Abstract
Conditional expression of hairpin constructs in Drosophila is a powerful method to disrupt the activity of single genes with a spatial and temporal resolution that is impossible, or exceedingly difficult, using classical genetic methods. We previously described a method (Ni et al. 2008) whereby RNAi constructs are targeted into the genome by the phiC31-mediated integration approach using Vermilion-AttB-Loxp-Intron-UAS-MCS (VALIUM), a vector that contains vermilion as a selectable marker, an attB sequence to allow for phiC31-targeted integration at genomic attP landing sites, two pentamers of UAS, the hsp70 core promoter, a multiple cloning site, and two introns. As the level of gene activity knockdown associated with transgenic RNAi depends on the level of expression of the hairpin constructs, we generated a number of derivatives of our initial vector, called the “VALIUM” series, to improve the efficiency of the method. Here, we report the results from the systematic analysis of these derivatives and characterize VALIUM10 as the most optimal vector of this series. A critical feature of VALIUM10 is the presence of gypsy insulator sequences that boost dramatically the level of knockdown. We document the efficacy of VALIUM as a vector to analyze the phenotype of genes expressed in the nervous system and have generated a library of 2282 constructs targeting 2043 genes that will be particularly useful for studies of the nervous system as they target, in particular, transcription factors, ion channels, and transporters.
IN the past few years a number of constructs have been generated for transgenic RNAi. The first generation of vectors, referred to as “hairpin loop RNA,” was based on transgenes having an inverted-repeat configuration either driven from a single promoter (Fortier and Belote 2000; Kennerdell and Carthew 2000; Lam and Thummel 2000; Martinek and Young 2000) or symmetrically transcribed from opposing promoters (Giordano et al. 2002). A number of difficulties were observed with these constructs. First, the vectors were found to induce a variable RNAi silencing effect, resulting in incomplete penetrance. Second, in the case of the single promoter constructs, the cloning of the inverted repeats was complicated by the instability of the plasmid in Escherichia coli. The use of recombination-deficient bacterial strains as well as the introduction of a long spacer sequence between the inverted repeats was found to improve the cloning steps. However, the presence of a spacer was associated with weaker silencing activity (Piccin et al. 2001).
The second generation of vectors, referred to as “intron-spliced,” was inspired by the observation that in plants intron-spliced hairpin RNAs are more efficient at gene silencing than the hairpin loop RNA (Smith et al. 2000). A number of different strategies have been used whereby the inverted repeats, separated by a functional intron, behave as exons. These include genomic/cDNA hybrids (Kalidas and Smith 2002) or intron sequences from the mub (Reichhart et al. 2002), white (Lee and Carthew 2003), Ret (Pili-Floury et al. 2004), and fushi-tarazu (ftz) (Kondo et al. 2006) genes. In another case the ftz intron was placed at the end of the hairpin structure (Dietzl et al. 2007). Although a careful side-by-side comparison of multiple genes has not been reported, the intron-spliced vectors appear to give more robust RNAi phenotypes, possibly due to the enhanced formation of duplex dsRNAs following the splicing event and/or the enhanced export of the processed mRNAs from the nucleus. A number of improvements in the cloning of inverted repeats have also been reported. In particular, Bao and Cagan (2006) replaced the backbone of pWIZ (Lee and Carthew 2003) to generate a vector that exhibits a much higher efficiency in assembling inverted repeats. Further, to streamline the production of RNAi transgenes, Kondo et al. (2006) developed a novel transformation vector, prize, that uses an attR1-ccd-attR2 cassette for in vitro recombination. Finally, Haley et al. (2008) reported success in using a microRNA-based RNAi approach for in vivo transgenic RNAi. However, since only three genes were tested, it remains to be determined how reliable this method is.
We previously described a new construct for targeted RNAi, Vermilion-AttB-Loxp-Intron-UAS-MCS (VALIUM) (referred to here as VALIUM1), and documented that it was an effective vector for targeted transgenic RNAi (Ni et al. 2008). Importantly, we showed that the efficacy of the RNAi phenotypes depended on the level of expression provided by varying the amount of Gal4 protein as well as the number of UAS modules. VALIUM1 (Figure 1A) contains vermilion as a selectable marker (Fridell and Searles 1991); an attB sequence to allow for phiC31-targeted integration at genomic attP landing sites (Thomason et al. 2001; Groth et al. 2004); two pentamers of UAS, one of which can be excised using the Cre/loxP system (Siegal and Hartl 1996) to generate a 5XUAS derivative (and thus potentially reduce the level of expression); the hsp70 core promoter; a multiple cloning site (MCS) that allows a single PCR product to be cloned in both orientations to generate the hairpin construct; and two introns, the white intron located between the inverted DNA repeats and the ftz intron followed by the SV40 polyadenylation signal. In our efforts to further optimize the various features of this vector, we generated in parallel a number of derivatives (Figure 1) and tested them by a variety of functional assays. Here we report the optimization of this vector system as well as its application for large-scale transgenic RNAi studies. Further, we document the effectiveness of the VALIUM vectors for analyses of the nervous system and generate a resource of reagents to facilitate these studies.
Figure 1.—
(A) Strategy for optimization and (B) VALIUM vectors used in this study.
MATERIALS AND METHODS
Genetic manipulations:
The design of this novel Drosophila RNAi collection, as well as the selection of neuronal genes for selective targeting, was carried out as part of the Visitor Program at Janelia Farm in collaboration with Charles Zuker (University of California, San Diego/Howard Hughes Medical Institute) and Gerald Rubin (Janelia Farm). For the production of transgenic flies, DNA was injected into a y w, nanos integrase; attP2/attP2 stock as described in Ni et al. (2008) at Genetic Services, Inc. (GSI) (http://www.geneticservices.com) and G0 males were individually crossed to y v; attP2/attP2 virgin females. y+v+ male progeny were individually backcrossed to y v; Sb/TM3, Ser virgin females to establish homozygous stocks.
The following Gal4 lines were used: C96-Gal4; ms1096-Gal4; actin(act)5C-Gal4/CyO; act5C-Gal4/TM6B,Tb; tubulin(tub)-Gal4/TM6B,Tb; engrailed(en)-Gal4; and GMR-Gal4. A detailed list of the stocks used in this study can be found in supporting information, Table S1 and Table S2. A description of the lines is available from FlyBase (http://flybase.org/).
Phenotypic analyses of the wings and nervous system phenotypes were performed as described in Ni et al. (2008) and Achara et al. (1997), respectively.
Vector construction:
To construct VALIUM10, VALIUM1 was cut with HindIII and KpnI to remove both the vermilion and attB sequences and then was ligated with a small DNA fragment that contained the HindIII, KpnI, and SpeI sites (primers: F, 5′-AGCTTATCGAGTTAAAAGGCGCCACACTAGTAGTAC-3′; R, 5′-TACTAGTGTGGCGCCTTTTAACTCGATA-3′). The resulting vector was cut with MfeI and EcoRI and then was ligated with a fragment that contained the BglII and XbaI sites (primers: F, 5′-AATTCAGAAGAGCTAGCAGTTGCATC-3′; R, 5′-AATTGATGCAACTGCTAGCTCTTCTG-3′). gypsy insulator sequences were amplified using pCa4B2G (gift from Michele Markstein) as a template using specific primers (gypsy-SpeI-F, 5′-ATACTAGTTGGCCACGTAATAAGTGTGCGTTG-3′; gypsy-SpeI-R, 5′-ATACTAGTGTTGTTGGTTGGCACACCACA-3′; gypsy-SacI-F, 5′-GAGAGCTCTGGCCACGTAATAAGTGTGCGTTG-3′; gypsy-SacI-R, 5′-GAGAGCTCGTTGTTGGTTGGCACACCACA-3′). PCR products were digested with either SpeI or SacI and were cloned into the SpeI and SacI sites of the vector. prize (Drosophila Genomics Resource Center: https://dgrc.cgb.indiana.edu/) was cut with XbaI to release one attR fragment. The second attR fragment was cut by XbaI and BglII. After purification, these two fragments were subsequently cloned into the insulator-containing vector. The resulting vector was released with HindIII and KpnI, ligated with the attB sequence, and then further cut with HindIII and ligated with a HindIII-cut fragment containing the vermilion gene. The resulting vector is designated VALIUM10.
To construct VALIUM3, the Drosophila synthetic core promoter was amplified from pBPGUw (Pfeiffer et al. 2008) by specific primers (SalI-F, 5′-GGTCGACGAGCTCGCCCGGGGATCG-3′; EcoRI-R, 5′-GGAATTCGTTTGGTATGCGTCTTGTGATTC-3′). After confirmation by sequencing, the correct DNA fragment was isolated and ligated into VALIUM1 that had been linearized by SalI and EcoRI; the resulting vector was called VALIUM3. VALIUM1 was cut with MfeI and EcoRI and then ligated with a small DNA fragment that had BglII and XbaI sites (primers used: F, 5′-AATTCAGAAGAGCTAGCAGTTGCATC-3′; R, 5′-AATTGATGCAACTGCTAGCTCTTCTG-3′). After cutting with BglII and XbaI, two attR fragments were cloned into this site, and the resulting vector was called VALIUM9. VALIUM1 was cut with NheI and SacII to remove the ftz intron and then was ligated with an oligonucleotide fragment generated by annealing the following two primers (primers: F, 5′-CTAGCATCTAGAACATATGCAGATCTG-3′; R, 5′-GGTTCAATTGTCTAGCAGATCTGCATATGTTCTAGATG-3′); the resulting vector was named VALIUM13. To generate VALIUM14, the ftz intron was amplified by specific primers (AvrII-F, 5′-AACCTAGGCTAGAAGGTAGGCATCACAC-3′; NheI-R, 5′-AAGCTAGCACAAAGTGGTCACAGTCGAC-3′), and the resulting PCR product was confirmed by DNA sequencing. It was then cut with AvrII and NheI and, after purification, was cloned into VALIUM1 that had been linearized by AvrII and NheI, resulting in VALIUM14. VALIUM14 was further cut with NheI and SacII to remove the ftz intron and then was ligated with an oligonucleotide fragment generated by annealing two primers (F, 5′-CTAGCATCTAGAACATATGCAGATCTG-3′; R, 5′-GGTTCAATTGTCTAGCAGATCTGCATATGTTCTAGATG-3′) to generate VALIUM15. VALIUM10 was cut with EcoRI and XbaI to remove the two attR fragments and then was ligated with a DNA fragment containing BglII, XbaI, AvrII, and NheI sites (primers: F, 5′-AATTGAGATCTGTTCTAGAGTGGACATATGCACCTAGGA-3′; R, 5′-CTAGTCCTAGGTGCATATGTCCACTCTAGAACAGATCTC-3′). The resulting vector was linearized with AvrII and NheI and then ligated with the white intron-containing fragment that was liberated from VALIUM1 with AvrII and NheI. The resulting vector was cut with XbaI and BglII and ligated with one attR fragment isolated from VALIUM10 by digestion with XbaI and BglII. A second attR fragment was amplified from VALIUM10 by PCR (primers: NheI-F, 5′-AAGCTAGCCAAGTTTGTACAAAAAAGCTGAAC-3′; NheI-R, 5′-AAGCTAGCACCACTTTGTACAAGAAAGCT-3′) and, after confirmation by DNA sequencing, was cloned into the NheI site of the previously resulting vector to generate VALIUM17. pENTR-TOPO vector [Invitrogen (Carlsbad, CA), cat. no. k240020] was ligated with an oligonucleotide fragment generated by annealing two primers (F, 5′-CACCACTAGTCTCTAGAGTGGCAGAAAGAAGCTACCAATTGTGAATTCC-3′; R, 5′-GGAATTCACAATTGGTAGCTTCTTTCTGCCACTCTAGAGACTAGTGGTG-3′) to generate the vector mENTRY.
Construction of su(Hw)attP lines:
Addition of gypsy insulators and a transcriptionally neutral “spacer” fragment from pH-Pelican (Barolo et al. 2000) to pCaryP (gift of Michele Calos) was performed as follows: First, pH-Pelican was cut with HindIII (New England BioLabs, Beverly, MA), resulting in three fragments, two of which, a 383-bp spacer fragment and a 10-kb backbone piece that included the gypsy insulator, were gel extracted. The latter piece was subsequently cut with PstI and the 430-bp gyspy insulator was then made blunt using PfuUltra High-Fidelity Polymerase (Stratagene, La Jolla, CA). pCaryP was cut with XhoI, filled in, and then ligated to the gypsy fragment described above to create pCaryiP. Following SacI digestion and filling in of pCaryiP, the 430-bp blunt gypsy fragment was ligated a second time to create pCaryiPi. Finally, to improve germline transformation efficiency the 383-bp HindIII spacer fragment from the neomycin phototransferase gene was made blunt and ligated to NotI-cut and filled-in pCaryiPi, resulting in pCaryIP. All steps were verified for orientation and fidelity by PCR and restriction digest analysis. In addition, pCaryIP was sequence verified prior to P-element-mediated germline transformation. In this study we used two different lines, su(Hw)attP1 and su(Hw)attP4, located at 87B–87C and 67E2, respectively.
Luciferase assay:
Luciferase activity was measured using the Steady-Glo Luciferase Assay Kit [Promega (Madison, WI), cat. no. E2520] as described in Markstein et al. (2008). Briefly, five wandering L3 female larvae, or 10 female flies from 0 to 3 hr of age, or 10 2-day-old adult female flies were collected in 300 μl Glo Lysis Buffer (Promega, cat. no. E266A) for each sample; five independent samples were used for each luciferase assay. Samples were homogenized and then centrifuged at 20,000 rcf for 15 min. Forty microliters of supernatant were transferred to 1.5-ml transparent Eppendorf tubes and mixed with the same volume of luciferase reagent. After incubation in the dark for 20 min, luminescence was measured on a luminometer (Turner Biosystems Instrument, model 2030-101).
Large-scale production of transgenic RNAi constructs:
Primers were ordered in a 96-well plate format (all the following steps, such as PCR, purifications, enzyme digestions, and ligations, as well as minipreps, were done in 96-well plates). PCR was performed using either Drosophila genomic DNA or cDNA as a template; PCR products were verified by gel electrophoresis and then were purified using the QIAquick Biorobot kit following the manufacturer's specifications. The purified PCR products were digested with EcoRI (or MfeI) and XbaI (or SpeI, NheI, or AvrII). If the restriction enzyme recognition site is not present for these enzymes, the PCR product can be directly cloned into the pENTR-TOPO vector (Invitrogen, cat. no. k240020). The digestion was terminated and the resulting product purified. The purified DNA fragments were ligated with the mENTRY vector that had been linearized by digestion with SpeI and EcoRI. After transformation, the correct clones were selected by PCR, using specific primers (forward, 5′-CAAAAAAGCAGGCTCCGCGG-3′; reverse, 5′-GTACAAGAAAGCTGGGTCGG-3′). Plasmid DNA was prepared from the appropriate clones [for details see handbook, QIAGEN (Valencia, CA), cat. no. 27191] and then was recombined with the destination vector. Following transformation, the correct hairpin constructs were selected by PCR using specific primers (forward, 5′-ACCAGCAACCAAGTAAATCAAC-3′; reverse, 5′-CTAGACTGGTACCCTCGAATC-3′). Hairpin constructs were further confirmed by restriction enzyme digestion before germline transformation.
Primers for hairpins were designed as described in Ni et al. (2008), using the DRSC amplicon design tool SnapDragon (http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl <http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl>). Detailed information on the constructs can be found in the Table S2 and Table S3.
RESULTS AND DISCUSSION
In an attempt to improve the efficiency of VALIUM1, we tested whether the efficiency of RNAi-induced phenotypes could be affected by the following parameters: (1) the directionality of the hairpin sequences, (2) the choice of the basal promotor, (3) the number and nature of introns, (4) cloning vs. the recombination method, and (5) the presence of insulator sequences.
Hairpin directionality:
Because hairpin sequences can be cloned either head-to-head, i.e., sense 3′–5′ and reverse 5′–3′, or in the reverse tail-to-tail orientation, we tested whether the orientation affects the severity of the phenotype. Thus, we generated a number of hairpin constructs for the Notch, discs-large (dlg1), decapentaplegic (dpp), RacGap50C (RacGap), domeless (dome), cubitus interruptus (ci), son of sevenless (sos), and Epidermal Growth Factor Receptor (EGFR) genes. In five cases (Notch, dlg1, dpp, RacGap, and dome; Figure 2, A and B), the phenotypes generated from the head-to-head orientation were stronger than those from the tail-to-tail orientation. In two other cases, ci and Sos, the phenotypes derived from either orientation were similar (data not shown). Finally, in the case of EGFR, the phenotype generated from the tail-to-tail orientation was more severe than that generated from the head-to-head configuration (Figure 2B). On the basis of these observations, we decided to use the head-to-head orientation in all of our subsequent hairpin constructs.
Figure 2.—
Head-to-head orientation produces a more potent knockdown than tail-to-tail orientation. (A) The phenotype associated with C96-Gal4, Notch-hp flies is more severe in the head-to-head orientation than in the tail-to-tail orientation. We used the phenotypic classification described in Ni et al. (2008) to quantify the severity of the wing phenotypes. Class 1: wild type or a few bristles missing. Class 2: margin bristles missing but no notches. Class 3: moderate wing notching. Class 4: extensive wing notching. Class 5: most of the wing margin is missing. Flies were raised at different temperatures and the phenotypes in males vs. females were scored separately. (B) Comparison of the en-Gal4, hp phenotypes associated with head-to-head vs. tail-to-tail orientation for dlg1, dpp, RacGap, dome, and egfr. Flies were raised at 25°.
Basal promoter:
We initially used the hsp70 core promoter in VALIUM1 as it has been shown to be an effective promoter in UAS vectors (Brand and Perrimon 1993). However, to test whether a different basal promoter would improve expression of the hairpin constructs, we tested the Drosophila synthetic core promoter (DSCP) described in Pfeiffer et al. (2008), which contains the TATA, Inr, MTE, and DPE sequence motifs. In two different tests that use either a Notch hairpin (Notch-hp) (Figure 3A; Ni et al. 2008) or a luciferase assay (Figure 3B; Markstein et al. 2008), the hsp70 promotor generated better results and was incorporated into all subsequent vectors.
Figure 3.—
hsp70 is an effective core promotor. (A) In the C96-Gal4, Notch-hp assay, the hsp70 core promotor generates stronger Notch RNAi phenotypes than the DSCP promotor. (B) Similar conclusions were obtained using a luciferase assay. Luciferase lines were crossed with two different act5C-Gal4 insertions [(1) act5C-Gal4/CyO and (2) act5C-Gal4/TM6B,Tb] and with (3) tub-Gal4/TM6B,Tb. Luciferase activity was measured in 2-day-old adult males at 25°. Note that both act5C-Gal4 drivers have similar strengths and that the tub-Gal4 driver is ∼2.5 times stronger than the act5C-Gal4 drivers.
Type and number of introns:
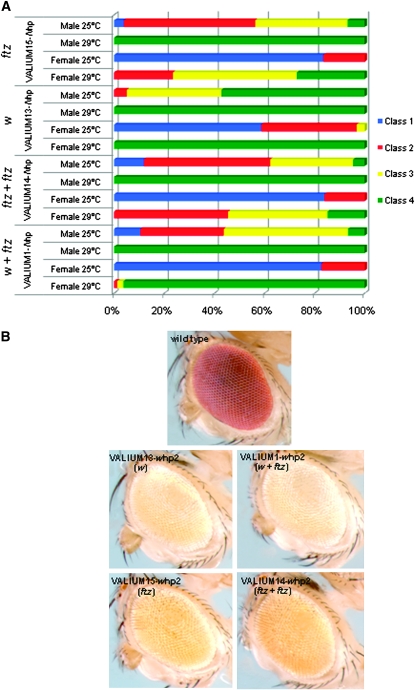
VALIUM1 contains both the white intron, located between the inverted DNA repeat, which has been shown to reduce toxicity in bacteria, and the ftz intron between hairpin and SV40 poly(A) tail, which has been proposed to facilitate hairpin-RNA processing and export from the nucleus (Dietzl et al. 2007). To test the role of these introns, we generated three additional vectors with different combinations of introns and tested them in the context of both the Notch and the white hairpins (Figure 4, A and B, respectively). Interestingly, in both assays the presence of the white, rather than the ftz, intron in the middle of the hairpins was more effective. Further, the absence of the 3′-end ftz intron slightly enhanced the severity of the RNAi phenotypes with either the white or the ftz intron in the middle of the hairpin.
Figure 4.—
The white intron alone is sufficient for effective knockdown by RNAi. We compared the efficacy of hairpins against Notch and white, respectively (A and B), in vectors that contain both the white and ftz introns (VALIUM1, w + ftz), only the white intron (VALIUM13, w), two ftz introns (VALIUM14, ftz + ftz), and only one ftz intron (VALIUM15, ftz). C96-Gal4;Notch-hp flies were grown at 25° and at 29°. GMR-Gal4; white-hp flies were raised at 25° and the eye color was examined after adult flies eclosed.
Cloning vs. recombination method:
To generate hairpin constructs in VALIUM1, we used a multiple-cloning site (MCS) that allows a single PCR product to be cloned in both orientations (Ni et al. 2008). However, to simplify the cloning strategy, we decided to use the att recombination method as it is more accurate, faster, and less expensive (Kondo et al. 2006). Further, unlike the MCS system, every gene can be cloned using the att system, as there are no limitations associated with the choice of restriction enzymes. Because the two methods lead to differences in the final vector sequences (Figure 5A), we compared the phenotypes generated using both approaches for hairpins against the Notch (Figure 5B) and white (Figure 5C) genes. Interestingly, both hairpins generated with the recombination system performed better than those generated using the MCS. It is possible that the addition of paired sequences may enhance formation of the duplex dsRNA following the splicing event and/or export of the processed mRNAs from the nucleus. Importantly, the additional sequences resulting from the recombination system do not show 19-nt homology to any Drosophila genes; thus any potential siRNAs derived from the additional sequences should not lead to sequence-specific off-target effects.
Figure 5.—
The recombination method improves vector efficiency. (A) The recombination method introduces additional sequences into the vector, which is not the case when using the MCS ligation approach. (B) Phenotypes of C96-Gal4; Notch-hp flies grown at either 25° or 29°. Introduction of the Notch-hp sequence using the recombination system leads to stronger RNAi phenotypes than using the MCS strategy. (C) Eye phenotypes of GMR-Gal4;white-hp flies at 25° soon after emergence. The phenotype obtained with VALIUM9-white-hp is stronger than that obtained with VALIUM14-white-hp. Similar results were reached when two different hairpins against the white gene were tested. Data are shown here for only the white-hp2 sequence.
Insulator sequences:
Insulator sequences have been shown to increase the level of expression of the insulated genes (Markstein et al. 2008). To test whether adding such sequences to VALIUM vectors can generate more potent RNAi phenotypes, we added gypsy insulator sequences to the vector to create VALIUM10 and tested their effect in the context of both the Notch and the white hairpins (Figure 6, A and B, respectively). In both cases, addition of insulators significantly enhanced the hairpin phenotypes (compare results obtained with VALIUM10 vs. the VALIUM9 control, as well as those obtained with VALIUM1). Similar enhancements were seen when the RNAi phenotypes of hairpins against the EGFR, Sos, dpp, ci, dome, dlg1, and RacGap50 genes were compared side-by-side with all three vectors (data not shown).
Figure 6.—
Insulator sequences significantly improve vector efficacy. (A and B) VALIUM 10 is a better vector than either VALIUM1 or VALIUM9 using either the C96-Gal4;Notch-hp or the GMR-Gal4;white-hp assays. Note that in the Notch assay, a class 5 phenotype (see Figure 2A) characterized by very reduced wing size is observed. The RNAi phenotypes of Notch-hp and white-hp were much stronger with VALIUM10, demonstrating the potency of the insulators. Interestingly, VALIUM17 behaves as well as VALIUM10 in the Notch-hp assay but not in the white-hp assay (see text). (C) To monitor both the basal and the induced levels of transgene expression in the various VALIUM vectors, we examined the expression of UAS-luciferase introduced into VALIUM1, -9, or -10, either in the absence of Gal4 or in the presence of act5C-Gal4. Note that the VALIUM1 and VALIUM9 luciferase constructs differ only by a few base pairs (see DataS1) and as expected behave similarly in all assays.
Previously, Markstein et al. (2008) reported that insulation of UAS-driven transgenes could lead to an increase in their basal level of expression. Because such leaky expression could potentially be deleterious to the animal, we determined both the level of basal expression associated with VALIUM10 using the luciferase assay and that induced by act5C-Gal4 (Figure 6C). VALIUM10 showed a low level of basal activity during the third larval instar stage due to salivary gland expression (data not shown, see also Markstein et al. 2008). On the other hand, basal levels of activity with VALIUM1 and VALIUM9, but not VALIUM10, were unexpectedly high soon after eclosion (0- to 3-hr-old whole flies). Importantly, in the presence of Gal4, luciferase activity was two- to threefold higher using the insulated VALIUM10 backbone, which is consistent with the RNAi experiments.
Finally, on the basis of the results shown in Figure 4 showing that, in a VALIUM1 context, a single white intron behaves better than two ftz introns, we generated VALIUM17 (Figure 1B). This vector is a derivative of VALIUM10 that possesses only the white intron and was expected to behave as well as, or better than, VALIUM10. Interestingly, while both VALIUM10 and VALIUM17 behaved similarly when tested with a hairpin against Notch, VALIUM17 did not perform as well when other hairpins against white (Figure 6B), EGFR, dpp, dlg1, ci, Sos, dome, and RacGap (data not shown) were tested. Altogether, among the VALIUM series, VALIUM10 is the best-performing vector for in vivo RNAi.
We also tested whether increasing the number of gypsy sequences from two to four improved the severity of the phenotypes generated by the hairpin construct. To do this, we generated by P-element transformation a number of attP docking sites flanked by gypsy sequences, referred to as su(Hw)attP (see materials and methods). We then integrated a Notch hairpin in either VALIUM9 or VALIUM10 into two of these sites [su(Hw)attP1 and su(Hw)attP4] and compared the resulting phenotypes with those generated using Notch hairpins in VALIUM9 or VALIUM10 integrated into the attP2 site (Figure 7). Interestingly, while the gypsy sequences present at the docking site were able to boost expression from the VALIUM9 hairpin, they appeared to be slightly less effective than when they are an integral part of VALIUM10. Further, since the presence of four vs. two gypsy sequences did not significantly increase the efficacy of the hairpin, we decided to use only two gyspy sequences in the final method design.
Figure 7.—
Increasing the number of Insulator sequences does not significantly improve vector efficacy. We compared the efficacy of a Notch hairpin when insulated by zero (VALIUM9 at attP2), two [VALIUM10 at attP2, VALIUM9 at su(Hw)attP1 and su(Hw)attP4], and four [VALIUM10 at su(Hw)attP1 and su(Hw)attP4] gypsy sequences. Flies were raised at 25°.
Generation of a transgenic RNAi resource for neurogenetics:
Because of Janelia Farm's research focus on understanding the structure and function of the fly nervous system, we chose to test these methods on a set of genes selected on the basis of their likely functions in neurons. We optimized the various steps in the cloning protocol (see materials and methods) and generated 2282 constructs targeting 2043 genes that encompass transcription factors, ion channels, transporters, and other relevant genes, and we designed hairpin sequences using Snapdragon (see materials and methods). A complete list of the constructs and lines that have been generated can be found in Table S3 and at http://flyrnai.org/TRiP-HOME.html. Note that the current collection includes 729 constructs in VALIUM1 and 1553 in VALIUM10. Note also that all lines will be available from either the Bloomington Drosophila Stock Center or the TRiP. In addition, additional lines are continually produced as part of the TRiP project.
Screening transgenic RNAi lines for nervous system defects:
To assess the performance of these lines with respect to the nervous system, we analyzed in detail 18 lines that target 13 genes with specific functions in the eye and nervous system (Table 1).
TABLE 1.
Phenotypic analyses of transgenic RNAi lines in the eye
| CG no. | Line no. | Gene name | Phenotype |
|---|---|---|---|
| Eye morphology | |||
| CG7245 | TR00021A.1 | eys/spam | Same as null allele by EM examination |
| CG7245 | TR00022A.1 | eys/spam | Same as null allele by EM examination |
| CG1744 | TR00610A.1 | chp | Same as null allele by EM examination |
| CG18085 | TR00604A.1 | sev | Same as hypomorph by EM examination |
| CG5996 | TR00660A.1 | TRPgamma | Defective eye morphology by EM |
| CG5996 | TR00661A.1 | TRPgamma | Defective eye morphology by EM |
| Photoreceptor function | |||
| CG6518 | TR00601A.1 | inaC | Same as null on ERGs |
| CG5962 | TR00603A.1 | arr2 | Same as strong hypomorph on ERGs |
| CG17759 | TR00593A.1 | Galpha | Protein null on Western blots |
| CG4574 | TR00595A.1 | PLC21c | Protein null on Western blots |
| Other | |||
| CG15860 | TR00016A.1 | pain | Same as hypomorph in behavior |
| CG2647 | TR00624A.1 | per | Same as hypomorph in behavior (M. Rosbash, personal communication) |
| Negative results | |||
| CG15860 | TR00015A.1 | pain | No phenotype |
| CG10609 | TR00615A.1 | OR83B | No phenotype |
| CG10609 | TR00616A.1 | OR83B | No phenotype |
| CG13948 | TR00431A.1 | GR21a | No phenotype |
| CG13948 | TR00619A.1 | GR21a | No phenotype |
| CG11020 | TR00018A.1 | nompC | No phenotype |
A description of the mutant phenotypes is available from FlyBase (http://flybase.org/).
As null mutations in these genes are homozygous viable, we first analyzed whether they show phenotypes when crossed with ubiquitous Gal4 drivers. Flies were scored with three different Gal4 lines (act5C-Gal4/CyO; act5C-Gal4/TM6B,Tb; and tub-Gal4/TM6B,Tb) and at different temperatures (Table S4). Five lines (28%) showed consistent significant lethality with all these drivers, and lethality was more severe as temperature increased. These observations are consistent with the data reported by Dietzl et al. (2007) (Table S4), who found that 15 of the 63 lines (26%) that should be viable with act5C-Gal4 showed some lethality. Dietzl et al. (2007) suggested that these effects were associated with sequence-specific off-target effects. However, as the design of our hairpins specifically avoids the presence of predicted off-target sequences at ≥19 nt (Kulkarni et al. 2006; Ma et al. 2006; Ni et al. 2008), the lethality observed with ubiquitous drivers possibly reflects some general toxicity associated with dsRNA/siRNA production or nonspecific interference with the miRNA pathway. Further studies will be needed to distinguish between these and other possibilities.
To avoid the complication associated with the use of ubiquitous drivers, we tested the lines with more specific drivers. All lines exhibit normal viability at 25° and 29° using the elav-Gal4 and/or GMR-Gal4 drivers, and none of the lines display rough eye morphologies, indicating that the overproduced dsRNAs do not have either nonspecific effects on viability or large off-target effects. Transgenic lines for 10 of the 13 genes gave us the expected phenotypes demonstrating the specificity of the hairpin lines (see Table 1 and examples in Figure 8). False negative results were obtained with one of the two hairpin lines against pain and with hairpins against OR83B, GR21a, and nompC, possibly reflecting the fact that the GMR-Gal4 driver is not very strong in adults or that the dsRNA sequences used to target those genes were not optimal. Also, note that, for historical reasons, the lines we chose for these studies were in VALIUM1, which we have shown is a less efficient vector than VALIUM10. Altogether, we expect that most of the lines that we have generated will prove to perform extremely well for phenotypic analyses in the nervous system and other tissues (for example, see phenotypes generated with these lines in the wing in Figure S1).
Figure 8.—
RNAi phenotypes in the nervous system. Electron micrograph (EM) of (A) control cn bw, (B) cn bw; GMR-Gal4, (C) eyes shut (eys) mutant, (D) eys RNAi, (E) chaoptic (chp) mutant, and (F) chp RNAi. Note the similarity of the phenotypes generated from either the null mutations or expression of the RNAi construct. (G) Electroretinogram (ERG) following a 20-sec white light pulse of Canton S (control), arrestin2 (arr2) RNAi, and inactivation no afterpotential C (inaC) RNAi. Note that the termination of the ERG response is much slower in either arr2 or inaC RNAi flies, which is consistent with the previously described mutant phenotypes (http://flybase.org/).
Acknowledgments
We thank Charles Zuker [Howard Hughes Medical Institute(HHMI)/University of California, San Diego] and Gerald Rubin (Janelia Farm) for advice, discussion, and encouragement throughout the course of this work and for providing support for the participating members of their laboratories; Karen Hibbard, Don Hall, Monti Mercer, Megan Hong, Jessica Keating, and Grace Zheng of the Janelia Farm Fly Facility for help with establishing the transgenic lines; Susan Zusman and Michael Tworoger of Genetic Services, Inc., who generated most of the transgenic lines described here; and Michael Rosbash (HHMI/Brandeis) for the characterization of the per mutant phenotype. The major support for this work was provided by the Janelia Farm Visitor Program; additional support was provided by National Institutes of Health grants GM067761 and GM084947 to N.P.
Supporting information is available online at: http://www.genetics.org/cgi/content/full/genetics.109.103630/DC1.
References
- Achara, J. K., K. Jalink, R. W. Hardy, V. Hartenstein and C. S. Zuker, 1997. InsP3 receptor is essential for growth and differentiation but not for vision in Drosophila. Neuron 18 881–887. [DOI] [PubMed] [Google Scholar]
- Bao, S., and R. Cagan, 2006. Fast cloning inverted repeats for RNA interference. RNA 12 2020–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo, S., L. A. Carver and J. W. Posakony, 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29 726–732. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Fortier, E., and J. M. Belote, 2000. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis 26 240–244. [DOI] [PubMed] [Google Scholar]
- Fridell, Y. W., and L. L. Searles, 1991. Vermilion as a small selectable marker gene for Drosophila transformation. Nucleic Acids Res. 19 5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano, E., R. Rendina, I. Peluso and M. Furia, 2002. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics 160 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth, A. C., M. Fish, R. Nusse and M. P. Calos, 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, B., D. Hendrixa, V. Tranga and M. Levine, 2008. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 321 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalidas, S., and D. P. Smith, 2002. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron 33 177–184. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J. R., and R. W. Carthew, 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18 896–898. [DOI] [PubMed] [Google Scholar]
- Kondo, T., S. Inagaki, K. Yasuda and Y. Kageyama, 2006. Rapid construction of Drosophila RNAi transgenes using pRISE, a P-element-mediated transformation vector exploiting an in vitro recombination system. Genes Genet. Syst. 81 129–134. [DOI] [PubMed] [Google Scholar]
- Kulkarni, M. M., M. Booker, S. J. Silver, A. Friedman, P. Hong et al., 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 3 833–838. [DOI] [PubMed] [Google Scholar]
- Lam, G., and C. S. Thummel, 2000. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr. Biol. 10 957–963. [DOI] [PubMed] [Google Scholar]
- Lee, Y. S., and R. W. Carthew, 2003. Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30 322–329. [DOI] [PubMed] [Google Scholar]
- Ma, Y., A. Creanga, L. Lum and P. A. Beachy, 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443 359–363. [DOI] [PubMed] [Google Scholar]
- Markstein, M., C. Pitsouli, C. Villalta, S. E. Celniker and N. Perrimon, 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek, S., and M. W. Young, 2000. Specific genetic interference with behavioral rhythms in Drosophila by expression of inverted repeats. Genetics 156 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J. Q., M. Markstein, R. Binari, B. Pfeiffer, L. P. Liu et al., 2008. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods 5 49–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, B. D., A. Jenett, A. S. Hammonds, T. T. Ngo, S. Misra et al., 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccin, A., A. Salameh, C. Benna, F. Sandrelli, G. Mazzotta et al., 2001. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 29 E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pili-Floury, S., F. Leulier, K. Takahashi, K. Saigo, E. Samain et al., 2004. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 279 12848–12853. [DOI] [PubMed] [Google Scholar]
- Reichhart, J. M., P. Ligoxygakis, S. Naitza, G. Woerfel, J. L. Imler et al., 2002. Splice-activated UAS hairpin vector gives complete RNAi knockout of single or double target transcripts in Drosophila melanogaster. Genesis 34 160–164. [DOI] [PubMed] [Google Scholar]
- Siegal, M. L., and D. L. Hartl, 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green et al., 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320. [DOI] [PubMed] [Google Scholar]
- Thomason, L. C., R. Calendar and D. W. Ow, 2001. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage phiC31 site-specific recombination system. Mol. Genet. Genomics 265 1031–1038. [DOI] [PubMed] [Google Scholar]