Abstract
Rice blast, caused by Magnaporthe oryzae, is one of the most devastating diseases. The two major subspecies of Asian cultivated rice (Oryza sativa L.), indica and japonica, have shown obvious differences in rice blast resistance, but the genomic basis that underlies the difference is not clear. We performed a genomewide comparison of the major class of resistant gene family, the nucleotide-binding site–leucine-rich repeat (NBS–LRR) gene family, between 93-11 (indica) and Nipponbare (japonica) with a focus on their pseudogene members. We found great differences in either constitution or distribution of pseudogenes between the two genomes. According to this comparison, we designed the PCR-based molecular markers specific to the Nipponbare NBS–LRR pseudogene alleles and used them as cosegregation markers for blast susceptibility in a segregation population from a cross between a rice blast-resistant indica variety and a susceptible japonica variety. Through this approach, we identified a new blast resistance gene, Pid3, in the indica variety, Digu. The allelic Pid3 loci in most of the tested japonica varieties were identified as pseudogenes due to a nonsense mutation at the nucleotide position 2208 starting from the translation initiation site. However, this mutation was not found in any of the tested indica varieties, African cultivated rice varieties, or AA genome-containing wild rice species. These results suggest that the pseudogenization of Pid3 in japonica occurred after the divergence of indica and japonica.
THE two elite rice varieties with sequenced genomes, Nipponbare from Japan and 93-11 from China, represent the two major Asian cultivated rice subspecies, Oryza sativa L. japonica and indica, respectively. Sequencing these two genomes (Goff et al. 2002; Yu et al. 2002, 2005; IRGSP 2005) has opened an avenue for exploring the genomic basis of agricultural trait differences between these two subspecies (Garris et al. 2005). Rice blast caused by Magnaporthe oryzae is one of the most devastating diseases in both indica and japonica varieties. Many rice breeders have observed the difference in blast resistance between the two subspecies. Genetic characterization of blast resistance has suggested that indica varieties have more complicated blast resistance patterns than japonica varieties (Mackill et al. 1985; Yu et al. 1987; Mackill and Bonman 1992; Pan et al. 1996; Imbe et al. 1997). In contrast to other Asian countries, China has a long history of growing both indica and japonica varieties (Normile 1997; Zong et al. 2007). When Chinese rice varieties were tested with different Japanese M. oryzae strains, most of the japonica varieties could be classified into groups according to their respective disease-response patterns to the M. oryzae strains. However, when Chinese indica varieties were tested with the same stains, it was difficult to analyze their disease-response patterns because some were resistant to all the strains (Kiyosawa 1984; Ling 1984). This suggests that the indica and japonica varieties have diverged significantly in resistance to M. oryzae strains.
Plants face frequent threats of many potential pathogens, including viruses, bacteria, and fungi, in their environments. During the long coevolution with pathogens, plants have evolved sophisticated mechanisms to perceive and defend against a pathogen's attack (Dangl and Jones 2001). Innate immunity, the most important of the plant defense responses, can be classified into “basal” and “R-gene-mediated” defenses (Abramovitch et al. 2006). The R-gene-mediated defense was originally proposed by H. H. Flor (Flor 1956). This type of resistance is highly effective and easily manipulated in breeding programs to improve crop resistance. The majority of R proteins contain a nucleotide-binding site and a carboxy-terminal leucine-rich repeat domain (Martin et al. 2003). Notably, almost all the functionally identified nucleotide-binding site–leucine-rich repeat (NBS–LRR) genes are R genes with only one exception (Faigon-Soverna et al. 2006) and thus this class of genes is regarded as the core component of the plant immune system (Nimchuk et al. 2003).
Thus far, 10 rice blast R genes [Pib (Wang et al. 1999), Pita (Bryan et al. 2000), Pi9 (Qu et al. 2006), Pid2 (Chen et al. 2006), Pi2 (Zhou et al. 2006), Piz-t (Zhou et al. 2006), Pi36 (Liu et al. 2007), Pi37 (Lin et al. 2007), Pikm (Ashikawa et al. 2008), and Pi5 (Lee et al. 2009)] have been identified via map-based cloning methods. All the cloned blast resistance genes encode NBS–LRR proteins except for Pid2, which encodes a receptor-like kinase (Chen et al. 2006). The large number of published blast R gene isolations reveals the importance of the NBS–LRR gene family in rice blast disease resistance.
Recently, a genomewide investigation of the NBS–LRR genes of the two sequenced rice genomes was reported (Yang et al. 2006). However, that study focused only on genetic variations among common R gene sequences in the two genomes, but did not associate the genetic variations with the differential disease resistance between indica and japonica rice. In view of the difference in blast resistance between indica and japonica varieties described above, we performed a genomewide comparison of NBS–LRR gene sequences between 93-11 and Nipponbare. Our investigation focused on the systemic changes in the NBS–LRR gene sequences between the two genomes. In the survey, we found many potential NBS–LRR pseudogenes in both 93-11 and Nipponbare genomes due to premature stop codons or frameshift mutations in the ORFs. Notably we found great differences in constitution and distribution of pseudogenes between the two genomes.
The rice genome has been estimated to carry ∼500 NBS–LRR genes (Bai et al. 2002; Monosi et al. 2004; Zhou et al. 2004). Although most of the annotated rice NBS–LRR genes have not been functionally identified, we can expect that if a complete NBS–LRR gene functions as a blast R gene in one genome, its allelic pseudogene in the other genome might lose the function of blast resistance. Thus, we believe that the NBS–LRR pseudogene alleles can be used as cosegregation markers for blast susceptibility in an appropriate rice segregation population. In this study, we demonstrated the feasibility of this approach. We applied this novel strategy to the indica variety, Digu, and successfully identified a new blast R gene, Pid3.
MATERIALS AND METHODS
Plant materials:
The indica and japonica varieties used in this study are listed in supporting information, Table S1. The accessions and geographic origins of the 36 lines of wild rice and 4 varieties of African cultivated rice are listed in Table S2.
Fungal inoculation:
The M. oryzae strains used in this study were collected and isolated from indica- and japonica-cultivated regions in China or Japan. The M. oryzae stain Zhong-10-8-14 was isolated from a japonica-cultivated region in northern China. We expected to use this strain to screen useful R genes from the highly resistant indica variety, Digu. The isolated R genes might be used to improve the blast resistance of Chinese japonica varieties.
Rice seedlings at the four-leaf stage were inoculated by spraying a spore suspension (5 × 104 spores/ml) of the M. oryzae strain onto the leaves. Subsequently, the inoculated plants were placed into a chamber maintained at 25° and 100% humidity in the dark for 24 hr and then transferred to a greenhouse. The disease reaction was examined 1 week after inoculation with the susceptible variety, TP309, as a control.
Identification of NBS–LRR seed sequences of rice:
By searching with the keyword “NBS,” we retrieved 68 protein sequences from the Swiss-Prot database (http://www.ebi.ac.uk/swissprot/). A set of 32,127 rice full-length cDNA sequences was downloaded from the KOME database (http://cdna01.dna.affrc.go.jp/cDNA/). The cDNA sequences were translated in six-frame and the longest ORF was reported as the putative protein sequence. A set of 153 sequences with the NBS motif was selected from the complete set of predicted full-length (fl)-cDNA supported rice proteins, using a hidden Markov model (HMM) (Eddy 1998) for the NBS domain from the Pfam database (PF0931; http://pfam.wustl.edu) (Bateman et al. 2002). The 153 protein sequences were aligned only on the basis of the NBS domain, using hmmalign. This alignment was then used to develop a rice-specific HMM model to identify related sequences. The refined HMM was compared again with the complete set of fl-cDNA sequences, using estwisedb in the wise2 software package (ftp://ftp.ebi.ac.uk/pub/software/unix/wise2/) (Birney et al. 2004). All sequences that matched the model with a score of ≥25 were extracted (http://www.ebi.ac.uk/Tools/Wise2/doc_wise2.html#SECTION00055000000000000000). In total, we obtained 166 nonredundancy cDNA sequences that encoded the NBS domain. We identified a subset of them as seed sequences according to three criteria: (1) The cDNA sequences were aligned against the closest matching protein among 68 known NBS proteins, using estwise without obvious ORF-disrupted mutations (such as premature stop codons or frameshifts); (2) the noncoding regions of cDNA were initially searched against 68 known NBS protein sequences, using tblastn with an expectation score of >0.1; and (3) all sequences that met the previous criteria were verified by cross-validation, using estwise without obvious disablements. We found a total of 102 seed proteins, including 99 fl-cDNA-supported proteins and the 3 proteins encoded by Pib (Wang et al. 1999), Pita (Bryan et al. 2000), and Pi9 (Qu et al. 2006). Alignment of protein amino acid sequences with nucleotide sequences was performed with the wise2 package.
Identification of NBS–LRR genes and pseudogenes in rice:
Nipponbare and 93-11 genome sequences were downloaded from the TIGR Rice Genome Annotation Database (version 4.0, http://rice.plantbiology.msu.edu/) and RISE Database (http://rise.genomics.org.cn), respectively. To identify the regions that contain the NBS domain, the genomes were scanned with a rice-specific NBS HMM model, using Genewisedb with a score of ≥25. A set of 653 regions was detected from the database of Nipponbare, and a set of 553 regions was detected from that of 93-11.
Using the BGI FGF pipeline (http://fgf.genomics.org.cn/), we compared the genome regions containing the NBS domain with the closest matching seed protein, using Genewise. If there was a continuous span of sequence that was ≥70% of the length of the closest matching seed protein, we designated that sequence as an NBS–LRR gene; otherwise it was designated as an NBS–LRR gene segment. If an NBS gene contained a premature stop codon or frameshift mutation when compared with the seed protein, it was designated as a pseudogene.
Identification of Nipponbare and 93-11 NBS–LRR allelic pairs:
We generated the Nipponbare and 93-11 NBS–LRR allelic pairs by comparing the gene sequences from the two genomes. We conducted a multiple sequence alignment of the 1206 protein sequences that contained an NBS domain from the Nipponbare and 93-11 genomes using ClustalW (Chenna et al. 2003), including 267 NBS–LRR protein segments. Then the least divergent NBS–LRR gene pairs between the Nipponbare and 93-11 genomes were extracted from the phylogenetic tree, which was generated by using PAUP 4.0 (Swofford 2000) based on the neighbor-joining (NJ) method. If they met all the following criteria, the least divergent pairs were assigned as NBS–LRR allelic pairs: (1) neither gene in the pair is an NBS–LRR segment, (2) both genes are located at the corresponding locus of the homologous chromosomes, and (3) the nucleotide sequence identity of the pair is ≥82%.
Expression analysis of Pid3:
The four-leaf stage plants of the resistant indica variety Digu and the susceptible japonica variety TP309 were inoculated with M. oryzae strain Zhong-10-8-14. Total RNAs were isolated from the leaf tissue at 0, 3, 6, 12, 24, 48, and 72 hr, respectively, after inoculation. An equal amount of total RNA was pretreated with RNase-free DNase I (Promega, Madison, WI) and subjected to reverse transcription, using the Reverse Transcription System (Promega). The semiquantitative RT–PCR with the Pid3-specific primers 5′-TACTACTCATGGAAGCTAGTTCTC-3′ and 5′-ACGTCACAAATCATTCGCTC-3′ was performed for 30 cycles. The RT–PCR with Actin-specific primers 5′-CCTCGTCTCGACCTTGCTGGG-3′ and 5′-GAGAACAAGCAGGAGGACGGC-3′ was performed for 25 cycles as a control.
Sequence analysis of Pid3:
The cDNA sequence of Pid3 was obtained through RACE–PCR and RT–PCR analysis. RACE–PCR was carried out using the SMART RACE cDNA Amplification kit (Clontech) according to the manufacturer's instructions. The Pid3-specific primers 5′-CCATCTTGCACATCCTCTTGAGTG-3′ and 5′-ACATCTTATTGGAGAAGCAC-3′ were used in 5′-RACE and 3′-RACE, separately. The primers 5′-GAAGCTAGCAAGCTATGGCGGAG-3′ and 5′-ACGTCACAAATCATTCGCTC-3′ were used in RT–PCR. The 5′- and 3′-RACE ends and RT–PCR products were cloned into the pGEM-T-easy vector (Promega) prior to sequencing.
The coding regions of Pid3 alleles in Digu, 93-11, Zaiyeqing8, LTH, Nipponbare, TP309, and the wild rice A4 were amplified from the respective genomic DNAs by using the primers 5′-AGTAACACCCAAGGATAGGATAG-3′ and 5′-GAACGACAAGTGCGACATGATTG-3′. The PCR products were cloned into the pGEM-T-easy vector (Promega) and confirmed by DNA sequencing.
Analysis of the pseudogenization of the Pid3 alleles:
To examine the Pid3 nonsense mutation locus, we designed two derived cleaved amplified polymorphic sequence (dCAPS) markers. With dCAPS marker Pid3-dCAPS-1, a 178-bp fragment was amplified using the primers 5′-TACTACTCATGGAAGCTAGTTCTC-3′ and 5′-GCAGCACTTCTTGACTACTGTCTGT-3′ and then digested with XbaI. With dCAPS marker Pid3-dCAPS-2, a 178-bp fragment was amplified using the primers 5′-TACTACTCATGGAAGCTAGTTCTC-3′ and 5′-AGCACTTCTTGACTACTGTCTGCCT-3′ and then digested with BamHI.
Vector construction and rice transformation:
The 6235-bp genomic sequence of Pid3 containing the promoter region and the full coding region was amplified from genomic DNA of Digu, using the forward primer 5′-ACCGAATTCCACACATTGTACACCTACGACCAC-3′ (EcoRI recognition site is underlined) and the reverse primer 5′-ACCGTCGACGAACGACAAGTGCGACATGATTG-3′ (SalI recognition site is underlined), and then cloned into the binary vector pCAMBIA1300 through the EcoRI and SalI cloning sites. After sequence verification the final construct was introduced into Agrobacterium tumefaciens LBA4404. The callus of susceptible japonica variety TP309 was transformed according to published methods (Hiei et al. 1994). The resistance of the primary transgenic lines (T0) and of the self progenies (T1) of the T0 lines was challenged by inoculation with the M. oryzae strain Zhong-10-8-14. Segregation of the transgene in T1 plants was confirmed by dCAPS marker Pid3-dCAPS-1; double bands containing 178- and 153-bp fragments can be checked on the agarose gel if the T1 plants contained the transgenic fragment.
RESULTS
Genomewide comparison of the NBS–LRR genes between 93-11 and Nipponbare:
To understand the genomic difference in blast resistance between indica and japonica varieties, we compared the NBS–LRR gene sequences between 93-11 and Nipponbare with a focus on the intact genes in one genome and their allelic pseudogenes in the other. We surveyed both 93-11 and Nipponbare genomes and reannotated all the NBS–LRR genes and their pseudogene counterparts, using a modified homology-based method for human pseudogene annotation (Zhang and Gerstein 2003a; Zhang et al. 2006). First, using an optimized HMM for the rice NBS domain, we identified 653 and 553 gene loci with the NBS domain from the 93-11 and Nipponbare genomes, respectively. Second, we defined a set of 102 seed protein sequences as intact NBS–LRR proteins (see details in materials and methods). The seed sequences were derived from 99 fl-cDNA (Kikuchi et al. 2003) supported genes and 3 cloned NBS–LRR genes (Wang et al. 1999; Bryan et al. 2000; Qu et al. 2006). Finally, we selected the other NBS–LRR genes from the NBS domain loci in either the 93-11 or the Nipponbare genome if they had a continuous homology span on the closest matching seed protein sequence in at least 70% of its length. The homology search in the 93-11 genome resulted in 471 NBS–LRR genes among the 653 NBS domain loci. However, 184 of them were further identified as pseudogenes due to frameshift mutation or nonsense mutation or both. Meanwhile, in the Nipponbare genome, we identified 468 NBS–LRR genes from 553 gene loci, of which 150 were considered to be pseudogenes. The detailed information on this analysis is available in Table S3. All of the annotated NBS–LRR genes and pseudogenes have been submitted to GenBank as third party annotation (accession nos.: annotated Nipponbare NBS–LRR intact genes, FJ771051–FJ771368; annotated Nipponbare NBS–LRR pseudogenes, FJ771369–FJ771518; annotated 93-11 NBS–LRR intact genes, FJ771519–FJ771802; and annotated 93-11 NBS–LRR pseudogenes, FJ771803–FJ771989).
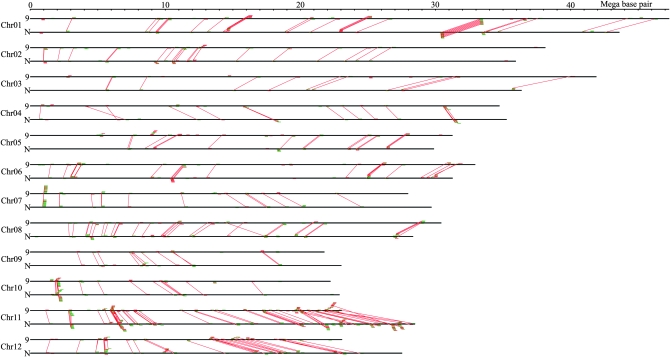
Next, we identified the NBS–LRR gene pairs by the most similarity criterion between the two genomes. Each NBS–LRR gene in one genome was paired only once to the closest homolog in the other. However, because of chromosome DNA rearrangements in some regions where some of the NBS–LRR clustered gene family locates, not every NBS–LRR gene of either Nipponbare or 93-11 has its closest match at the same loci in the other genome. Of the total 337 pairs from the two rice genomes, 301 were considered allelic, while the other 36 were nonallelic due to their different chromosome locations (Figure 1). Further analysis showed that there are 147 allelic pairs that contain at least one pseudogene. These 147 allelic pairs are grouped as follows: (1) both alleles are pseudogenes (53 pairs), (2) only the allele in 93-11 is a pseudogene (59 pairs), and (3) only the allele in Nipponbare is a pseudogene (35 pairs). The positions of the 94 allelic NBS–LRR gene pairs with a pseudogene allele either in 93-11 or in Nipponbare on the chromosomes are shown in Figure S1.
Figure 1.—
Genomewide comparison of NBS–LRR genes between 93-11 and Nipponbare. The horizontal bars represent the physical maps of the chromosomes of 93-11 (marked with “9”) and Nipponbare (marked with “N”). The green boxes represent the intact NBS–LRR genes, and the red boxes represent the NBS–LRR pseudogenes. Genes above and below the chromosomes correspond to forward and reverse orientations, respectively. The linking lines between chromosomes of 93-11 and Nipponbare indicate allelic loci.
We compared our annotated 150 NBS–LRR pseudogenes in Nipponbare with the corresponding genes annotated by TIGR (Yuan et al. 2005). In the corresponding TIGR models, the fragments that contain frameshift or nonsense mutation sites in the 90 pseudogenes we annotated and those in the other 34 pseudogenes were taken as introns and untranslated regions (UTRs), respectively. The pseudogenization ratio of the KOME fl-cDNA NBS–LRR genes (27%, 41/152) is similar to that of the genomewide annotated NBS–LRR genes (32%, 150/468) in Nipponbare. These results also suggest that most NBS–LRR pseudogenes are expressed as intact genes.
Identifying an unknown blast R gene locus using an NBS–LRR pseudogene as a molecular marker:
To analyze the genetic relationship between the pseudogenization of NBS–LRR genes and rice blast resistance, an F2 population was developed from a cross between the blast-resistant indica variety, Digu, and the susceptible japonica variety, TP309. A total of 473 individuals of the F2 population were inoculated with the M. oryzae strain Zhong-10-8-14. This M. oryzae strain was isolated from a japonica-cultivated area in northern China, and it is compatible to the previously cloned blast resistance gene Pid2 from the variety, Digu. In the F2 population, most plants were resistant, but we were able to clearly distinguish 24 susceptible individuals. Thus, the segregation ratio of resistance to susceptibility is 15:1 (resistant:susceptible, 449:24; χ215:1 = 0.04), indicating that Digu may contain two genes that confer resistance to the strain Zhong-10-8-14.
We designed the PCR-based molecular markers specific to the 35 Nipponbare pseudogenes in the above-described 35 allelic NBS–LRR gene pairs in the third group. (See the structures of the allelic gene pairs in Figure S2 and see the PCR primers and restriction enzymes used in these markers in Table S4.) We used these primers to survey the 24 susceptible individuals and the parent varieties Digu and TP309. We found a candidate pseudogene on chromosome 6 that could be detected in all 24 susceptible individuals and TP309 but not in Digu. This suggests that the intact NBS–LRR allele in Digu corresponding to this pseudogene is most likely one of the two functional blast resistance genes against Zhong-10-8-14. We named this blast R gene Pid3, as we found it has a different chromosomal location from the two previously identified Pid(t)1 and Pid2 from Digu (Chen et al. 2004, 2006).
Cloning and functional analysis of Pid3:
We amplified and sequenced the candidate NBS–LRR gene Pid3 and its alleles from the genomes of the rice varieties Digu, 93-11, TP309, and Nipponbare. Both the ORFs of the Digu Pid3 candidate and the allele from 93-11 consist of the same 2772-nucleotide (nt) sequence, encoding a protein with 924 amino acids. However, the alleles in japonica varieties TP309 and Nipponbare contain the same premature stop codon due to a C to T nonsense mutation. This mutation leads to the production of a truncated 737-amino-acid polypeptide with a disrupted LRR region (Figure 2A). The allelic pid3 candidate in Nipponbare corresponds to the gene Loc_Os06g22460 in TIGR's annotation (http://www.jcvi.org/). On the basis of the annotation, Loc_Os06g22460 contains a 135-nt intron that includes the nonsense mutation to avoid the ORF disruption.
Figure 2.—
Sequence analysis of Pid3 protein. (A) Pid3 encodes a 924-amino-acid protein. The susceptible alleles in japonica varieties contain a premature stop codon due to a C to T nonsense mutation. This mutation leads to the production of a truncated polypeptide of 737 amino acids, resulting in a disrupted LRR region. (B) Pid3 protein sequence. The NBS domain from amino acids 158 to 466 is shown in italics. The conserved motifs are underlined. The C-terminal LRR region is composed of 13 imperfect LRR repeats that are shown separately.
Using the DNA or protein sequences of the candidate Pid3 as queries, BLAST searches in GenBank (http://www.ncbi.nih.gov/blast) and KOME rice fl-cDNA databases (http://cdna01.dna.affrc.go.jp/cDNA/) were performed, respectively. No corresponding mRNA or fl-cDNA could be found. A 521-nt EST sequence (GenBank accession no. CK083629) was found to match the 3′ region of the candidate Pid3, indicating that the candidate Pid3 is transcribed. We performed semiquantitative RT–PCR analysis of Pid3 in Digu and TP309 infected with M. oryzae Zhong-10-8-14. The expression analysis indicated that Pid3 was constitutively expressed in both the resistant indica variety Digu and the susceptible japonica variety TP309 (see Figure S3).
Through RT–PCR and RACE–PCR we obtained the mRNA sequence of the candidate Pid3, including a 2772-nt coding sequence, a 63-nt 5′-UTR, and a 135-nt 3′-UTR, with the TIGR-predicted intron around the nonsense mutation also being included in the mature mRNA. The candidate Pid3 encodes a 924-amino-acid polypeptide that contains a conserved NBS domain in positions 158–466 from the translation initiation site. The NBS domain includes four sequence motifs, GMGGIGKTA (positions 202–210), KRYVLVLDDVW (positions 280–290), IGRIILTSRNYDV (positions 307–319), and GLPIAI (positions 373–378), corresponding to kinase 1a (p-loop), kinase 2, kinase 3a, and GLPL motifs, respectively. At the C terminus is the LRR region that comprises 13 imperfect LRR repeats. The MHD motif, MHDILRV (positions 502–508), and the NBS–LRR linker motif, EQNFCIVVNHS (positions 516–526), are present between the NBS domain and the LRR region. At the N terminus, there is a conserved motif, RSLALSIEDVVD (positions 78–89), but no TIR or coiled-coil motif is found (Figure 2B).
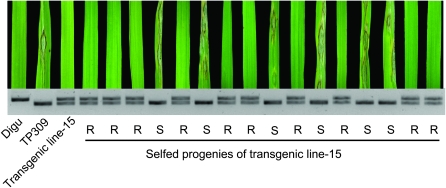
To confirm that Pid3 confers blast resistance to the strain Zhong-8-10-14, we performed a genetic complementation test. Using Digu's genomic DNA as template, a 6235-bp DNA fragment, including the entire 2772-bp coding region, a 3010-bp upstream region, and a 451-bp downstream region, was amplified by high-fidelity PCR. After sequencing verification, this fragment was subcloned into the binary vector pCAMBIA1300, which was then introduced into susceptible variety TP309 by Agrobacterium-mediated transformation. We obtained 21 independent primary transgenic lines (T0), all with resistance to the M. oryzae strain Zhong-10-8-14. Cosegregation of the transgene and blast resistance were confirmed in selfed progenies (T1) of the T0 lines (Figure 3). We then inoculated the transgenic plants with eight indica M. oryzae strains and eight japonica strains collected from China. As shown in Table 1, TP309 shows susceptibility to all 16 strains, whereas Pid3 confers resistance to strains CH43, Sichuan36, JS2001-108, CH706, Zhong-10-8-14, ZK-10-2, 97-3-1, and 99-20-2, of which Pid2 shows susceptibility to Sichuan36 and Zhong-8-10-14. This result suggests that Pid3 confers a race-specific resistance to M. oryzae. Thus, we successfully identified a new blast R gene, Pid3, using the NBS–LRR pseudogene markers. (The sequence of Pid3 has been deposited in GenBank, accession no. FJ745364.)
Figure 3.—
Complementation test with Pid3. The primary transgenic line and the selfed progenies were inoculated by M. oryzae Zhong-10-8-14 and the genotypes were analyzed by a Pid3 dCAPS marker. The resistant variety Digu and the susceptible variety TP309 are shown as controls.
TABLE 1.
Inoculation results of Pid2 and Pid3 transgenic lines with 16 M. oryzae strains
| ZB15 | CH43 | Sichuan36 | JS2001-108 | Sichuan26 | CH706 | CH45 | CH704 | Zhong10-8-14 | ZK-10-2 | 91-65-1 | 97-3-1 | 97-27-2 | 99-20-2 | 99-26-1 | 99-26-2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP309 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Pid3 transgenic line | S | R | R | R | S | R | S | S | R | R | S | R | S | R | S | R |
| Pid2 transgenic line | R | R | S | R | R | R | R | R | S | R | S | R | S | R | S | R |
S, susceptible; R, resistant.
Comparison of the Pid3 nonsense mutation among indica, japonica, and wild rice:
To test if the ORF-disrupted mutation of Pid3 occurs in other rice varieties, we designed specific dCAPS markers that can distinguish the mutation alleles (see the primers and restriction enzymes used in the dCAPS markers in Table S4). In the japonica collection, 29 of 32 varieties contain the same C to T mutation, whereas none of the varieties in the indica collection contains this mutation. This result indicates that Pid3 is replaced by its pseudogene in most tested japonica varieties (Figure S4).
The genus Oryza includes 2 cultivated species and 21 wild species (Khush 1997). O. sativa, Asian cultivated rice, is cultivated all over the world while O. glaberrima, African cultivated rice, is grown on a small scale in western Africa (Khush 1997). On the basis of gene transfer, the 2 cultivated species, O. sativa and O. glaberrima, and 6 wild species, O. rufipogon, O. nivara, O. glumaepatula, O. meridionalis, O. breviligulata, and O. longistaminata, have been grouped into a primary gene pool (Khush 1997). These 8 Oryza species all share the AA genome and the gene transfer among them can be accomplished through conventional hybridization and selection procedures (Khush 1997). Using the above-mentioned dCAPS markers, we examined whether the nonsense mutation in Pid3 occurs in four African rice varieties and a collection of 36 wild rice lines belonging to the 6 AA genome wild species. The accessions and geographic origins of the wild rice and African cultivated rice are listed in Table S2. Notably, we found that none of the tested African cultivated varieties or the tested wild rice lines contains the nonsense mutation. These results suggest that the mutation creating the nonfunctional allele of Pid3 occurred in O. sativa subspecies japonica after its split from subspecies indica.
Next we sequenced the respective allelic Pid3 coding regions of the O. rufipogon wild rice line A4, the indica variety Zhaiyeqing 8 (ZYQ8) that had shown resistance to Zhong-10-8-14 (Zhu et al. 1993), and the japonica variety LTH that had been regarded as an universal blast-susceptible variety lacking all known major blast R genes (Ling et al. 2000; Tsunematsu et al. 2000). Protein sequence comparison revealed a few amino acid changes among the Pid3 proteins of A4, Digu, ZYQ8, 93-11, Nipponbare, TP309, and LTH (see the protein sequence alignment in Figure S5). The sequences of the Pid3 alleles were submitted to GenBank [accession nos. FJ745365 (Pid3-A4), FJ745366 (Pid3-ZYQ8), FJ745367 (Pid3-TP309), FJ745368 (Pid3-LTH), FJ773285 (Pid3-9311), and FJ773286 (Pid3-Nip)].
DISCUSSION
As a major R gene family in plants, NBS–LRR genes have been studied extensively. These investigations, however, have been focused mainly on the functional genes with a complete NBS–LRR coding sequence. Genomewide analyses of NBS–LRR genes in Arabidopsis (Columbia) and rice (Nipponbare) have revealed that a large number of NBS–LRR genes encoded in these two genomes are pseudogenes due to frameshift mutations or premature stop codons (Meyers et al. 2003; Monosi et al. 2004). At least 12 of 149 (∼8%) NBS–LRR genes in Arabidopsis (Columbia) (Meyers et al. 2003) and >100 of the ∼500 (20%) NBS–LRR genes in rice (Nipponbare) (Monosi et al. 2004) were predicted as pseudogenes. In these previous studies, manual reannotation was used to verify the results obtained from automated annotation programs. However, pseudogenes are often misincorporated into gene collections due to their high level of sequence homology to their real gene counterparts (Nelson 2004). It has been demonstrated that either a frameshift mutation or a nonsense mutation in an internal exon may force gene-prediction programs to adjust their splicing patterns and thus transform the pseudogenes into variable gene models (Harrison et al. 2002; Zhang and Gerstein 2004; Zhang et al. 2004, 2006; Zhu et al. 2006). In our study, we used a homology-based method derived from a pipeline that has been successfully used in human pseudogene annotation (Zhang and Gerstein 2003b; Zhang et al. 2006) to reannotate the complete sets of NBS–LRR genes and pseudogenes in 93-11 and Nipponbare genomes. According to our annotation, >30% of the NBS–LRR genes are pseudogenes in either the 93-11 or the Nipponbare genomes, greater than the 20% that was reported previously (Monosi et al. 2004).
The number of annotated NBS–LRR genes in 93-11 (471) is similar to that in Nipponbare (468). However, the number of NBS-domain-containing loci in 93-11 (653) is more than that in Nipponbare (553). The higher number of NBS-domain-containing loci in 93-11 could be artificiality inflated from the “whole-genome shotgun” strategy applied to the 93-11 genome sequencing. Sequence assembly errors in this method might put fragments from a single locus into multiple loci. Although much effort has been made to obtain a high-quality genome assembly during sequencing of 93-11 and Nipponbare, sporadic mistakes in the genome databases are inevitable and would result in misannotated genes and overestimation of the number of pseudogenes during our analysis using these databases.
Thus far, >70 blast R-gene loci have been identified and mapped on rice chromosomes (Ballini et al. 2008). Some of the genes have been successfully used in blast resistance breeding. When a map-based cloning strategy is used in the identification of R genes, resistance gene analog (RGA) markers that are designed according to the conserved NBS domain could be used to identify the corresponding candidate NBS–LRR resistance gene (Mago et al. 1999; Chauhan et al. 2002; Zhuang et al. 2002; Lee et al. 2003; Sallaud et al. 2003; Chen et al. 2004). In practice, however, successful examples of isolation of an NBS–LRR resistance gene with RGA markers have rarely been reported. The ineffectiveness of this method is possibly due to the complex nature of NBS–LRR genes in the genome. In contrast to the use of RGA markers, we designed molecular markers on the basis of the pseudogenization of NBS–LRR genes. We analyzed the relationship between the NBS–LRR pseudogenes and blast susceptibility in an appropriate population derived from resistant and susceptible varieties. In our method, when cosegregation of a tested pseudogene with the susceptible phenotype is observed, the intact allele can be regarded as the candidate blast R gene. This strategy has two advantages. First, only a small number of susceptible individual plants need to be analyzed with the landing markers. Second, scoring susceptible plants is more precise than scoring resistant plants. In this study, we demonstrated the effectiveness of this strategy by identifying a previously unknown R gene, Pid3, from the blast-resistant variety, Digu, in contrast to our labor-intensive cloning of the Pid2 gene from the same variety (Chen et al. 2006). In fact, we also tried to identify the blast R gene other than Pid3 revealed in the genetic analysis of the F2 population by using the same strategy, but we could not find any other pseudogene marker associated with the susceptible individuals. This implies that not all of the susceptible alleles of blast R genes are pseudogenes. For example, one amino acid change resulted in the loss of resistance of Pita (Bryan et al. 2000) and Pi36 (Liu et al. 2007).
Acknowledgments
We thank Li Heng and Zheng Hongkun (Beijing Institute of Genomics) for their helpful suggestions and discussions. We thank Laura Bartley and Dante F. Placido for their careful revision of the manuscript. This research was supported by grants from the National High Technology Research and Development Program of China (863 Program) (2006AA10A101, 2006AA10Z192, and 20060110Z1142), the National Key Basic Research Science Foundation of China (2006CB101904), the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-N-005), and the National Science Foundation of China (30640095 and 30550005).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102871/DC1.
Sequence data from this article have been deposited with the GenBank Data Library under accession no. FJ745364.
References
- Abramovitch, R. B., J. C. Anderson and G. B. Martin, 2006. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell. Biol. 7 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa, I., N. Hayashi, H. Yamane, H. Kanamori, J. Wu et al., 2008. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer pikm-specific rice blast resistance. Genetics 180 2267–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, J., L. A. Pennill, J. Ning, S. W. Lee, J. Ramalingam et al., 2002. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 12 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini, E., J. B. Morel, G. Droc, A. Price, B. Courtois et al., 2008. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 21 859–868. [DOI] [PubMed] [Google Scholar]
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller et al., 2002. The Pfam protein families database. Nucleic Acids Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., M. Clamp and R. Durbin, 2004. Genewise and genomewise. Genome Res. 14 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, G. T., K. S. Wu, L. Farrall, Y. Jia, H. P. Hershey et al., 2000. tA single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, R. S., M. L. Farman, H. B. Zhang and S. A. Leong, 2002. Genetic and physical mapping of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene AVR1–CO39 of Magnaporthe grisea. Mol. Genet. Genomics 267 603–612. [DOI] [PubMed] [Google Scholar]
- Chen, X., J. Shang, D. Chen, C. Lei, Y. Zou et al., 2006. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46 794–804. [DOI] [PubMed] [Google Scholar]
- Chen, X. W., S. G. Li, J. C. Xu, W. X. Zhai, Z. Z. Ling et al., 2004. Identification of two blast resistance genes in a rice variety, Digu. J. Phytopathol. 152 77–85. [Google Scholar]
- Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson et al., 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Eddy, S. R., 1998. Profile hidden Markov models. Bioinformatics 14 755–763. [DOI] [PubMed] [Google Scholar]
- Faigon-Soverna, A., F. G. Harmon, L. Storani, E. Karayekov, R. J. Staneloni et al., 2006. A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. Plant Cell 18 2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H., 1956. The complementary genic systems in flax and flax rust. Adv. Genet. 8 29–54. [Google Scholar]
- Garris, A. J., T. H. Tai, J. Coburn, S. Kresovich and S. McCouch, 2005. Genetic structure and diversity in Oryza sativa L. Genetics 169 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., D. Ricke, T. H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100. [DOI] [PubMed] [Google Scholar]
- Harrison, P. M., H. Hegyi, S. Balasubramanian, N. M. Luscombe, P. Bertone et al., 2002. Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 12 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., S. Ohta, T. Komari and T. Kumashiro, 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Imbe, T., S. Oba, M. J. T. Yanoria and H. Tsunematsu, 1997. A new gene for blast resistance in rice cultivar, IR24. Rice Genet. Newsl. 14 60–62. [Google Scholar]
- IRGSP, 2005. The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Khush, G. S., 1997. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35 25–34. [PubMed] [Google Scholar]
- Kikuchi, S., K. Satoh, T. Nagata, N. Kawagashira, K. Doi et al., 2003. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379. [DOI] [PubMed] [Google Scholar]
- Kiyosawa, S., 1984. Establishment of differential varieties for pathogenicity test of rice blast fungus. Rice Genet. Newsl. 1 95–97. [Google Scholar]
- Lee, S. Y., J. S. Seo, M. Rodriguez-Lanetty and D. H. Lee, 2003. Comparative analysis of superfamilies of NBS-encoding disease resistance gene analogs in cultivated and wild apple species. Mol. Genet. Genomics 269 101–108. [DOI] [PubMed] [Google Scholar]
- Lee, S. K., M. Y. Song, Y. S. Seo, H. K. Kim, S. Ko et al., 2009. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two CC-NB-LRR Genes. Genetics 181 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F., S. Chen, Z. Que, L. Wang, X. Liu et al., 2007. The blast resistance gene Pi37 encodes an NBS-LRR protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 177 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, Z., 1984. Blast resistance classification of some rice varieties in China (in Chinese). Scientia Agricultura Sinica 2 19–28. [Google Scholar]
- Ling, Z., T. Mew, J. Wang, C. Lei and N. Huang, 2000. Development of Chinese near isogenic lines of rice and their differential ability to pathogenic races of Pyricularia grisea (in Chinese). Scientia Agricultura Sinica 33 1–8. [Google Scholar]
- Liu, X., F. Lin, L. Wang and Q. Pan, 2007. The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 176 2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackill, D. J., and J. M. Bonman, 1992. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82 746–749. [Google Scholar]
- Mackill, D. J., J. M. Bonman, H. S. Suh and R. Srilingam, 1985. Genes for resistance to Philippine isolates of the rice blast pathogen. Rice Genet. Newsl. 2 80–81. [Google Scholar]
- Mago, R., S. Nair and M. Mohan, 1999. Resistance gene analogues from rice: cloning, sequencing and mapping. Theor. Appl. Genet. 99 50–57. [Google Scholar]
- Martin, G. B., A. J. Bogdanove and G. Sessa, 2003. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54 23–61. [DOI] [PubMed] [Google Scholar]
- Meyers, B. C., A. Kozik, A. Griego, H. Kuang and R. W. Michelmore, 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosi, B., R. J. Wisser, L. Pennill and S. H. Hulbert, 2004. Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 109 1434–1447. [DOI] [PubMed] [Google Scholar]
- Nelson, D. R., 2004. ‘Frankenstein genes’, or the Mad Magazine version of the human pseudogenome. Hum. Genomics 1 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., T. Eulgem, B. F. Holt, 3rd and J. L. Dangl, 2003. Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Normile, D., 1997. Archaeology: Yangtze seen as earliest rice site. Science 275 309–310. [Google Scholar]
- Pan, Q., L. Wang, H. Ikehashi and T. Tanisaka, 1996. Identification of a new blast resistance gene in the indica rice cultivar kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 86 1071–1075. [Google Scholar]
- Qu, S., G. Liu, B. Zhou, M. Bellizzi, L. Zeng et al., 2006. The broad-spectrum blast resistance gene pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud, C., M. Lorieux, E. Roumen, D. Tharreau, R. Berruyer et al., 2003. Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Theor. Appl. Genet. 106 794–803. [DOI] [PubMed] [Google Scholar]
- Swofford, D., 2000. PAUP*: Phylogenetic Analysis Using Parsimony. Sinauer Associates, Sunderland, MA.
- Tsunematsu, H., M. J. T. Yanoria, L. A. Ebron, N. Hayashi, I. Ando et al., 2000. Development of monogenic lines of rice for blast resistance. Breed. Sci. 50 229–234. [Google Scholar]
- Wang, Z. X., M. Yano, U. Yamanouchi, M. Iwamoto, L. Monna et al., 1999. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 19 55–64. [DOI] [PubMed] [Google Scholar]
- Yang, S., Z. Feng, X. Zhang, K. Jiang, X. Jin et al., 2006. Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol. Biol. 62 181–193. [DOI] [PubMed] [Google Scholar]
- Yu, J., S. Hu, J. Wang, G. K. Wong, S. Li et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Yu, J., J. Wang, W. Lin, S. Li, H. Li et al., 2005. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z. H., D. J. Mackill and J. M. Bonman, 1987. Inheritance of resistance to blast in some traditional and improved rice cultivars. Phytopathology 77 323–326. [Google Scholar]
- Yuan, Q., S. Ouyang, A. Wang, W. Zhu, R. Maiti et al., 2005. The institute for genomic research Osa1 rice genome annotation database. Plant Physiol. 138 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and M. Gerstein, 2003. a The human genome has 49 cytochrome c pseudogenes, including a relic of a primordial gene that still functions in mouse. Gene 312 61–72. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and M. Gerstein, 2003. b Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 31 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and M. Gerstein, 2004. Large-scale analysis of pseudogenes in the human genome. Curr. Opin. Genet. Dev. 14 328–335. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., N. Carriero and M. Gerstein, 2004. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 20 62–67. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., N. Carriero, D. Zheng, J. Karro, P. M. Harrison et al., 2006. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 22 1437–1439. [DOI] [PubMed] [Google Scholar]
- Zhou, T., Y. Wang, J. Q. Chen, H. Araki, Z. Jing et al., 2004. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271 402–415. [DOI] [PubMed] [Google Scholar]
- Zhou, B., S. Qu, G. Liu, M. Dolan, H. Sakai et al., 2006. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 19 1216–1228. [DOI] [PubMed] [Google Scholar]
- Zhu, L., Y. Chen, Y. Xu, J. Xu, H. Cai et al., 1993. Construction of a molecular map of rice and gene mapping using a double haploid population of a cross between Indica and Japonica varieties. Rice Genet. Newsl. 10 132–135. [Google Scholar]
- Zhu, X., M. Gerstein and M. Snyder, 2006. ProCAT: a data analysis approach for protein microarrays. Genome Biol. 7 R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, J.-Y., W.-B. Ma, J.-L. Wu, R.-Y. Chai, J. Lu et al., 2002. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica 128 363–370. [Google Scholar]
- Zong, Y., Z. Chen, J. B. Innes, C. Chen, Z. Wang et al., 2007. Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449 459–462. [DOI] [PubMed] [Google Scholar]