. Cagan, H. Krämer and S. L. Zipurpsky, 1991. Induction in the developing compound eye of Drosophila: multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell 67 1145–1155. [DOI] [PubMed] [Google Scholar]
- Voas, M. G., and I. Rebay, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 229 162–175. [DOI] [PubMed] [Google Scholar]
- Wang, S., S. Younger-Shepherd, L. Y. Jan and Y. N. Jan, 1997. Only a subset of the binary cell fate decisions mediated by Numb/Notch signaling in Drosophila sensory organ lineage requires Suppressor of Hairless. Development 124 4435–4446. [DOI] [PubMed] [Google Scholar]
- Weng, L., C. Zhu, J. Xu and W. Du, 2003. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 22 3865–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer, F., A. van der Straten, K. Keleman, B. J. Dickson and E. Hafen, 2001. Lilliputian: an AF4/FMR2-related protein that controls cell identity and cell growth. Development 128 791–800. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1993. Pattern formation in the Drosophila retina, pp. 1277–1325 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Yu, Y. B., 2002. Coiled-coils: stability, specificity, and drug delivery potential. Adv. Drug Deliv. Rev. 54 1113–1129. [DOI] [PubMed] [Google Scholar]
- Zhu, M. Y., R. Wilson and M. Leptin, 2005. A screen for genes that influence fibroblast growth factor signal transduction in Drosophila. Genetics 170 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]