Abstract
Adaptation to cold is one of the greatest challenges to forest trees. This process is highly synchronized with environmental cues relating to photoperiod and temperature. Here, we use a candidate gene-based approach to search for genetic associations between 384 single-nucleotide polymorphism (SNP) markers from 117 candidate genes and 21 cold-hardiness related traits. A general linear model approach, including population structure estimates as covariates, was implemented for each marker–trait pair. We discovered 30 highly significant genetic associations [false discovery rate (FDR) Q < 0.10] across 12 candidate genes and 10 of the 21 traits. We also detected a set of 7 markers that had elevated levels of differentiation between sampling sites situated across the Cascade crest in northeastern Washington. Marker effects were small (r2 < 0.05) and within the range of those published previously for forest trees. The derived SNP allele, as measured by a comparison to a recently diverged sister species, typically affected the phenotype in a way consistent with cold hardiness. The majority of markers were characterized as having largely nonadditive modes of gene action, especially underdominance in the case of cold-tolerance related phenotypes. We place these results in the context of trade-offs between the abilities to grow longer and to avoid fall cold damage, as well as putative epigenetic effects. These associations provide insight into the genetic components of complex traits in coastal Douglas fir, as well as highlight the need for landscape genetic approaches to the detection of adaptive genetic diversity.
A fundamental goal of molecular population and quantitative genetics is to discover polymorphisms that underlie adaptive phenotypic traits. Elucidation of the genetic components for ecologically relevant traits within natural populations has been slow, due mostly to the disconnect between organisms with detailed genomic resources and those that have phenotypes with ecological relevance (Stinchcombe and Hoekstra 2008). Rapid advances and applications of high-throughput marker technologies are beginning to amend this disconnect for forest trees. Several applications of association mapping approaches using functional marker data have been fruitful in identifying putatively causal single-nucleotide polymorphisms (SNPs) for an array of adaptive phenotypes across different forest tree species. The importance of these associations is clear, with putative applications ranging from marker-assisted breeding to gene conservation in the face of climate change (Walther et al. 2002; Aitken et al. 2008).
Adaptation to cold is one of the greatest challenges to forest trees and is highly synchronized with environmental cues, primarily photoperiod and temperature (Saxe et al. 2001; Howe et al. 2003). The annual growth cycle of temperate forest trees involves a trade-off between the timing of initiation and cessation of growth that takes full advantage of favorable climatic conditions, while avoiding cold damage from late frosts in the spring and early frosts in the fall. Timing of bud flush is predominantly influenced by temperature following adequate chilling, while bud set is influenced by photoperiod (short days), as well as temperature, soil moisture, nutrition, and light quality (Sakai and Larcher 1987; Howe et al. 2003). The first stage of cold hardiness is also induced by short days, while low temperatures induce the second stage (Weiser 1970; Sakai and Larcher 1987).
Here, we take an association genetic approach to the dissection of cold-hardiness related traits within natural populations of coastal Douglas fir [Pseudotsuga menziesii (Mirb.) Franco var. menziesii]. The range of this species extends from the Pacific Coast of North America to the eastern slope of the Rocky Mountains, with trees from the Pacific Coast classified as P. menziesii var. menziesii and those from the Rocky Mountains classified as P. menziesii var. glauca (Bessin.) Franco. The success of Douglas fir across this highly heterogeneous landscape is due largely to its ability to maximize growth during favorable climatic conditions, balanced with tolerance to low temperatures (Rehfeldt 1989; St. Clair et al. 2005; St. Clair 2006).
Genetic variation for cold hardiness in coastal Douglas fir is well documented among geographic sources and among families within sources (Campbell and Sorensen 1973; White 1987; Loopstra and Adams 1989; Aitken and Adams 1996, 1997; O'Neill et al. 2001; St. Clair 2006). Most of these traits are also heritable, with h2 values ranging from 0.10 to 0.85. Population differences in cold adaptation across the range of Douglas fir are strongly influenced by geographic and climatic variables (Howe et al. 2003). Differences in cold season temperature and associated geographic variables (e.g., latitude, elevation, and distance from the ocean) are important selective forces driving local adaptation of populations (St. Clair et al. 2005). For example, population differentiation at quantitative traits (QST) related to fall cold hardiness is eightfold greater than differentiation at anonymous and presumably neutral markers (FST), suggesting the action of natural selection acting upon these traits (St. Clair 2006). The genes underlying cold hardiness, however, have remained elusive, despite numerous efforts to map quantitative trait loci (QTL) (Jermstad et al. 2001a,b, 2003) and to analyze patterns of collocation between QTL and candidate genes (Wheeler et al. 2005).
Expression studies support the hypothesis that similar types of genes to those identified in Arabidopsis are involved with cold adaptation in conifers (Guy et al. 1985; Thomashow 1999; Fowler and Thomashow 2002; Sekai et al. 2002; Lee et al. 2005; Yakovlev et al. 2006; Holliday et al. 2008). Population genetic investigations into patterns of diversity and divergence at candidate genes for cold adaptation, as well as a suite of other adaptive phenotypes, however, often find few loci consistent with the action of natural selection (Brown et al. 2004; Krutovsky and Neale 2005; González-Martínez et al. 2006; Heuertz et al. 2006; Ingvarsson et al. 2006; Hall et al. 2007; Pyhäjärvi et al. 2007; Eveno et al. 2008) (reviewed by Neale 2007; Savolainen and Pyhäjärvi 2007; Neale and Ingvarsson 2008). Even the low power of the methods employed in these investigations (Zhai et al. 2009) and the theoretical expectations that selected loci may be unable to be detected using outlier approaches (Le Corre and Kremer 2003) are unlikely to account for the paucity of results. Larger sets of candidate genes are crucial, therefore, for the continued investigation and identification of major portions of the adaptive genetic diversity in forest trees.
Similar patterns have been found in association genetic analyses, where only a small number of markers all of small effect are detected (Neale and Savolainen 2004; Thumma et al. 2005; González-Martínez et al. 2007, 2008; Ingvarsson et al. 2008) (reviewed by Neale 2007; Grattapaglia and Kirst 2008; Grattapaglia et al. 2009). Much of this work has focused on point mutations within coding regions, thus ignoring regulatory regions affecting gene expression. Seminal work has illuminated the possibility that many of the adaptive responses by forest trees to their environments, however, may stem from epigenetic effects (Johnsen et al. 1996; Hänninen et al. 2001; Saxe et al. 2001; Johnsen et al. 2005a,b; Webber et al. 2005; Kvaalen and Johnsen 2008). The prevalence of such effects modifies the expectation of the quantity, type, and effect size of genes involved with adaptation by forest trees.
The segregation of adaptive genetic diversity by coastal Douglas fir along environmental gradients is clearly established. Surveys of molecular diversity and divergence across 139 candidate genes have documented a set of those genes that deviate from the standard neutral model (Krutovsky and Neale 2005; Eckert et al. 2009b). These are prime candidates for the further dissection of cold-hardiness related traits using association mapping (cf. Wright and Gaut 2005). Here, we aim to bridge the gap between molecular population and quantitative genetics, using an association mapping approach. Our primary goal is to identify single-marker associations with 21 cold-hardiness traits. In doing so, we highlight the need for future investigations into landscape approaches to the description of adaptive genetic diversity, as well as studies of epigenetic effects in coastal Douglas fir.
MATERIALS AND METHODS
Association population and phenotypic data:
Association population:
The association population consisted of 700 of the 1338 unrelated families that were assessed in the genecology study of St. Clair et al. (2005). They represent an extensive rangewide sample covering 6.8° of latitude, 4.1° of longitude, and a diversity of environmental conditions (Figure 1; supporting information, Table S1). Wind-pollinated seed was collected from trees that originated from naturally regenerated stands throughout the range of Douglas fir in western Oregon and Washington. Twenty progeny were grown in raised nursery beds that were located in Corvallis, Oregon. Families were randomly assigned to five-tree row plots in each of the four raised beds, with each bed treated as a block. The term family is used to refer to source trees (i.e., mothers) because the phenotypic values we use are breeding values, and this is the terminology used in the original studies in which the phenotypes were measured (cf. St. Clair et al. 2005; St. Clair 2006).
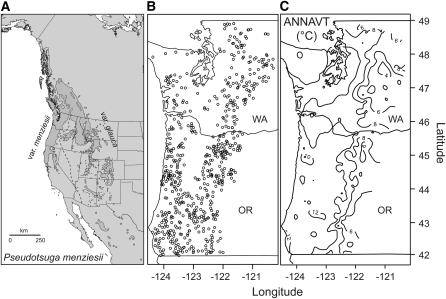
Figure 1.—
Descriptive information about the distribution, sampling localities, and climate across the range of coastal Douglas fir. (A) Range map for coastal Douglas fir. (B) Sample locations for coastal Douglas fir across Oregon and Washington. Each point denotes a single tree (n = 700) that was sampled, and since the phenotypes represent breeding values, these samples are referred to as families. (C) The annual average temperature (ANNAVT) gradient across the sample localities. Contours are isotherms, ranging from 6° to 12°.
Phenotypes:
Seedlings were grown for 2 years, during which they were measured for 21 traits related to cold injury, emergence, bud phenology, growth, and resource partitioning (Table 1). The data for cold-tolerance traits were obtained from St. Clair (2006). Emergence was determined following procedures described by Campbell and Sorensen (1979). Height and bud set were measured at the end of the first growing season, and bud burst was measured at the beginning of the second growing season. Bud set was also measured at the end of the second growing season. Samples were frozen in a programmable freezer and fall cold hardiness was assessed on needle, stem, and bud tissues after the second growing season following the methods of Aitken and Adams (1996). Whole seedlings, including roots, were then harvested and were measured for stem diameter, height from root collar to terminal bud, height to bud scar resulting from second flushing, and length of the longest root. Dry weights of roots and shoots were determined after drying the seedlings at 80° for 24 hr. Values for each phenotypic trait were calculated as the grand mean of family plot means. Year-to-year environmental variation was removed by standardizing the plot mean data, so that the means and standard deviations of control plots were equal across years.
TABLE 1.
Description of measured traits listed by phenotypic categories
| Trait | Abbreviation | Description | Unit |
|---|---|---|---|
| Emergence | |||
| Rate of emergence | EMEAN | Cumulative no. of seedlings that emerged in a plot | Probits d−1 |
| Standard deviation | EMSTDa | Standard deviation of the rate of emergence | Probits d−1 |
| Growth and resource partitioning | |||
| Stem diameter | DIAM | At 1 cm above root collar after 2 yr | mm |
| Propensity to 2nd flush | FLUSH | Proportion of 2-yr seedlings with lammas growth of terminal leader | Proportion |
| Length of 2nd flush | FLUSHLG | Distance from visible bud scar to base of terminal bud | cm |
| Height | HT1 | From root collar to base of terminal bud after 1 yr | cm |
| Height | HT2a | From root collar to base of terminal bud after 2 yr | cm |
| Height increment | HTINC | HT2–HT1 | cm |
| Root length | RTLG | From root collar to tip of longest root | mm |
| Root-to-shoot ratio | RTSH | Ratio of dry weights after 2 yr | g g−1 |
| Root weight | RTWT | Root dry weight after 2 yr | g |
| Shoot weight | SHWT | Shoot dry weight after 2 yr | g |
| Taper | TAPER | DIAM/HT2 | mm cm−1 |
| Total weight | TOTWTa | Sum of shoot and root weights | g |
| Phenology and cold tolerance | |||
| Bud burst | BB2 | First green needles from terminal bud | Days since Jan. 1 |
| Bud set | BS1 | First visible terminal bud scales at end of first growing season | Days since Jan. 1 |
| Bud set | BS2 | First visible terminal bud scales at end of second growing season | Days since Jan. 1 |
| Bud cold injury | Budcold | Percentage of cold injury | % |
| Needle cold injury | Ndlcold | Percentage of cold injury | % |
| Stem cold injury | Stmcold | Percentage of cold injury | % |
| Other | |||
| Seed weight | SDWTb | Weight per 100 seeds | g |
Loadings of derived traits from PCA are listed in Table S2.
Variables not included in PCA since the trait is a linear combination of other traits.
Nonseedling trait, not included in PCA.
Individual phenotypic traits were highly correlated (Pearson's r: −0.71–0.94). To account for these correlations, multivariate traits were constructed with a principal components analysis (PCA), using PROC PRINCOMP with the correlation matrix in the SAS software (SAS system for Windows, version 9.1, Copyright 2002; SAS Institute, Cary, NC). We retained all components with eigenvalues greater than one. In total these components accounted for 80.0% of the variance. Factor loadings for each component are located in Table S2.
SNP genotyping:
DNA isolation:
Total genomic DNA was isolated using the DNeasy plant mini kit (QIAGEN, Valencia, CA) and quantified using the PicoGreen assay (Invitrogen, Carlsbad, CA). For each maternal tree, haploid megagametophyte tissue excised from 10 seeds was combined and ground under liquid nitrogen. Inferring the diploid maternal genotype from haploid tissues can result in a bias against detection of heterozygotes. Using 10 megagametophytes, however, results in only a ∼0.2% expected probability of this type of error (Morris and Spieth 1978). All DNA extractions were carried out at the U.S. Department of Agriculture Forest Service, National Forest Gel Electrophoresis Laboratory at the Institute of Forest Genetics (Placerville, CA).
Candidate gene selection:
Candidate genes with a putative role in conferring tolerance to cold temperatures were selected according to three criteria: (i) genes found to collocate with QTL for cold hardiness in Douglas fir, (ii) genes with physiological roles in cold tolerance response, and (iii) genes showing differential expression in microarray studies of Arabidopsis. A full description of the candidate gene selection process can be found elsewhere (Krutovsky and Neale 2005; Eckert et al. 2009b). In brief, we used the 939 genes identified by Lee et al. (2005) as cold regulated in Arabidopsis to mine Douglas fir expressed sequence tag (EST) libraries using standard BLAST tools. Putative homologs were sequence validated prior to construction of the final candidate gene list.
SNP discovery and selection:
The discovery of SNPs was conducted previously by direct sequencing of haploid megagametophyte DNA samples in a diversity panel of 23–32 trees for 18 (Krutovsky and Neale 2005) and 121 (Eckert et al. 2009b) cold-hardiness and wood-related candidate genes. From those sets (400 SNPs from Krutovsky and Neale 2005; 933 SNPs from Eckert et al. 2009b) we selected 384 SNPs from 117 genes with which to construct a GoldenGate genotyping assay (Illumina, San Diego). This platform has been shown previously to work well for conifer genomes (Pavy et al. 2008; Eckert et al. 2009a). Selection of SNPs was based on four criteria: (i) gene function, (ii) SNP annotation, (iii) Illumina designability score, and (iv) minor allele frequency. One to 12 SNPs per gene were selected to capture most of the haplotypic variation within candidate genes (Table S3).
SNP genotyping:
Genotyping was carried out using the Illumina GoldenGate SNP genotyping platform (Landegren et al. 1988; Oliphant et al. 2002; Fan et al. 2003). In brief, this assay involves generating hundreds of templates with specific target and address sequences, using allele-specific extension followed by ligation and amplification with universal primers. Fluorescent products are hybridized to precoded beads on an array matrix from which the signal intensities are subsequently determined using the BeadArray Reader (Illumina). This is followed by quantification and matching of those intensities to specific alleles using BeadStudio ver. 3.1.14 (Illumina). Manual adjustments to genotypic clusters were made when necessary. For inclusion of SNPs into the final data set, we used conservative thresholds of 0.35 and 0.85 for the GenCall50 (GC50) and call rate (CR) indexes, respectively. These are common quality metrics with which to evaluate the successfulness of Illumina genotyping data (cf. Pavy et al. 2008; Eckert et al. 2009a) and represent the reliability of samples to be clustered into genotypic categories (GC50) and the fraction of the 700 samples that had a genotype called for a given SNP (CR). Genotyping was conducted at the DNA Technologies Core Facility located at the University of California, Davis (http://www.genomecenter.ucdavis.edu). Primer sequences used for SNP discovery and Illumina genotyping are available in File S1 and File S2).
Tests for association:
Population structure:
Population structure is the leading cause of false positives in genetic association studies. Populations of coastal Douglas fir are not differentiated strongly from one another using allozymes (Li and Adams 1989), RAPDs (Aagaard et al. 1998), or chloroplast and nuclear microsatellites (Viard et al. 2001; Krutovsky et al. 2009). The average level of population differentiation (GST) was ∼0.02–0.07 in all studies, with Li and Adams (1989) noting weak to moderate (r < 0.30) isolation-by-distance effects in the coastal populations. Low levels of population structure can be observed in widespread species when populations as defined a priori are not meaningful biologically (Waples and Gaggiotti 2006). Applications of the Bayesian clustering algorithm in the program STRUCTURE (cf. Falush et al. 2003) produce results that concur with the observed low values of GST, with most individuals being assigned equally well to all of the assumed clusters (K) across values of K ranging from 2 to 18 (Krutovsky et al. 2009). For association analyses, we utilized a Q-matrix defined by 15 clusters, because this was the smallest value of K producing a large (i.e., >100 log units) change in the log probability of the data. This matrix was estimated using 25 isozymes and six nuclear microsatellite markers for the same families as those presented here.
Common garden studies indicate that a set of 57 families sampled east of the Cascade crest in northeastern Washington resembled the interior variety more so than the coastal variety for cold tolerance, phenology, and growth phenotypes (St. Clair et al. 2005). Genetic differentiation of these families was assessed using hierarchical analysis of molecular variance (AMOVA) with 25 allozyme markers (data from Krutovsky et al. 2009). We defined populations according to 20 ecological regions and then placed those populations into groups corresponding to populations located to the west or the east of the Cascade crest in northeastern Washington (Figure S1). Confidence intervals (95% C.I.'s) for global fixation indexes corresponding to FCT (between groups), FSC (among populations within groups), and FIS (within individuals) across loci were determined by bootstrapping (n = 20,000 replicates). Global fixation indexes were obtained by summing variance components across loci. All analyses were conducted in Arlequin ver. 3.11 (Excoffier et al. 2005). These analyses were used to investigate further population structure putatively not captured by patterns in the Q-matrix obtained from Krutovsky et al. (2009).
Statistical models:
Single-marker models were conducted for all SNP–trait combinations. We preferred single-marker relative to haplotype-based tests due to their simplicity, as well as to their similar statistical power to that of haplotype-based tests (Long and Langley 1999). A general linear model (GLM) was fitted to each trait–SNP combination (cf. Yu et al. 2006), with SNP markers as fixed effects and elements of the Q-matrix as covariates. We removed the first cluster in the Q-matrix because if included it would make an unnecessary linear dependence among the covariates when we performed F-tests of the covariates. All GLM analyses were conducted using Tassel ver. 2.0.1 (released April, 2007). The positive false discovery rate (FDR) method was used to correct for multiple testing (Storey 2003). All the necessary data to perform these analyses are available in File S1, File S2, File S3, File S4, and File S5).
Modes of inheritance and LD:
The prevalence of nonadditive effects was quantified using the ratio of dominance (d) to additive (a) effects. Partial or complete dominance was defined as values in the range of 0.50 < |d/a| < 1.25, while additive effects were defined as values in the range −0.50 ≤ d/a ≤ 0.50. Values of |d/a| > 1.25 were equated with over- or underdominance. We also investigated patterns of linkage disequilibrium (LD) among SNPs that were associated significantly with the same trait, using the maximum-likelihood approach implemented within the GENETICS package available in R (Warnes and Leisch 2006). We quantified patterns of LD using the squared allelic correlation coefficient (r2) and tested the significance of the inferred level of disequilibrium using Fisher's exact tests with a Bonferroni correction to account for multiple testing.
RESULTS
Data summary:
The 384 SNPs chosen for genotyping using the Illumina GoldenGate platform represent 117 unique candidate genes with 1–12 SNPs per gene (Table S3). Of the 384 SNPs, 228 (59%) yielded data consistent with our quality thresholds. The median GC50 score across all usable SNPs was 0.742, with the average CR being 94%. The majority of the 228 successfully genotyped SNPs were silent, with nonsynonymous SNPs accounting for 25% of the total. This did not deviate greatly from the original fraction in the full 384-SNP set (28%).
Population structure:
Strong patterns of population stratification were not apparent in the results obtained from Krutovsky et al. (2009). Most individuals were assigned equally well to one of the K clusters (i.e., Q ∼ 1/K). Latitude and longitude were largely uncorrelated to the Q-values for each of the 15 clusters, but patterns were apparent. Four clusters illustrated a correlation of Q-values to geography, with one of the 15 clusters corresponding to the 57 families located east of the Cascade crest in northeastern Washington (Figure S2). Differentiation across 25 allozyme markers for these families is moderate (global FCT = 0.035; 95% C.I., 0.015–0.082) and accounts for >90% of the differentiation among populations. Inbreeding within populations, however, was not significant (FIS = 0.026; 95% C.I., −0.005–0.055). The trait means for these families also differ significantly for 23 of the 25 traits (Table S4). We focus on association analyses that have these 57 families removed and then compare the results to those obtained when they are included.
Summary of significant associations:
A total of 5700 (228 SNPs × 25 traits) association tests were performed. Of these, 455 were significant at the nominal threshold of P = 0.05. Multiple test corrections using the FDR method reduced this number to 30 at a significance threshold of Q = 0.10. Of these, four marker–trait pairs remain significant after a conservative Bonferroni correction (Table 2). The number of significant associations varied across traits, ranging from 0 to 6. The 30 significant associations represent 15 unique SNPs from 12 candidate genes that affect 10 different traits. We discuss these associations in further detail below.
TABLE 2.
List of significant marker–trait pairs after a correction for multiple testing (FDR Q ≤ 0.10), using the 643 families located west of the Cascade crest
| Trait | Locus | Gene product | SNPa | ASb | n | F | r2 | P | Q |
|---|---|---|---|---|---|---|---|---|---|
| Emergence | |||||||||
| EMEAN | 60s RPL31a-418 | 60s ribosomal protein L31a | [A/G]c | — | 570 | 9.501 | 0.032 | 0.0001 | 0.0372 |
| EMEAN | CN639236.1-518 | Guanine nucleotide-binding protein | [A/G]d | A | 617 | 8.092 | 0.025 | 0.0003 | 0.0765 |
| Growth and resource partitioning | |||||||||
| RTLG | ES421311.1-369 | Hypothetical protein | [A/G]c | G | 642 | 23.841 | 0.036 | 1.3 × 10−6 | 0.0038 |
| RTSH | Pm_CL783Contig1-212 | SOUL heme-binding family protein | [A/G]e | C | 641 | 13.696 | 0.021 | 0.0002 | 0.0573 |
| RTSH | 4CL1-363 | 4-coumarate:CoA ligase 1 | [A/G]e | — | 643 | 12.936 | 0.019 | 0.0003 | 0.0765 |
| TAPER | LEA-EMB11-263 | Late embryogenesis abundant protein | [A/C]c | — | 614 | 13.832 | 0.022 | 0.0002 | 0.0561 |
| Phenology and cold tolerance | |||||||||
| BB2 | LEA-EMB11-263 | Late embryogenesis abundant protein | [A/C]c | — | 614 | 16.748 | 0.025 | 4.9 × 10−5 | 0.0342 |
| BB2 | 60s RPL31a-295 | 60s ribosomal protein L31a | [A/G]d | — | 641 | 12.834 | 0.019 | 0.0004 | 0.0765 |
| BB2 | 60s RPL31a-418 | 60s ribosomal protein L31a | [A/G]c | — | 570 | 7.607 | 0.024 | 0.0006 | 0.0913 |
| BS1 | CN637339.1-337 | Hypothetical protein | [A/G]e | A | 599 | 9.029 | 0.029 | 0.0001 | 0.0429 |
| BS1 | Pm_CL783Contig1-212 | SOUL heme-binding family protein | [A/G]e | C | 641 | 12.485 | 0.019 | 0.0004 | 0.0841 |
| BS2 | 60s RPL31a-418 | 60S ribosomal protein L31a | [A/G]c | — | 570 | 7.752 | 0.026 | 0.0005 | 0.0841 |
| Budcold | 4CL1-520 | 4-coumarate:CoA ligase 1 | [A/G]e | — | 641 | 9.118 | 0.028 | 0.0001 | 0.0427 |
| Budcold | CN638489.1-116 | α-expansin | [A/G]d | C | 628 | 7.607 | 0.024 | 0.0005 | 0.0913 |
| Ndlcold | sSPcDFD040B03103-274 | MADS-box transcription factor | [A/G]d | C | 641 | 20.392 | 0.03 | 7.5 × 10−6 | 0.0071 |
| Ndlcold | CN637306.1-381 | MYB-like transcription factor | [A/G]d | T | 642 | 8.563 | 0.026 | 0.0002 | 0.0561 |
| Ndlcold | 4CL1-520 | 4-coumarate:CoA ligase 1 | [A/G]e | — | 641 | 7.875 | 0.024 | 0.0004 | 0.0841 |
| Ndlcold | f3h2-54 | Flavanone-3-hydroxylase | [A/C]c | — | 642 | 7.749 | 0.023 | 0.0005 | 0.0841 |
| Stmcold | CN637306.1-381 | MYB-like transcription factor | [A/G]d | T | 642 | 11.485 | 0.034 | 1.3 × 10−5 | 0.0101 |
| Stmcold | f3h2-54 | Flavanone-3-hydroxylase | [A/C]c | — | 642 | 9.427 | 0.028 | 0.0001 | 0.0372 |
| Stmcold | Pm_CL234Contig1-156 | Rab GTPase | [A/T]c | T | 594 | 8.791 | 0.029 | 0.0002 | 0.0513 |
| Stmcold | CN637339.1-337 | Hypothetical protein | [A/G]e | A | 599 | 8.593 | 0.028 | 0.0002 | 0.0561 |
| Stmcold | CN638489.1-116 | α-expansin | [A/G]d | C | 628 | 8.036 | 0.025 | 0.0004 | 0.0765 |
| Stmcold | sSPcDFD040B03103-274 | MADS-box transcription factor | [A/G]d | C | 641 | 12.34 | 0.019 | 0.0005 | 0.0841 |
| Multivariate traits | |||||||||
| Prin1 | Pm_CL783Contig1-212 | SOUL heme-binding family protein | [A/G]e | C | 641 | 15.878 | 0.024 | 0.0001 | 0.0372 |
| Prin1 | sSPcDFD040B03103-274 | MADS-box transcription factor | [A/G]d | C | 641 | 14.843 | 0.023 | 0.0001 | 0.0427 |
| Prin2 | 60s RPL31a-418 | 60s ribosomal protein L31a | [A/G]c | — | 570 | 12.648 | 0.043 | 4.3 × 10−6 | 0.0048 |
| Prin2 | 60s RPL31a-55 | 60s ribosomal protein L31a | [A/G]d | — | 571 | 9.836 | 0.033 | 0.0001 | 0.0372 |
| Prin3 | LEA-EMB11-263 | Late embryogenesis abundant protein | [A/C]c | — | 614 | 22.475 | 0.034 | 2.7 × 10−6 | 0.0038 |
| Prin3 | 4CL1-520 | 4-coumarate:CoA ligase 1 | [A/G]e | — | 641 | 9.512 | 0.028 | 0.0001 | 0.0372 |
SNPs in boldface type were not consistent with Hardy–Weinberg equilibrium (HWE) expectations, as tested using Fisher's exact tests, at a Bonferroni-corrected significance threshold of P = 0.00022 (i.e., 0.05/228).
The ancestral state as determined by comparison to a single sequence of bigcone Douglas fir (Pseudotsuga macrocarpa). Dashes indicate that an outgroup sequence was unavailable.
Noncoding polymorphism.
Synonymous polymorphism.
Nonsynonymous polymorphism.
Individual phenotypic traits:
Growth and resource partitioning traits:
Twelve of the 21 traits were related to growth and resource partitioning (Table 1). These traits had a total of four significant marker–trait associations located within four unique candidate genes. One of these associations, the effect of marker ES421311.1-369 on root length (RTLG), survives a Bonferroni correction. These markers explain a small portion of the phenotypic variance in our sample, with effects ranging from 1.9 to 3.6%.
Phenology and cold-tolerance traits:
Six of the 21 traits were related to phenology and cold tolerance (Table 1). These traits had a total of 18 significant associations representing 11 unique SNPs located within 10 different candidate genes. One of these 18 associations survives a Bonferroni correction (Table 2). The majority of these SNPs illustrated patterns of gene action consistent with nonadditive effects (Table 3). For example, heterozygotes for the CN637339.1-367 marker set bud 3 days later on average than either homozygote class (274.3 for A/A, 277.7 for A/G, 275.1 for G/G). This marker is also associated with cold damage to stems and illustrated a similar mode of gene action, with heterozygotes having 0.35% more cold damage on average (2.1 for A/A, 2.4 for A/G, 1.9 for G/G). These 2 traits, however, are correlated with one another. The G allele at this marker is the derived state and causes an isoleucine (Ile) → valine (Val) amino acid substitution. Interestingly, all 5 additional marker–trait associations for cold damage to the stem phenotype illustrate a similar pattern, with heterozygotes having significantly more damage.
TABLE 3.
List of marker effects for significant marker–trait pairs using the 643 families located west of the Cascade crest
| Trait | Locusa | Gene product | 2ab | dc | d/a | 2a/σpd | Frequencye | αf |
|---|---|---|---|---|---|---|---|---|
| Emergence | ||||||||
| EMEAN | 60s RPL31a-418 | 60s ribosomal protein L31a | 0.002 | 0.001 | 1.01 | 0.69 | 0.24 (G) | −0.0004 |
| Phenology and cold tolerance | ||||||||
| BB2 | 60s RPL31a-418 | 60s ribosomal protein L31a | 3.26 | −1.21 | −0.73 | 0.74 | 0.24 (G) | 0.6916 |
| BS1 | CN637339.1-337 | Hypothetical protein | 0.73 | 2.80 | 7.68 | 0.10 | 0.21 (G) | 0.9737 |
| BS2 | 60s RPL31a-418 | 60s ribosomal protein L31a | 7.36 | −1.54 | −0.42 | 0.62 | 0.24 (G) | 2.0844 |
| Budcold | 4CL1-520 | 4-coumarate:CoA ligase 1 | 1.10 | −0.04 | −0.07 | 0.85 | 0.36 (G) | −0.1740 |
| Budcold | CN638489.1-116 | α-expansin | 0.39 | 0.30 | 1.54 | 0.30 | 0.49 (A) | −0.0900 |
| Ndlcold | 4CL1-520 | 4-coumarate:CoA ligase 1 | 1.78 | −0.11 | −0.12 | 0.85 | 0.36 (G) | −0.2854 |
| Stmcold | Pm_CL234Contig1-156 | rab GTPase | 0.50 | 0.24 | 0.96 | 0.36 | 0.76 (A) | −0.0874 |
| Stmcold | CN637339.1-337 | Hypothetical protein | 0.31 | 0.35 | 2.23 | 0.22 | 0.21 (G) | 0.2791 |
| Stmcold | CN638489.1-116 | α-Expansin | 0.45 | 0.34 | 1.49 | 0.32 | 0.49 (A) | −0.1044 |
| Multivariate traits | ||||||||
| Prin2 | 60s RPL31a-418 | 60s ribosomal protein L31a | 1.36 | 0.47 | 0.70 | 0.83 | 0.24 (G) | 0.0251 |
| Prin2 | 60s RPL31a-55 | 60s ribosomal protein L31a | 1.03 | 0.32 | 0.62 | 0.63 | 0.35 (A) | 0.0895 |
| Prin3 | 4CL1-520 | 4-coumarate:CoA ligase 1 | 0.72 | 0.09 | 0.23 | 0.51 | 0.36 (G) | −0.2366 |
Markers with only two observed genotypic classes or those that deviated significantly from HWE expectations are not included.
Calculated as the difference between the phenotypic means observed within each homozygous class (2a = |GBB − Gbb|, where Gij is the trait mean in the ijth genotypic class).
Calculated as the difference between the phenotypic mean observed within the heterozygous class and the average phenotypic mean across both homozygous classes [d = GBb – 0.5(GBB + Gbb), where Gij is the trait mean in the ijth genotypic class].
σp, standard deviation for the phenotypic trait under consideration. Prior to calculating this measure, the observed distributions of the trait values were tested for normality using Kolmogorov–Smirnov tests and a Bonferroni-corrected significance threshold of P = 0.002 (i.e., 0.05/25).
Allele frequency of either the derived or the minor allele. SNP alleles corresponding to the frequency listed are given in parentheses.
The additive effect was calculated as α = pB(GBB) + pb(GBb) – G, where G is the overall trait mean, Gij is the trait mean in the ij th genotypic class, and pi is the frequency of the ith marker allele. These values were always calculated with respect to the derived allele. When this was unknown, the values listed are for the minor allele. Values calculated in the latter manner are in boldface type.
Polymorphisms located within different candidate genes that were associated to the same trait were largely in linkage equilibrium. Departures from this pattern, however, were apparent. Significant pairwise estimates of LD were documented between markers located in the CN637306.1, f3h2, Pm_CL234Contig1-156, and CN638489.1 candidate gene loci. These three genes have SNPs associated significantly with cold damage to the stems, with 2.5–3.4% of the phenotypic variance being explained by each marker. The departures from linkage equilibrium are small (r2 < 0.20, P < 0.0001), yet define a set of candidate genes whose products are a transcription factor (CN63730.1), a rab GTPase (Pm_CL234Contig1), a cell wall architecture protein (α-expansin), and a flavanoid pathway protein (f3h2). Marker CN63730.1-381, however, deviates significantly from Hardy–Weinberg equilibrium (HWE) expectations (P < 0.0001).
Emergence:
Two of the 21 traits were related to emergence. These traits had a total of two significant associations representing two unique candidate genes. Both markers explained ∼3% of the phenotypic variance. Mean values of emergence (EMEAN) across genotypic classes at marker 60s RPL31a-418 indicate that the G allele is dominant to the A allele (0.0435 for A/A, 0.0459 for A/G, and 0.0458 for G/G). A similar analysis for the CN639236.1-518 locus was not performed, because the G/G genotype was not observed in our sample.
Multivariate phenotypic traits:
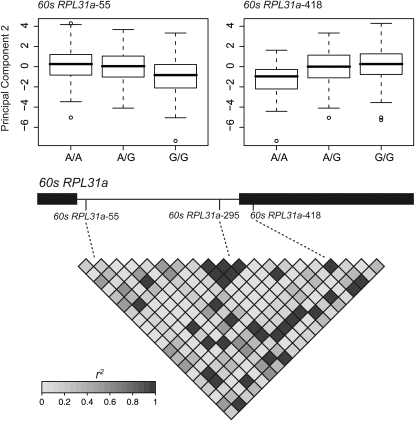
Marker–trait associations with principal components mirrored those detected for individual traits. The first principal component accounted for a large fraction of the phenotypic variance (44.8%). This component was primarily composed of growth traits, with phenology and cold-tolerance traits also strongly affecting its composition (Table S2). Two SNPs located within two different candidate genes were associated significantly with this component. Both loci explained a small fraction of the phenotypic variance (r2 < 0.025) and were associated primarily with individual traits related to phenology and cold tolerance. The second principal component was related largely to cold damage traits. Two markers located within the 60s RPL31a locus were associated significantly with this component (Table 2). These markers were also in significant LD with one another and explained similar fractions of the phenotypic variance (Figure 2). The average values of this component across genotypic classes for each SNP are suggestive of partial dominance, with the A allele at the 60s RPL31a-55 locus and the G allele at the 60s RPL31a-418 locus being partially dominant (Figure 2). These are also the alleles that are associated with less cold damage to stems, needles, and buds. The third principal component was related to bud burst and emergence and cold damage to needles. Two markers were associated significantly with this component, with each of those mirroring their effects on the single traits that are correlated with this component. No marker–trait associations survived multiple-test corrections for the fourth principal component.
Figure 2.—
An example of marker effects on the second principal component. This component is related largely to phenology and cold-tolerance traits. Each marker explains a small portion of the phenotypic variance (r2 ∼ 3%) and is consistent with an additive model of gene action. Whiskers in the box plots represent 1.5 times the interquartile range. The 2 associated SNPs are in LD with one another. Illustrated are the 21 SNPs discovered for the 60s RPL31a locus relative to the inferred gene model, as well as 3 of those 21 that were genotyped (dashed lines). Solid boxes denote exons in the gene model.
Modes of gene action and marker effects:
Many of the 30 SNPs associated significantly with at least one trait were consistent with modes of gene action other than codominance (Table 3). Four of the 13 marker–trait pairs (31%) for which dominance and additive effects could be calculated were consistent with over- or underdominance (|d/a| > 1.25). The majority of these markers were related to cold damage phenotypes, where the heterozygote had higher damage on average. The remaining 9 markers were split between modes of gene action that were additive [|d/a| < 0.50, n = 4 (31%)] or partially to fully dominant [0.50 < |d/a| < 1.25, n = 5 (38%)]. Most effects were small to moderate and accounted for only 10–85% of the phenotypic standard deviation.
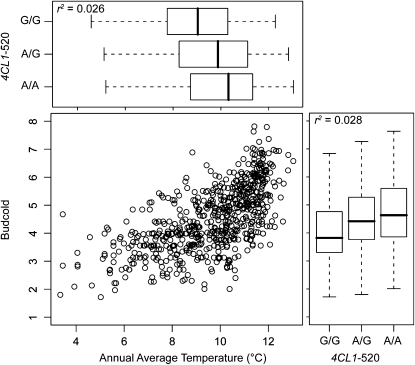
The derived allele was typically the minor allele. The additive effects of these derived alleles varied by trait, but were often small to moderate in size (Table 3). The additive effects on the cold damage to stems phenotype (stmcold), for example, varied from −0.10% (CN637339.1-337) to 0.28% (Pm_CL234Contig1-156). An analysis using the minor allele for those loci without an outgroup sequence found similar effects, with the minor allele typically conveying changes in trait consistent with cold hardiness (Table 3). The relationship between the genotypic classes of a marker associated to a phenotype and the environmental gradient that affects that phenotype is consistent with this pattern (Figure 3). For example, the G allele at the 4CL1-520 marker is the minor allele and genotypes containing this allele are found at sites with significantly lower annual average temperatures. Trees at these sites also had significantly lower cold damage to buds. The additive effect of the G allele at this marker was estimated to be −0.17% (Table 3).
Figure 3.—
An illustration of the relationship between phenotype, genotype, and the annual average temperature gradient (ANNAVT). Cold damage to buds is related to the annual average temperature of the locality where the source tree was sampled. This phenotype is also associated with a SNP at the 4CL1 locus. Genotypes at this SNP vary along the temperature gradient as expected, with the G allele being located at lower annual average temperatures. This SNP fits well with an additive model of gene action and is nonsynonymous.
Comparison to analyses using the full data set:
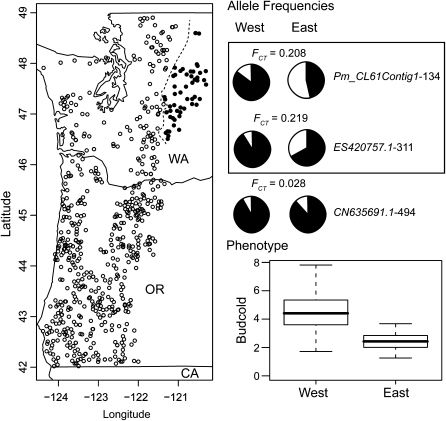
Inclusion of the 57 families located east of the Cascade crest in northeastern Washington changed the association analyses dramatically. The number of significant associations increased to 668 at a nominal significance threshold of P = 0.05. Multiple-test corrections using the FDR method reduced this number to 164 at a significance threshold of Q = 0.10 (Table S5). These represent increases in the number of detected associations of 47 and 447%, respectively. These 164 associations represent 44 unique SNP markers located within 35 candidate genes. Surprisingly, the vast increase in significant results is caused by a set of 24 candidate genes, with markers in 7 of those 24 accounting for 40% of the increased number of significant tests. These 7 markers also exhibit increased levels of differentiation as measured by FCT (Figure S3). The Pm_CL61Contig1-134 and ES420757.1-311 markers, for example, have values of FCT ∼10-fold higher than SNPs that remained unassociated with a phenotype regardless of whether or not the 57 families were included (Figure 4). Each of these markers was associated to ∼65% of the individual phenotypic traits when the 57 families in question were included in the analysis.
Figure 4.—
The families located east of the Cascade crest in northeastern Washington affect the outcome of the association testing. Inclusion of these 57 families inflates the number of significant associations from 30 to 164. This is due mainly to a set of seven markers, each within a unique candidate gene, that show elevated levels of differentiation (FCT = 0.08–0.22) across this region. Illustrated are two examples (markers enclosed in the box), with these SNPs being associated to ∼65% of the traits when the 57 eastside families are included in the analysis. The global FCT estimate for unassociated SNP and allozyme markers is in the range of 0.019–0.035 for comparison. Pie charts denote SNP allele frequencies east (open sections) and west (solid sections) of the Cascade crest.
DISCUSSION
Associations and marker effects:
Temperature is the most important environmental variable with respect to adaptation by coastal Douglas fir to the Pacific Northwest environments (St. Clair et al. 2005; St. Clair 2006). We have identified a set of 12 candidate genes that are associated significantly with cold hardiness in coastal Douglas fir. Four of the 15 polymorphisms within these genes are nonsynonymous substitutions, with the remainder being located largely in synonymous positions. These associations represent a refined list of candidate genes for further analysis, as well as provide insight into the genetic components of complex traits in coastal Douglas fir.
All 30 significant associations accounted for a small proportion of the phenotypic variance and were within the range of those published previously for forest trees (<1%−4%; Thumma et al. 2005; González-Martínez et al. 2007, 2008; Ingvarsson et al. 2008), as well as other wind-pollinated species (<1%–5%; Weber et al. 2007, 2008). Patterns such as these reflect a polygenic quantitative model, which is supported by a long history of quantitative genetic studies in forest trees (Namkoong 1979). Several marker alleles deviate from this quantitative model, however, when effects were measured in terms of phenotypic standard deviations (Table 3). Most of the associated markers accounted for less than half of a phenotypic standard deviation. Two markers in the 60s RPL31a candidate gene explain upward of 80% of the standard deviation for the second principal component (60s RPL31a-55-Prin2, 0.63σp; 60s RPL31a-418-Prin2, 0.83σp). The additive effect of the latter marker on bud set, moreover, was large, with a substitution of the G allele producing a 2-day increase in the time to set bud. Such effects compounded across a few loci could explain large portions of the development of cold hardiness in coastal Douglas fir.
The cumulative effects across loci each with a small effect, however, can also be large. For example, ∼20% of the phenotypic variance in early wood specific gravity was explained by a handful of SNPs in loblolly pine (González-Martínez et al. 2007). Here, six SNPs explain ∼17% of the phenotypic variance in cold damage to stems (Table 2). Thus, analyses based on expanded sets of candidate genes have the potential to identify markers that explain a large fraction of the phenotypic variance across numerous traits.
Seventeen candidate genes were shown previously to collocate with clonally replicated spring and fall cold-hardiness QTL in coastal Douglas fir (Wheeler et al. 2005). Of those, 9 had SNPs genotyped within the association population. Marker–trait associations within these 9 genes did not survive multiple-test corrections when the families east of the Cascade crest were excluded from the analysis. When these families were included, however, 2 of those 9 genes harbored markers associated with at least one cold-hardiness related trait: α-tubulin and erd15. Both candidate genes were mapped to linkage group seven and collocated with a QTL for spring needle cold hardiness (cf. Figure 1 in Wheeler et al. 2005). Markers within these candidate genes were associated with bud-set and cold-tolerance phenotypes, respectively.
More than half of the SNPs associated with cold tolerance and bud set showed modes of gene action consistent with over- or underdominance (Table 3). A trade-off exists between growth and fall cold hardiness and these two traits are often negatively correlated, both phenotypically and genetically in Douglas fir (Rehfeldt 1979; Aitken et al. 1996). There is also a positive genetic correlation between bud set and growth (Rehfeldt 1979, 1983; Campbell 1986; Li and Adams 1993) and a negative genetic correlation between bud set and fall cold hardiness (O'Neill et al. 2001). The relationships among these traits indicate that trees that set bud later have better growth (likely due to extending their growing season), but experience higher fall cold injury (lower cold hardiness), a pattern that is observed in many forest trees.
The phenotypic correlations of stem cold damage with bud set were moderate and highly significant (BS1, F1,863 = 613.8, P < 0.001, r = 0.64; BS2, F1,863 = 396.5, P < 0.001, r = 0.56; data from St. Clair et al. 2005 and St. Clair 2006). The correlation with height was weaker, but there was a significant correlation with height increment (HTINC: F1,863 = 63.5, P < 0.001, r = 0.26). The inferiority of heterozygotes for cold hardiness may thus be a correlated response that is a trade-off for higher growth. This explanation is consistent with the additive effects estimated for the CN637339.1-337 marker, with the G allele producing a later date of bud set (∼1 day later) while at the same time being correlated with ∼0.28% higher cold damage to stems (Table 3).
The large number of loci consistent with nonadditive modes of gene action is also consistent with epigenetic effects. These effects have been identified primarily in Norway spruce [Picea abies (L.) Karst.], where the timings of bud burst in spring, cessation of leader shoot growth in summer, and bud set in autumn are processes that are modified according to the temperature during female meiosis (Johnsen et al. 1996; Hänninen et al. 2001; Saxe et al. 2001; Johnsen et al. 2005a,b). Conditions colder than normal advance the onset of these effects, while temperatures above normal delay their onset (Kvaalen and Johnsen 2008). Further work has shown that this is not a genotypic-selection scenario and is likely to be a long-lasting epigenetic phenomenon tied to the temperature and photoperiod of the maternal tree during seed production (Besnard et al. 2008). The same traits as those shown to have epigenetic effects in Norway spruce have been implicated in coastal Douglas fir as showing strong evidence for phenotypic adaptation. Given that our sampling design was sensitive primarily to additive effects (i.e., we used breeding values for the phenotypes) and that many of the deviations from additive effects are strong (cf. Table 3), epigenetic factors warrant further investigation. This is especially true because of the importance of this species to forest ecosystems in western North America and the inferred impacts of global climate change on its abundance and distribution.
Effects of population structure on association analyses:
Strong population differentiation along an environmental gradient provides indirect evidence of the role of natural selection in shaping adaptive phenotypic variation (Endler 1986). This has aptly been demonstrated for a variety of cold-hardiness and growth-related phenotypes in coastal Douglas fir. Similar trends for the molecular genetic variation underlying these traits have been identified here, with most SNP genotypes tracking variation along the same gradient as their associated phenotypes (Figure 3). These gradients may be confounded with a contact zone between the coastal and the interior varieties of Douglas fir. Of particular interest are the 57 families located east of the Cascade crest in northeastern Washington. Inclusion of these families increased dramatically the number of significant associations from 30 to 164.
The disparity between these numbers could be due to neutral population structure not captured by the Q-matrix or strong population differences reflecting adaptive differentiation between varieties or intervarietal hybrids. The two varieties are differentiated strongly at 20 isozyme loci (Li and Adams 1989). If the families located east of the Cascade crest are the interior variety or intervarietal hybrids, unaccounted for population structure could inflate the number of false positives. If this was the case, we expect many loci to be differentiated strongly between the eastside families and those located west of the Cascade crest. This is not what is observed when considering those SNPs not associated strongly when all or just the westside families are included in the analyses (n = 184 SNPs; global FCT, 0.019; 95% C.I., 0.010–0.035). This value is much lower in magnitude than that estimated between the varieties using allozymes (GST = 0.116; Li and Adams 1989). This suggests that the Q-matrix utilized in the GLMs is controlling correctly for background levels of population structure. The nonsignificant global estimate of FIS, moreover, supports our avoidance of a mixed linear model (MLM) (cf. Yu et al. 2006) approach using kinship estimates. Such an analysis was conducted for loblolly pine, with the general conclusion that the MLM and GLM approaches produced similar results due to the fact that individuals were related less than second cousins on average (González-Martínez et al. 2007).
It is more likely that the effects the eastside families have on the association results are those due to adaptive differentiation between populations. Seven markers cause the majority of the increase in significant association results due to the correlation between allele frequency and phenotypic differentiation across the Cascade crest. These seven markers have values of FCT in the range of 0.08–0.22, which is an order of magnitude greater than the background level of differentiation. Further interpretation regarding the effects of these genes relative to the phenotypes is complicated given that most of the phenotypes also differ between these families and those located west of the Cascade crest.
Palynological data support a postglacial contact between varieties emerging from distinct southern Pleistocene refugia occurring ∼7000 years ago (Tsukada 1982; Wells 1983). Given that the coastal variety is more similar to the northern populations of the interior variety at allozyme loci (Li and Adams 1989), it is probable that introgression has and is occurring between varieties east of the Cascade crest. This also suggests that cold-adapted alleles identified here may have originated in the interior variety. Consistent with this hypothesis are common garden studies of intervarietal hybrids, which yield data consistent with largely nonadditive allelic effects for growth and cold-hardiness traits in the hybrids (Rehfeldt 1977). Many of the associations detected here may also reflect the correlation between freezing and drought-stress tolerance (cf. Blödner et al. 2005), especially given that water is one of the major limiting factors across the northern ranges of coastal and interior Douglas fir (Littell et al. 2008). Regardless of the hypothesis, it is clear that further phylogeographic investigations and association studies are needed across the entire range of coastal and interior Douglas fir.
Functions of candidate genes:
Inferred functionality of associated candidate genes varied across trait categories. Genes associated to cold-tolerance traits had homology to loci encoding proteins involved with lignin biosynthesis and cell wall architecture, transcription regulation, and signal transduction in Arabidopsis. Similar types of genes were upregulated in Sitka spruce in response to cold temperatures (Holliday et al. 2008). There was a surprising lack of calcium-signaling related genes producing significant marker–trait associations. This is not the case with the inclusion of the 57 families located east of the Cascade crest. Inclusion of those families resulted in markers located within loci encoding a cysteine proteinase (Pm_CL135Contig1) and a cyclophilin (Pm_CL61Contig1) being associated with several cold-tolerance and phenology traits. Cyclophilins are involved with cysteine biosynthesis in plants and in calcium–calmodulin-activated serine/threonine-specific protein phosphatase calcineurin in humans and yeast (Wang and Heitman 2005; Dominguez-Solis et al. 2008).
Genes associated with phenology and emergence traits were homologous to loci encoding proteins involved with ribosome biogenesis (60s RPL31a) and stress tolerance (LEA-EMBL11). Notably, a SOUL heme-binding protein (Pm_CL783Contig1) had significant marker–trait associations with bud-set and root-shoot ratio. A homologous protein has been localized to vacuoles in Arabidopsis and linked putatively with hemoprotein synthesis (Jaquinod et al. 2007).
Integration with population genetic results:
Patterns of polymorphism and divergence for 2 of the 12 candidate genes identified here were characterized previously as deviant from null models incorporating genetic drift and some forms of historical demography (Eckert et al. 2009b). The Pm_CL234Contig1 locus has an excess of high-frequency derived polymorphisms (Fay and Wu's normalized H = −2.35, P < 0.05), a pattern that is consistent with hitchhiking, while the ES421311.1 locus has an excess of rare alleles (Tajima's D = −1.70, P < 0.05). Both of these results, however, did not survive multiple-test corrections or incorporation of severe bottlenecks into hypothesis tests of neutrality. The association reported here for the Pm_CL234Contig1 locus is with a marker segregating a high-frequency derived allele. This allele conveys a 1.11% reduction in cold damage to stems. A simplistic interpretation of this pattern is that the association detected here is due to the recent fixation of an advantageous haplotype containing the derived SNP allele. Future research combining population, quantitative, and landscape genetics, therefore, will offer new insights into the genetic architecture of cold-hardiness related traits in coastal Douglas fir.
Conclusions:
We identified 30 marker–trait associations across 12 candidate genes and 10 cold-hardiness related traits. Marker effects were small (1% < r2 < 3.6%), consistent with a polygenic quantitative model, and tracked similar environmental gradients to those affecting the phenotypes to which the markers were associated. Higher-order effects (e.g., dominance) were prevalent throughout the association results, with 86% of the significant markers having nonadditive effects. Such results would benefit directly from validation in additional association populations (cf. Weber et al. 2008), especially since significant QTL-by-environment interactions have been documented for several adaptive traits (Jermstad et al. 2003). Nonetheless, these results represent a further step toward the dissection of cold hardiness in coastal Douglas fir.
Acknowledgments
We thank Valerie Hipkins and her staff at the National Forest Genetics Electrophoresis Laboratory located at the Institute for Forest Genetics (Placerville, CA) for performing the DNA extractions, Charles Nicolet and Vanessa Rashbrook for performing the SNP genotyping, and John Liechty and Benjamin Figueroa for bioinformatics support. This article was also much improved by incorporating comments from two anonymous reviewers. Funding for this project was made available through a U.S. Department of Agriculture National Research Initiative Plant Genome Grant (04-712-0084).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102350/DC1.
References
- Aagaard, J. E., K. V. Krutovskii and S. H. Strauss, 1998. RAPDs and allozymes exhibit similar levels of diversity and differentiation among populations and races of Douglas-fir. Heredity 81 69–78. [Google Scholar]
- Aitken, S. N., and W. T. Adams, 1996. Genetics of fall and winter cold hardiness of coastal Douglas-fir in Oregon. Can. J. For. Res. 26 1828–1837. [Google Scholar]
- Aitken, S. N., and W. T. Adams, 1997. Cold hardiness under strong genetic control in Oregon populations of Pseudotsuga menziesii var. menziesii. Can. J. For. Res. 27 1773–1780. [Google Scholar]
- Aitken, S. N., W. T. Adams, N. Schermann and L. H. Fuchigami, 1996. Family variation for fall cold hardiness in two Washington populations of coastal Douglas-fir (Pseudotsuga menziesii var. menziesii [Mirb.] Franco). For. Ecol. Manage. 80 187–195. [Google Scholar]
- Aitken, S. N., S. Yeaman, J. A. Holliday, T. Wang and S. Curtis-McLane, 2008. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard, G., V. Acheré, S. Jeandroz, Ø. Johnsen, P. Faivre Rampant et al., 2008. Does maternal environmental condition during reproductive development induce genotypic selection in Picea abies? Ann. For. Sci. 65 109–114. [Google Scholar]
- Blödner, C., T. Skrøppa, Ø. Johnsen and A. Polle, 2005. Freezing tolerance in two Norway spruce (Picea abies (L.) Karst.) progenies is physiologically correlated with drought tolerance. J. Plant Physiol. 162 549–558. [DOI] [PubMed] [Google Scholar]
- Brown, G. R., G. P. Gill, R. J. Kuntz, C. H. Langley and D. B. Neale, 2004. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc. Natl. Acad. Sci. USA 101 15255–15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. K., 1986. Mapped genetic variation of Douglas-fir to guide seed transfer in southwest Oregon. Silvae Genet. 35 85–96. [Google Scholar]
- Campbell, R. K., and F. C. Sorensen, 1973. Cold acclimation in seedling Douglas-fir related to phenology and provenance. Ecology 54 1148–1151. [Google Scholar]
- Campbell, R. K., and F. C. Sorensen, 1979. A new basis for characterizing germination. J. Seed. Tech. 4 24–34. [Google Scholar]
- Dominguez-Solis, J. R., Z. He, A. Lima, J. Ting, B. B. Buchanan et al., 2008. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl. Acad. Sci. USA 105 16386–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert, A. J., B. Pande, E. S. Ersöz, M. H. Wright, V. K. Rashbrook et al., 2009. a High-throughput genotyping and mapping of single nucleotide polymorphisms in loblolly pine (Pinus taeda L.). Tree Genet. Genomes 5 225–234. [Google Scholar]
- Eckert, A. J., J. L. Wegrzyn, B. Pande, K. D. Jermstad, J. M. Lee et al., 2009. b Multilocus patterns of nucleotide diversity and divergence reveal positive selection at candidate genes related to cold-hardiness in coastal Douglas fir (Pseudotsuga menziesii var. menziesii). Genetics 183 (in press). [DOI] [PMC free article] [PubMed]
- Endler, J. A., 1986. Natural Selection in the Wild. Princeton University Press, Princeton, NJ.
- Eveno, E., C. Collada, M. A. Guevara, V. Léger, A. Soto et al., 2008. Contrasting patterns of selection at Pinus pinaster Ait. drought stress candidate genes as revealed by genetic differentiation analyses. Mol. Biol. Evol. 25 417–437. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., G. Laval and S. Schneider, 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1 47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush, D., M. Stephens and J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. B., A. Oliphant, R. Shen, B. G. Kermani, F. Garcia et al., 2003. Highly parallel SNP genotyping. Cold Spring Harbor Symp. Quant. Biol. 68 69–78. [DOI] [PubMed] [Google Scholar]
- Fowler, S., and M. F. Thomashow, 2002. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González–Martínez, S. C., E. S. Ersöz, G. R. Brown, N. C. Wheeler and D. B. Neale, 2006. DNA sequence variation and selection of tag SNPs at candidate genes for drought–stress response in Pinus taeda L. Genetics 172 1915–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González–Martínez, S. C., N. C. Wheeler, E. Ersöz, C. D. Nelson and D. B. Neale, 2007. Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González–Martínez, S. C., D. Huber, E. Ersöz, J. M. Davis and D. B. Neale, 2008. Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 101 19–26. [DOI] [PubMed] [Google Scholar]
- Grattapaglia, D., and M. Kirst, 2008. Eucalyptus applied genomics: from gene sequences to breeding tools. New Phytol. 179 911–929. [DOI] [PubMed] [Google Scholar]
- Grattapaglia, D., C. Plomion, M. Kirst and R. R. Sederoff, 2009. Genomics of growth traits in forest trees. Curr. Opin. Plant Biol. 12 148–156. [DOI] [PubMed] [Google Scholar]
- Guy, C. L., K. J. Niemi and R. Brambl, 1985. Altered gene expression during cold acclimation of spinach. Proc. Natl. Acad. Sci. USA 82 3673–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D., V. Luquez, M. V. Garcia, K. R. St. Onge, S. Jansson et al., 2007. Adaptive population differentiation in phenology across a latitudinal gradient in European aspen (Populus tremula L.): a comparison of neutral markers, candidate genes and phenotypic traits. Evolution 61 2849–2860. [DOI] [PubMed] [Google Scholar]
- Hänninen, H., E. Beuker, Ø. Johnsen, I. Leinonen, M. Murray et al., 2001. Impacts of climate change on cold hardiness of conifers, pp. 305–333 in Conifer Cold Hardiness, edited by F. Bigras and S. Colombo. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Heuertz, M., E. De Paoli, T. Källman, H. Larsson, I. Jurman et al., 2006. Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce [Picea abies (L.) Karst]. Genetics 174 2095–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, J. A., S. Ralph, R. White, J. Bohlmann and S. N. Aitken, 2008. Global monitoring of gene expression during autumn cold acclimation among rangewide populations of Sitka spruce [Picea sitchensis (Bong.) Carr.]. New Phytol. 178 103–122. [DOI] [PubMed] [Google Scholar]
- Howe, G. T., S. N. Aitken, D. B. Neale, K. D. Jermstad, N. C. Wheeler et al., 2003. From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can. J. Bot. 81 1247–1266. [Google Scholar]
- Ingvarsson, P., M. V. Garcia, D. Hall, V. Luquez and S. Jansson, 2006. Clinal variation in phyB2, a candidate gene for day-length-induced growth cessation and bud set, across a latitudinal gradient in European aspen (Populus tremula). Genetics 182 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson, P., M. V. Garcia, V. Luquez, D. Hall and S. Jansson, 2008. Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod, M., F. Villiers, S. Kieffer-Jaquinod, V. Hugouvieux, C. Bruley et al., 2007. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell Proteomics 6 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, K. S. Jech, N. C. Wheeler and D. B. Neale, 2001. a Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. I. Timing of vegetative bud flush. Theor. Appl. Genet. 102 1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, N. C. Wheeler, T. S. Anekonda, S. N. Aitken et al., 2001. b Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. II. Spring and fall cold-hardiness. Theor. Appl. Genet. 102 1152–1158. [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, K. S. Jech, G. A. Ritchie, N. C. Wheeler et al., 2003. Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. III. Quantitative trait loci-by-environment interactions. Genetics 165 1489–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen, Ø., T. Skrøppa, O. Junttila and O. G. Dæhlen, 1996. Influence of the female flowering environment on autumn frost hardiness of Picea abies progenies. Theor. Appl. Genet. 92 797–802. [DOI] [PubMed] [Google Scholar]
- Johnsen, Ø., O. G. Dæhlen, G. Østreng and T. Skrøppa, 2005. a Daylength and temperature during seed production interactively affect adaptive performance of Picea abies progenies. New Phytol. 168 589–596. [DOI] [PubMed] [Google Scholar]
- Johnsen, Ø., C. G. Fossdal, N. E. Nagy, J. Mølmann, O. G. Dæhlen et al., 2005. b Climatic adaptation in Picea abies progenies is affected by the temperature during zygotic embryogenesis and seed maturation. Plant Cell Environ. 28 1090–1102. [Google Scholar]
- Kvaalen, H., and Ø. Johnsen, 2008. Timing of bud set in Picea abies is regulated by a memory of temperature during zygotic and somatic embryogenesis. New Phytol. 177 49–59. [DOI] [PubMed] [Google Scholar]
- Krutovsky, K. V., and D. B. Neale, 2005. Nucleotide diversity and linkage disequilibrium in cold-hardiness and wood-quality related candidate genes in Douglas-fir. Genetics 171 2029–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutovsky, K. V., J. B. St. Clair, R. Saich, V. D. Hipkins and D. B. Neale, 2009. Estimation of population structure in coastal Douglas-fir [Pseudotsuga menziesii (Mirb.) Franco var. menziesii] using allozyme and microsatellite markers. Tree Genet. Genomics (in press).
- Landegren, U., R. Kaiser, J. Sanders and L. Hood, 1988. A ligase-mediated gene detection technique. Science 241 1077–1080. [DOI] [PubMed] [Google Scholar]
- Le Corre, V., and A. Kremer, 2003. Genetic variability at neutral markers, quantitative trait loci and trait in a subdivided population under selection. Genetics 164 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.-H., D. A. Henderson and J.-K. Zhu, 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., and W. T. Adams, 1989. Range-wide patterns of allozyme variation in Douglas-fir (Pseudotsuga menziesii). Can. J. For. Res. 19 149–161. [Google Scholar]
- Li, P., and W. T. Adams, 1993. Genetic control of bud phenology in pole-size trees and seedlings of coastal Douglas-fir. Can. J. For. Res. 23 1043–1051. [Google Scholar]
- Littell, J., D. L. Peterson and M. Tjoelker, 2008. Douglas-fir growth in mountain ecosystems: water limits tree growth from stand to region. Ecol. Monogr. 78 349–368. [Google Scholar]
- Long, A. D., and C. H. Langley, 1999. The power of association studies to detect the contribution of candidate gene loci to variation in complex traits. Genome Res. 9 720–731. [PMC free article] [PubMed] [Google Scholar]
- Loopstra, C. A., and W. T. Adams, 1989. Patterns of variation in first-year seedling traits within and among Douglas-fir breeding zones in southwest Oregon. Silvae Genet. 38 235–243. [Google Scholar]
- Morris, R. W., and P. T. Spieth, 1978. Sampling strategies for using female megagametophytes to estimate heterozygosities in conifers. Theor. Appl. Genet. 51 217–222. [DOI] [PubMed] [Google Scholar]
- Namkoong, G., 1979. Introduction to quantitative genetics in forestry. Technical Bulletin 1588. USDA Forest Service, Washington, DC.
- Neale, D. B., 2007. Genomics to tree breeding and forest health. Curr. Opin. Genet. Dev. 17 539–544. [DOI] [PubMed] [Google Scholar]
- Neale, D. B., and P. K. Ingvarsson, 2008. Population, quantitative and comparative genomics of adaptation in forest trees. Curr. Opin. Plant Biol. 11 1–7. [DOI] [PubMed] [Google Scholar]
- Neale, D. B., and O. Savolainen, 2004. Association genetics of complex traits in conifers. Trends Plant Sci. 9 325–330. [DOI] [PubMed] [Google Scholar]
- Oliphant, A., D. L. Barker, J. R. Stuelpnagel and M. S. Chee, 2002. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques 56–8(Suppl.): 60–61. [PubMed] [Google Scholar]
- O'Neill, G. A., S. N. Aitken and W. T. Adams, 2001. Genetic selection for cold hardiness in coastal Douglas-fir seedlings and saplings. Can. J. For. Res. 30 1799–1807. [Google Scholar]
- Pavy, N., B. Pelgas, S. Beauseigle, S. Blais, F. Gagnon et al., 2008. Enhancing genetic mapping of complex genomes through the design of highly-multiplexed SNP arrays: application to the large and unsequenced genomes of white spruce and black spruce. BMC Genomics 9 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhäjärvi, T., M. R. García-Gil, T. Knürr, M. Mikkonen, W. Wachowiak et al., 2007. Demographic history has influenced nucleotide diversity in European Pinus sylvestris populations. Genetics 177 1713–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt, G. E., 1977. Growth and cold hardiness of intervarietal hybrids of Douglas-fir. Theor. Appl. Genet. 50 3–15. [DOI] [PubMed] [Google Scholar]
- Rehfeldt, G. E., 1979. Ecological adaptations in Douglas-fir (Pseudotsuga menziesii var. glauca) populations. I. North Idaho and north-east Washington. Heredity 43 383–397. [Google Scholar]
- Rehfeldt, G. E., 1983. Genetic variability within Douglas-fir populations: implications for tree improvement. Silvae Genet. 32 9–14. [Google Scholar]
- Rehfeldt, G. E., 1989. Ecological adaptations in Douglas-fir (Pseudotsuga menziesii var. glauca): a synthesis. For. Ecol. Manage. 28 203–215. [Google Scholar]
- Sakai, A., and W. Larcher, 1987. Frost Survival of Plants. Springer-Verlag, Berlin.
- Savolainen, O., and T. Pyhäjärvi, 2007. Genomic diversity in forest trees. Curr. Opin. Plant Biol. 10 162–167. [DOI] [PubMed] [Google Scholar]
- Saxe, H., M. G. R. Cannell, Ø. Johnsen, M. G. Ryan and G. Vourlitis, 2001. Tree and forest functioning in response to global warming. Tansley review no. 123. New Phytol. 149 369–400. [DOI] [PubMed] [Google Scholar]
- Sekai, M., M. Narusaka, J. Ishida, T. Nanjo, M. Fujita et al., 2002. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 279–292. [DOI] [PubMed] [Google Scholar]
- St. Clair, J. B., 2006. Genetic variation in fall cold hardiness in coastal Douglas-fir in western Oregon and Washington. Can. J. Bot. 84 1110–1181. [Google Scholar]
- St. Clair, J. B., N. L. Mandel and K. W. Vance-Borland, 2005. Genecology of Douglas-fir in western Oregon and Washington. Ann. Bot. 96 1199–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe, J. R., and H. E. Hoekstra, 2008. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100 158–170. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31 2013–2035. [Google Scholar]
- Thomashow, M. F., 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 571–599. [DOI] [PubMed] [Google Scholar]
- Thumma, B. R., M. F. Nolan, R. Evans and G. F. Moran, 2005. Polymorphisms in cinnamoly CoA reductase (ccr) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., 1982. Pseudotsuga menziesii (Mirb.) Franco: its pollen dispersal and late Quaternary history in the Pacific Northwest. Jpn. J. Ecol. 32 159–187. [Google Scholar]
- Viard, F., Y. A. El-Kassaby and K. Ritland, 2001. Diversity and genetic structure in populations of Pseudotsuga menziesii (Pinaceae) at chloroplast microsatellite loci. Genome 44 336–344. [DOI] [PubMed] [Google Scholar]
- Walther, G.-R., E. Post, P. Convey, A. Menzel, C. Parmesan et al., 2002. Ecological responses to recent climate change. Nature 416 389–395. [DOI] [PubMed] [Google Scholar]
- Wang, P., and J. Heitman, 2005. Protein family review: the cyclophilins. Genome Biol. 6 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples, R. S., and O. Gaggiotti, 2006. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 15 1419–1439. [DOI] [PubMed] [Google Scholar]
- Warnes, G., and F. Leisch, 2006. Genetics: population genetics. R Package, version 1.2.1. http://cran.r-project.org.
- Webber, J., P. Ott, J. Owens and W. Binder, 2005. Elevated temperature during reproductive development affects cone traits and progeny performance in Picea glauca x engelmannii complex. Tree Physiol. 25 1219–1227. [DOI] [PubMed] [Google Scholar]
- Weber, A. L., R. M. Clark, L. Vaughn, J. de Jesús Sánchez-Gonzalez, J. Yu et al., 2007. Major regulatory genes in maize contribute to standing variation in teosinte (Zea mays spp. parviglumis). Genetics 177 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A. L., W. H. Briggs, J. Rucker, B. M. Baltazar, J. de Jesús Sánchez-Gonzalez et al., 2008. The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): new evidence from association mapping. Genetics 180 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser, C. J., 1970. Cold resistance and injury in woody plants. Science 169 1269–1278. [DOI] [PubMed] [Google Scholar]
- Wells, P. V., 1983. Paleobiogeography of montane islands in the Great Basin since the last glaciopluvial. Ecol. Monogr. 53 341–382. [Google Scholar]
- Wheeler, N. C., K. D. Jermstad, K. V. Krutovsky, S. N. Aitken, G. T. Howe et al., 2005. Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. IV. Cold-hardiness QTL verification and candidate gene mapping. Mol. Breed. 15 145–156. [Google Scholar]
- White, T. L., 1987. Drought-tolerance of southwestern Oregon Douglas-fir. For. Sci. 33 283–293. [Google Scholar]
- Wright, S. I., and B. S. Gaut, 2005. Molecular population genetics and the search for adaptive evolution in plants. Mol. Biol. Evol. 22 506–519. [DOI] [PubMed] [Google Scholar]
- Yakovlev, I. A., C.-G. Fossdal, Ø. Johnsen, O. Junttlia and T. Skrøppa, 2006. Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genet. Genomes 2 39–52. [Google Scholar]
- Yu, J., G. Pressoir, W. H. Briggs, I. Vroh Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38 203–208. [DOI] [PubMed] [Google Scholar]
- Zhai, W., R. Nielsen and M. Slatkin, 2009. An investigation of the statistical power of neutrality tests based on comparative and population genetic data. Mol. Biol. Evol. 26 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]