Abstract
The interplay of balancing selection within a species and rapid gene evolution between species can confound our ability to determine the functional equivalence of interspecific and intergeneric pairs of alleles underlying reproduction. In crucifer plants, mating specificity in the barrier to self-fertilization called self-incompatibility (SI) is controlled by allele-specific interactions between two highly polymorphic and co-evolving proteins, the S-locus receptor kinase (SRK) and its S-locus cysteine rich (SCR) ligand. These proteins have diversified both within and between species such that it is often difficult to determine from sequence information alone if they encode the same or different SI specificity. The self-fertile Arabidopsis thaliana was derived from an obligate outbreeding ancestor by loss of self-incompatibility, often in conjunction with inactivation of SRK or SCR. Nevertheless, some accessions of A. thaliana can express self-incompatibility upon transformation with an SRK–SCR gene pair isolated from its self-incompatible close relative A. lyrata. Here we show that several additional and highly diverged SRK/SCR genes from A. lyrata and another crucifer plant, Capsella grandiflora, confer self-incompatibility in A. thaliana, either as intact genes isolated from genomic libraries or after manipulation to generate chimeric fusions. We describe how the use of this newly developed chimeric protein strategy has allowed us to test the functional equivalence of SRK/SCR gene pairs from different taxa and to assay the functionality of endogenous A. thaliana SRK and SCR sequences.
MATING reactions in plants, fungi, and animals are strongly influenced by molecular recognition machineries that act as gauges of genetic relatedness (Brown and Casselton 2001; Nasrallah 2005; Yamazaki and Beauchamp 2007). Many plants with hermaphroditic flowers have evolved inbreeding avoidance mechanisms, known as self-incompatibility (SI) systems. These systems are based on the ability of the female reproductive apparatus (the pistil) to discriminate among genetically distinct pollen grains, resulting in the failure of self-pollination despite functional female and male reproductive structures. In the Brassicaceae (crucifers), specific recognition of pollen by the epidermal cells of the stigma (a structure located at the tip of the pistil) is controlled by haplotypes of the S locus, and activation of the SI response leading to inhibition of pollen tube growth occurs if pollen and stigma are derived from plants that express the same S-locus haplotype (S haplotype). Within self-incompatible crucifer species, the number of S haplotypes and corresponding SI specificities is usually high, with >50 reported in some species (Watanabe et al. 2000), and SI dictates that self-incompatible plants are typically heterozygous and carry two S haplotypes. Each S haplotype is composed of two highly polymorphic genes that are the determinants of SI specificity in stigma and pollen (Stein et al. 1991; Schopfer et al. 1999). The S-locus receptor kinase (SRK) gene encodes a single-pass transmembrane serine/threonine kinase localized on the surface of stigma epidermal cells, and the S-locus cysteine-rich protein (SCR) gene encodes a small peptide localized in the pollen coat. SCR is the ligand for SRK and will bind to the extracellular domain of SRK (hereafter eSRK) only if both proteins are encoded by the same S-locus haplotype (Kachroo et al. 2001; Takayama et al. 2001; Chookajorn et al. 2004). The binding of SCR to its cognate eSRK triggers an intracellular phosphorylation cascade that results in pollen rejection by a poorly understood mechanism.
A mechanistic understanding of the recognition phase of SI requires detailed structure–function analyses of SRK and SCR aimed at identifying the amino acid residues that determine their allele-specific interaction and explaining the puzzling dominance/recessive interactions exhibited by different SRK alleles in the heterozygous stigmas of self-incompatible plants (Hatakeyama et al. 2001; Mable et al. 2003; Prigoda et al. 2005). Such structure–function studies require an experimental system that allows efficient in vivo functional analysis of large numbers of SRK and SCR sequence variants generated in vitro by site-directed mutagenesis or domain swapping between proteins that determine different SI specificities. The recent transfer of the SI trait into Arabidopsis thaliana has established this species as a model organism for mechanistic and evolutionary studies of mating systems in crucifers (Nasrallah et al. 2002, 2004). However, to date, only one SI specificity, that which is determined by the Sb haplotype of A. lyrata, has been successfully introduced into A. thaliana and shown to alter the plant's mating reaction from strict autogamy to full SI. To exploit fully the A. thaliana transgenic SI model, additional S haplotypes must be introduced into this species. In addition to facilitating mechanistic studies of the SRK–SCR interaction and dominance relationships, the expression of multiple SI specificities in A. thaliana promises to shed light on processes underlying the diversification of SRK and SCR genes. For example, expression in A. thaliana of SI specificities derived from different crucifer species will allow direct assays of the functional equivalence or nonequivalence of the corresponding S haplotypes, an issue that is difficult to resolve on the basis of sequence information alone.
Although conceptually simple, expressing different SI specificities by transformation with different SRK/SCR gene pairs is not a straightforward proposition. Difficulties stem largely from the availability of appropriate cloned SRK/SCR variants for use in transformation experiments. A large number of SRK/SCR gene pairs are available from Brassica species as a result of extensive and long-standing studies of SI. However, attempts to restore SI in transgenic A. thaliana using Brassica S-locus genes had met with failure (Bi et al. 2000; J. B. Nasrallah, unpublished data), possibly because of the inability of Brassica SRKs to interact productively with A. thaliana components of the SI signal transduction pathway. In the past few years, studies of SI were initiated in self-incompatible species more closely related to A. thaliana, such as A. lyrata, A. halleri, and Capsella grandiflora. However, with a few exceptions, these studies produced only partial SRK and SCR sequences amplified from genomic DNA (Schierup et al. 2001; Prigoda et al. 2005; Bechsgaard et al. 2006; Paetsch et al. 2006). The challenging task of cloning the very highly polymorphic SCR sequences and complete SRK and SCR genes, which requires genomic library construction and in many cases chromosome walking, has only been accomplished for two S haplotypes of A. lyrata, Sb (hereafter AlSb, which was used in previous transformation studies (Nasrallah et al. 2002, 2004), and Sa (AlSa; Kusaba et al. 2001), and for the S7 haplotype of C. grandiflora (CgS7; Nasrallah et al. 2007).
In this article, we report the isolation of two new SRK/SCR gene pairs from genomic libraries of A. lyrata and expression of the corresponding SI specificities in A. thaliana. We also describe a novel strategy for rapid and efficient transfer of several distinct SI specificities into A. thaliana, which only requires knowledge of the eSRK sequence and SCR second-exon sequences that encode the mature SCR protein.
MATERIALS AND METHODS
Our strategy to express different SI specificities in A. thaliana was to transform A. thaliana plants with two types of constructs: (1) intact SRK and SCR gene pairs, either previously described or newly isolated from genomic libraries; and (2) chimeric SRK and SCR genes, which circumvent the laborious task of constructing genomic libraries for isolating SRK and SCR alleles from different S haplotypes.
S haplotypes used in this study:
Several functional S haplotypes were used as sources of SRK and SCR genes. These include the C. grandiflora S7 haplotype and five functionally distinct S haplotypes from A. lyrata: the Sa and Sb haplotypes described in Kusaba et al. 2001 [also named S20 and S13, respectively, by Charlesworth et al. (2003)]; and the S6 (AlS6), S16 (AlS16), S25 (AlS25), and S37 (AlS37) haplotypes (Charlesworth et al. 2003; Bechsgaard et al. 2006). Additionally, we used the nonfunctional A. thaliana Cape Verde Islands (Cvi-0) pseudo-SB (ΨSB) haplotype, which is the likely ortholog of the AlS16 haplotype of A. lyrata (Bechsgaard et al. 2006).
Isolation of SRK and SCR genes from genomic libraries and construction of plant transformation vectors containing these genes:
Genomic libraries were constructed in λDASH II (Stratagene; La Jolla, CA) from DNA isolated from A. lyrata plants containing the S6 or S25 haplotypes. The libraries were screened with a a single-copy probe derived from sequences 5′ of At4g21380 (ARK3) (Kusaba et al. 2001), a gene located at one flank of the A. lyrata S locus, often very close to SRK. Chromosome walking was then performed until an SCR-like sequence containing eight cysteines was identified. The SRK and SCR genes were sequenced at the Cornell University Life Sciences Core Laboratories Center (Ithaca, NY).
To construct plant transformation plasmids, the AlSRK6 and AlSCR6 were inserted together into the pCAMBIA1300 vector. In the case of AlSRK25 and AlSCR25, fragments containing these genes were cloned separately into the pCAMBIA1300 vector, and introduced individually into A. thaliana, because repeated attempts to construct a plasmid containing both genes were unsuccessful. The C. grandiflora CgSRK7 and CgSCR7 genes had previously been isolated from a genomic library on one λ-clone containing an ∼13-kb insert (Nasrallah et al. 2007), and this insert was excised and cloned into the plant transformation vector pART27 (Gleave 1992).
Isolation of the complete SCR16 sequence from the A. lyrata S16 haplotype:
Previous analysis of the A. lyrata S16 and S37 haplotypes had identified SRK16, SRK37, and SCR37 sequences (Bechsgaard et al. 2006; Boggs et al. 2009), but not the SCR16 exon-2 sequence. To isolate the complete SCR16 sequence, we devised a polymerase chain reaction (PCR)-based strategy using genomic S16 DNA as follows.
AlSRK16 shares 92% amino acid sequence similarity with its ortholog, the A. thaliana Cvi-0 ΨSRKB allele. Furthermore, the ΨSRKB and ΨSCRB genes in Cvi-0 are oriented in a convergent manner and separated by very little intervening sequence (Shimizu et al. 2008). On the basis of these results and on the assumption that the AlS16 haplotype would have the same organization as the Cvi-0 ΨSB haplotype, we designed PCR primers on the basis of the ΨSB locus and used these primers to amplify a fragment starting from the second exon of AlSCR16, into the 3′ untranslated region of AlSCR16 and to the 3′ end of AlSRK16. Amplified fragments containing AlSCR16 sequences were cloned into pGemT-easy (Promega; Madison, WI), and inserts were sequenced.
Analysis of eSRK and SCR sequences:
The sequences of the following genes (with accession numbers) were reported previously (Kusaba et al. 2001; Bechsgaard et al. 2006; Nasrallah et al. 2007): AlSRKa (AB052755) and AlSCRa (AB052753); AlSRKb (AB052756) and AlSCRb (AB052754); CgSRK7 (EF530735) and CgSCR7 (EF530736); AlSRK16 (DQ520283); and AlSRK37 (DQ520289) and AlSCR37 (FJ752546). Sequences derived in this study have been deposited in GenBank under the following accession nos.: AlSRK6: GQ351354; AlSCR6: GQ351356; AlSRK25: GQ351355; AlSCR25: GQ351357; and AlSCR16: GQ351353. Amino acid alignments of the SRK extracellular domains and the mature SCR variants were performed using the sequence distance and tree function of MegAlign, a program in the Lasergene suite of applications (DNASTAR, Madison, WI; supporting information, Figure S1).
Construction of SRK and SCR chimeric genes:
SRK chimeric genes:
The SRK transcriptional unit consists of seven exons, the first of which encodes the signal peptide and the entire extracellular domain of SRK. Chimeric SRK genes were assembled in a pCAMBIA1300 derivative containing the AtS1 promoter, which drives expression specifically in the stigma epidermis (Dwyer et al. 1992), fused to the AlSRKb transcriptional unit from its initiating methionine codon and including the six introns and 3′-UTR of the gene. To generate this derivative plasmid,the major part of AlSRKb exon 1 was amplified from the start codon (using a primer that incorporated a KpnI restriction site) to the endogenous SacI site located 1230 bp after the start codon and cloned as a KpnI–SacI fragment into pCAtS1pr. The rest of the AlSRKb gene including introns and 3′-UTR, isolated as a SacI–XbaI fragment from a genomic clone containing AlSRKb, was then inserted downstream of AleSRKb exon-1 sequences. The resulting AtS1∷AlSRKb cassette was cloned into pCAMBIA1300 as an EcoRI–XbaI fragment. The resulting plasmid was subsequently used as a backbone for replacement of the eSRKb-containing KpnI–SacI fragment with eSRK sequences amplified from functional A. lyrata and C. grandiflora SRK alleles and from the A. thaliana ΨSB haplotype using specific forward and reverse primers that incorporated KpnI and SacI restriction sites, respectively.
Construction of the AlSRKa∷AlSRKb and CgSRK7∷AlSRKb chimeric genes was achieved by amplification of eSRK and SCR sequences from available genomic clones. In the case of AlSRK16 and AlSRK37, for which genomic clones are not available, SRK chimeras were constructed using eSRK sequences amplified from genomic DNA of A. lyrata plants harboring each of these S haplotypes.
SCR chimeric genes:
SCR genes consist of two exons, the first encoding the signal peptide and the second encoding the mature SCR protein. While the A. lyrata SCRa gene could be excised from a previously isolated Sa BAC clone (Kusaba et al. 2001) and cloned into the plant transformation vector pBIN-PLUS (van Engelen et al. 1995), constructs for expression of other SCR alleles corresponding to the SRKs described in the previous section, namely A. lyrata SCR16, SCR37, and SCR6, and C. grandiflora SCR7, were prepared as follows. An SCR expression cassette was constructed in pCAMBIA1300 using the promoter of the Brassica rapa SCR8 gene (Schopfer et al. 1999) and the octopine synthase (OCS) terminator, between which SCR coding regions consisting of the AlSCRb signal peptide fused to SCR exon-2 sequences and generated by recombinant PCR were inserted.
Two additional SCR expression constructs based on the ΨSCRB allele of the A. thaliana Cvi-0 accession were also prepared using the strategy described above: one construct was designed to express the SCRB protein predicted from the ΨSCRB sequence, and another construct was designed to express a modified version of SCRB in which an extra cysteine residue located at position 10 of the mature peptide was replaced by the phenylalanine codon found in the corresponding site of the orthologous SCR16 gene (see alignments in supporting information). Alignments of the predicted amino acid sequences of the SCR fusion proteins are shown in Figure S2.
Plant transformation and analysis of transgenic plants:
SRK and SCR constructs were transformed into Agrobacterium strain GV3101 (Koncz and Schell 1986) and subsequently used to transform A. thaliana C24 plants, which express robust and developmentally stable SI upon transformation with SRKb–SCRb (Nasrallah et al. 2004), or Col-0 plants in the case of the AlSCRa construct, by the floral dip method (Zhang et al. 2006). The independent derivation of primary (T1) transformants and number of T-DNA integration were assessed by DNA gel blot analysis using probes derived either from the hygromycin- or kanamycin-resistance gene. Transgene expression was assessed in CgS7 transformants by RT–PCR (reverse transcription–polymerase chain reaction) of total RNA isolated from stigmas as described previously (Nasrallah et al. 2002).
For each SRK and SCR construct, pollination responses were tested in an average of 12 plants per construct. Additionally, to confirm that the various SRK/SCR variants used in this study encode distinct SI specificities in transgenic A. thaliana, reciprocal cross-pollinations among the various SRK and SCR T2 plants transformants were performed in all combinations. In all cases, two sets of control pollinations were also performed: in one set, stigmas expressing each of the SRK chimeras were pollinated with pollen from untransformed A. thaliana plants, and in another set, pollen expressing each of the SCR chimeras was used to pollinate stigmas of untransformed A. thaliana plants. For all chimeric SRK- and SCR-expressing transformants, these control pollinations produced the expected large number of pollen tubes.
All of these pollination assays used pollen-free stigmas collected from buds just before anthesis, when the stigmas were receptive to pollen but before the pollen grains matured and were released from the anthers. Stigmas were manually pollinated, incubated for 2 hr, and subsequently stained with aniline blue and processed for observation by epifluorescence microscopy, as previously described (Kho and Baer 1968). Under these conditions, a strong incompatibility response is manifested by the absence or near-absence of pollen tubes (<5 tubes per pollinated stigma). A compatible response is evident by the growth of numerous pollen tubes (>50 pollen tubes per pollinated stigma).
Pollination assays were typically performed on at least five buds and repeated on at least two separate dates. For any construct that did not confer an incompatibility response, extensive pollination assays were performed on multiple stigmas with repetitions on separate dates. For example, for the CgS7 construct, seven independent transformants were assessed by pollination assays at least four times on 2 separate days, and four of those lines were analyzed throughout stigma development by pollination assays in floral buds ranging from the immature stage to full maturity. Independent SRK- and SCR-expressing lines were tested in 12 independent T1 plants on average and in the T2 generation (except for plants expressing AlSRKb, which were thoroughly tested in previous studies).
RESULTS
Isolation of novel A. lyrata SRK/SCR gene pairs that confer an incompatibility response in A. thaliana:
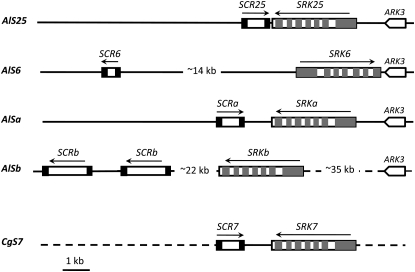
Two SRK/SCR gene pairs (see sequence alignments in Figure S1) were isolated from genomic libraries constructed from plants harboring the A. lyrata S6 and S25 haplotypes. Figure 1 shows the organization of the SRK and SCR genes in the AlS6 and AlS25 haplotypes in comparison to the structure of the previously reported AlSb, AlSa, and CgS7 haplotypes (Kusaba et al. 2001; Nasrallah et al. 2007). These comparisons highlight the extensive intraspecific structural heteromorphisms exhibited by S haplotypes, which are thought to contribute to reduced recombination in the region and maintenance of the tight genetic linkage of SRK and SCR (Boyes et al. 1997; Casselman et al. 2000; Nasrallah 2000). Indeed, the most structurally similar S haplotypes we analyzed are the A. lyrata AlSa and C. grandiflora CgS7 haplotypes (Figure 1). In contrast, the A. lyrata S haplotypes differ from one another in gene content and organization. AlSb, but not the other A. lyrata S haplotypes, contains duplicated copies of SCR (Shimizu et al. 2004). Furthermore, the haplotypes differ drastically in the distances separating SRK from SCR and from markers that flank the S locus (in this case, the ARK3 gene), as well as in the relative orientations of these genes. In particular, SRK can be separated from ARK3 by as little as a few hundred base pairs as in AlS6, or by as much as 35 kb as in AlSb. Similarly, the interval between SRK and SCR varies from <1 kb in AlS25 to ∼22 kb in AlSb.
Figure 1.—
Structure of the S haplotypes used in this study. The diagrams show the organization of the SRK and SCR genes in the A. lyrata AlS25, AlS6, AlSa, and AlSb haplotypes and the C. grandiflora CgS7 haplotype. The structures of the AlSa and AlSb haplotypes were determined in Kusaba et al. (2001) and that of the CgS7 in (Nasrallah et al. 2007). Note the drastic differences in the arrangements of the genes relative to each other and to the flanking gene ARK3. Intergenic distances that exceed 1 kb are indicated. The dashed lines in AlSb between ARK3 and SRKb and between SRKb and SCRb indicate that the distances are not drawn to scale. The dashed lines flanking SRK7 and SCR7 in the CgS7 haplotype indicate that these sequences have not been cloned. The ARK3 gene is not drawn to scale.
To determine if the newly isolated AlSRK and AlSCR genes, as well as the previously described C. grandiflora CgSRK7 and CgSCR7 genes, could confer an incompatibility response in A. thaliana, restriction fragments containing the SRK and SCR alleles were introduced into A. thaliana C24 plants. The AlS6- and CgS7-derived genes were introduced together on one plant transformation plasmid, but AlSRK25 and AlSCR25 were introduced individually on separate plasmids because plasmids containing both genes were unstable in bacteria (see materials and methods). Pollination assays of a minimum of 10 independent transformants per construct were performed by self-pollinating AlSRK6-AlSCR6 and CgSRK7-CgSCR7 transformants, or by pollinating the stigmas of AlSRK25 transformants with pollen from AlSCR25 transformants. Both the AlS6- and AlS25-derived genes conferred incompatibility in transgenic A. thaliana. Inhibition of self pollen was observed in 6 of 12 AlSRK6-AlSCR6 independent transformants analyzed, and these plants produced a negligible amount of seed (typically 0–5 seeds per plant). Similarly, the stigmas of 10 of 11 independent AlSRK25 transformants inhibited pollen from 7 of 10 independent AlSCR25 transformants. In contrast, the CgSRK7-CgSCR7 construct failed to confer SI: all 40 independent transformants analyzed produced wild-type levels of seed set, and self-pollination assays of seven of these plants failed to show inhibition of pollen tube development at any stage of stigma development.
Introduction of SI specificities into A. thaliana by expressing chimeric SRK and SCR genes:
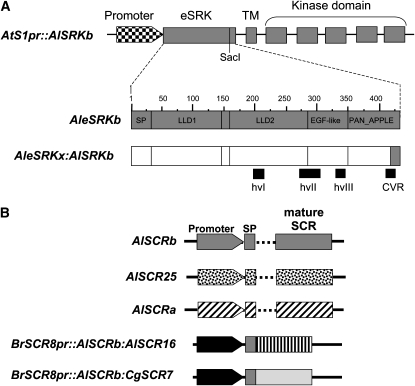
We used a chimeric SRK gene strategy designed to express, under control of the stigma-specific AtS1 promoter (Dwyer et al. 1992), a fusion protein, designated eSRKx:AlSRKb, in which a particular eSRK (minus the last 23 amino acids) is fused to the last 23 amino acids of AleSRKb followed by the AlSRKb transmembrane and kinase domains (Figure 2A). In parallel, and in cases where SCR genomic clones were not available, we used chimeric intronless SCR genes, designed to express, under control of a B. rapa SCR8 promoter (Schopfer et al. 1999), a mature SCR variant fused to the AlSCRb signal peptide (Figure 2B).
Figure 2.—
Diagrams of the chimeric SRK genes and the SCR constructs used in this study. (A) SRK chimeric genes. At top is a schematic of the AtS1pr∷AlSRKb backbone used for construction of SRK chimeric fusions, showing the AtS1 promoter (checkered arrowhead) driving the AlSRKb transcriptional unit with its seven exons, which encode the AlSRKb extracellular domain (eSRK; exon 1), the transmembrane domain (TM; exon 2), and the kinase domain (exons 3–7), followed by its native 3′ untranslated sequences (not shown). The unique SacI restriction site used for construction of chimeras is shown toward the 3′ end of the eSRK. Below are shown the structures of the AleSRKb (with numbers indicating amino acids) and of the eSRK of AleSRKbx:AlSRKb fusions. The vertical lines within the eSRKs delineate predicted structural subdomains in the eSRK (Naithani et al. 2007): SP, signal peptide; LLD1 and LLD2, lectin-like domains 1 and 2; EGF-like, epidermal growth factor-like domain; and PAN_APPLE domain. The location of hypervariable regions discussed in previous studies (Kusaba et al. 2001; Naithani et al. 2007) are indicated below the diagrams and correspond to the following regions in AlSRKb: 204–219 (hvI), 269–304 (hvII), 326–340 (hvIII), and 410–422 (C-terminal variable region/CVR). The unique SacI site corresponds to position 410 in the amino acid sequence, and the region C terminal to this position (shaded and spanning residues 411–434) was derived from AlSRKb in all AleSRKx:AlSRKb fusions. (B) SCR constructs. The AlSCRb, AlSCR25, and AlSCRa constructs were derived from λ-genomic fragments and contain the native SCR promoter, the two exons separated by an intron of variable length (dashed lines), and terminator. The intronless BrSCR8∷AlSCRb:AlSCR16 and BrSCR8∷AlSCRb:AlSCR7 constructs contain the B. rapa SCR8 promoter (Schopfer et al. 1999) followed by the signal peptide sequence of the AlSCRb gene (shaded box) fused to exon-2 sequences of AlSCR16 and CgSCR7 and the OCS terminator (not shown). SCR constructs are not drawn to scale.
The effectiveness of the chimeric SRK expression system was first confirmed for two variants known to confer an incompatibility response in A. thaliana. Specifically, expression of reconstituted AtS1pr∷AleSRKb:AlSRKb and AtS1pr∷AleSRK25:AlSRKb chimeric genes conferred on transgenic stigmas the ability to inhibit pollen expressing AlSCRb and AlSCR25, respectively. The SRK and SCR expression system was subsequently used to test the functionality of SRK and SCR variants that had not been previously assayed in A. thaliana. In particular, we tested A. lyrata variants derived from the AlSa haplotype, for which SRKa and SCRa genomic clones are available (Kusaba et al. 2001) but attempts to construct SRKa transformation vectors had not been successful (J. B. Nasrallah, unpublished data), as well as those derived from the AlS16 and AlS37 haplotypes (Kusaba et al. 2001; Bechsgaard et al. 2006), for which genomic clones are not available. Furthermore, the expression system was used to determine if chimeric SRK and SCR genes could confer incompatibility in cases where native genes were ineffective, as in the case of the C. grandiflora CgSRK7 and CgSCR7 genes.
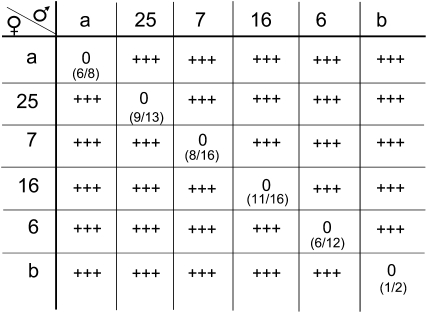
Each of the chimeric SRK and SCR genes was introduced individually into A. thaliana plants, and the stigmas of plants transformed with a particular AtS1pr∷eSRKx:AlSRKb gene were tested by application of pollen from plants transformed with the cognate SCR construct. With the exception of pollinations between AtS1pr∷eSRK37:AlSRKb and BrSCR8∷AlSCRb:SCR37 transformants, which were compatible, all other pollinations exhibited the expected incompatibility responses. As shown in Figure 3, stigmas expressing a particular eSRK:AlSRKb fusion inhibited pollen from plants expressing the eSRK's cognate SCR but not their own wild-type pollen. Similarly, pollen from plants expressing a particular SCR variant was inhibited on the stigmas of plants expressing the cognate eSRK:AlSRKb fusion, but produced many pollen tubes on wild-type stigmas (Figure 3). For each of the eSRK and SCR constructs tested, T1 plants were identified that exhibited a robust expression of the corresponding SI specificity and carried a single integration of the transgene. The T2 progenies of these plants recapitulated the pollination phenotype of T1 plants and were used for subsequent cross-pollination assays as described below.
Figure 3.—
Pollination phenotypes of A. thaliana plants transformed with the various eSRK:AlSRKb fusion constructs. The stigmas of first- (T1) and second- (T2) generation transgenic plants expressing each of the five SRK fusions and the AlSRKb gene were pollinated with pollen from plants expressing the cognate SCR and other SCRs. SRK variants are indicated in the column below the female symbol: a, AleSRKa:AlSRKb; 25, AleSRK25:AlSRKb; 7, CgeSRK7:AlSRKb; 16, AleSRK16:AlSRKb; 6, native AlSRK6; b, native AlSRKb. SCR variants are indicated in the row to the right of the male symbol: a, native AlSCRa; 25, native AlSCR25; 7, AlSCRb:CgSCR7; 16, AlSCRb:AlSCR16; 6, native AlSCR6; b, native AlSCRb. Ratios in parentheses indicate the number of T1 plants that expressed an incompatibility response toward pollen expressing cognate SCR over the total number of primary transformants analyzed. AlS6 and AlSb transformants were tested by self-pollination because they expressed both SRK and SCR genes. Pollen of AlSCR25 transformants was tested on the stigmas of plants expressing the AleSRK25:AlSRKb fusion and of plants expressing native AlSRK25. 0, an incompatible response (typically <5 pollen tubes per pollinated stigma); +++, a compatible response (typically >50 pollen tubes per pollinated stigma).
Expression of six distinct SI specificities in A. thaliana:
Although each of the AlSb-, AlSa-, AlS6-, AlS16-, AlS25-, and CgS7-derived SRK and SCR alleles conferred incompatibility in transgenic plants, it was important to determine if they bestowed distinct SI specificities in the A. thaliana genomic context. This is particularly critical in the case of the AlSa and CgS7 haplotypes, which are more similar to each other than to other A. lyrata and C. grandiflora S haplotypes, both in overall organization and sequence of their SRK and SCR genes (Nasrallah et al. 2007). Indeed, despite the divergence of A. lyrata and C. grandiflora, which are thought to have shared a common ancestor ∼10 million years ago, the exons of AlSRKa and CgSRK7 on the one hand and of AlSCRa and CgSCR7 on the other hand share an average of ∼86 and 74% amino acid sequence identity, respectively. This degree of sequence identity is similar to the 88 and 71% amino acid identity shared by SRK and SCR alleles from functionally equivalent S loci identified in studies of the very closely related B. rapa and B. oleracea or Brassica and its sister genus Raphanus (Kusaba et al. 2001; Kimura et al. 2002; Sato et al. 2003, 2004, 2006). Therefore, it is possible that the AlSa and CgS7 haplotypes might have been derived from a common ancestral haplotype and could be functionally equivalent.
To determine if the various SRK/SCR variants used in this study encode distinct SI specificities in transgenic A. thaliana, reciprocal cross-pollinations among SRK and SCR T2 transformants were performed. As shown in Figure 3, in all cases, transformants expressing a particular eSRKx:AlSRKb fusion inhibited pollen expressing the cognate SCR but not pollen expressing any one of the other five SCR variants tested. Conversely, pollen of transformants expressing one SCR variant was inhibited on stigmas expressing the cognate eSRK:AlSRKb fusion, but not on stigmas of plants expressing an independently derived eSRK:AlSRKb fusion. This pattern was observed not only among transformants expressing A. lyrata-derived variants, but also for transformants expressing the A. lyrata Sa- and C. grandiflora S7-derived genes. This result demonstrates that the AlSRKa/AlSCRa and CgSRK7/CgSCR7 gene pairs confer distinct SI recognition specificitites in A. thaliana. Interestingly, reciprocal pollinations of A. thaliana plants transformed with the CgS7 genomic fragment containing the native CgSRK7 and CgSCR7 genes with plants expressing the AtS1pr∷CgeSRK7:AlSRKb and BrSCR8∷AlSCRb:CgSCR7 chimeric genes demonstrated that the pollen of the CgS7 transformants expressed the S7 specificity (i.e., it was inhibited on the stigmas of AtS1pr∷CgeSRK7:AlSRKb transformants), while their stigmas did not (i.e., they did not inhibit pollen from BrSCR8∷AlSCRb:CgSCR7). Therefore, the lack of SI in CgS7 transformants was due to the inability of the native CgSRK7 gene to function in A. thaliana. The failure of this gene to confer SI was not due to lack of expression, however. Indeed, there was no significant difference between CgSRK7 transcript levels in transgenic A. thaliana CgS7 stigmas and C. grandiflora S7 stigmas (Figure S3).
Attempts to reconstitute functional alleles from A. thaliana SRK and SCR pseudogenes:
The chimeric gene expression system we developed allowed us to assess the extent of decay suffered by the nonfunctional ΨS locus of the A. thaliana Cvi-0 accession. The ΨSB haplotype was previously shown to contain a ΨSRK allele that encodes a truncated open reading frame containing a full-length eSRK and terminating at the end of exon 2 (Kusaba et al. 2001; Shimizu et al. 2004) and a ΨSCRB allele that encodes an apparently intact open reading frame, which had been suggested to have retained functionality. However, inspection of amino acid sequences of ΨSCRB with AlSCR16, AlSCRa, AlSCRb (Shimizu et al. 2008) and other SCRs used in this study (see Figure S1) shows that ΨSCRB is the only variant among these SCRs that contains an extra cysteine residue between the first and second of eight canonical cysteine residues (Chookajorn et al. 2004). These eight conserved cysteines form four disulfide bridges critical for SCR structure and function, and it is possible that this additional cysteine might disrupt the configuration of the SCRB protein and thus affect its function.
The functionality in A. thaliana of the AleSRK16:AlSRKb and AlSCRb:AlSCR16 chimeric genes, which are derived from the A. lyrata ortholog of Cvi-0 ΨSB, provided us with the necessary tools to determine if the ΨSCRB or ΨeSRKB sequences of the Cvi-0 ΨSB haplotype might have retained their mutual recognition or recognition of their A. lyrata S16 orthologs. An attempt was made to “correct” the obvious mutations in the A. thaliana ΨSRKB and ΨSCRB sequences. We transformed A. thaliana with a construct containing a full-length chimeric SRK open reading frame in which the Cvi-0 eSRKB sequence was fused to AlSRKb transmembrane and kinase domain sequences and with two ΨSCRB chimeric constructs, one containing unmodified exon-2 sequences and another containing a modified exon 2 in which the extra cysteine residue was replaced with a phenylalanine residue as occurs at the equivalent position in AlSCR16 (see Figure S2). None of the transformants exhibited an incompatibility response, either in reciprocal pollinations with plants transformed with the cognate “corrected” BrSCR8∷AlSCRb:AtSCRB or AtS1∷AteSRKB:AlSRKb genes, or with plants transformed with chimeric genes derived from their A. lyrata SCR16 or SRK16 orthologs.
DISCUSSION
This article describes the successful interspecific and intergeneric transfer of the self-incompatibility trait to A. thaliana by transformation with several newly isolated or previously described SRK/SCR allelic pairs from A. lyrata and C. grandiflora. This successful complementation was achieved using either the standard approach of transformation with intact SRK and SCR genes isolated from genomic libraries or the novel strategy of transformation with engineered genes designed to express chimeric proteins. Such a strategy is often used for analysis of receptor proteins in animals (i.e., Bergwitz et al. 1996) but it has not been previously applied to the analysis of SRK/SCR function. Together, these approaches bring to six the number of SI specificities that have been transferred to A. thaliana. The five newly transferred specificities, AlSa, AlS6, AlS16, AlS25, and CgS7, differ in the relatedness of their SRKs and SCRs to each other and to AlSRKb/AlSCRb, the only SRK/SCR pair previously shown to confer SI in A. thaliana, ranging from 60 to 82% for SRKs and from 40 to 60% for SCRs (Figure S1). The observation that highly diverged SRK and SCR variants are functional in transgenic A. thaliana indicates that the A. thaliana pollen–stigma interface provides an adequate molecular environment for transfer of highly diverged SCRs from pollen to stigma, their transport across the stigma epidermal cell wall, and their binding to highly diverged cognate eSRKs.
The functionality of the majority of chimeric SRK molecules assayed here also demonstrates that highly diverged eSRKs can effect their specific recognition function when fused to the transmembrane and kinase domains of AlSRKb, which can differ by as much as 25% (AlSRKb vs. CgSRK7) from their native kinase domains. These results prove that specificity in the SRK–SCR interaction is solely determined by the eSRK domain (excluding the last 23 amino acids), with no contribution from the transmembrane domain, which has been reported to be required for high-affinity ligand binding in vitro (Shimosato et al. 2007) or the cytoplasmic juxtamembrane domain.
The fact that the C. grandiflora S7 specificity could be expressed by transformation with the CgeSRK7:AlSRKb fusion, but not with an intact CgSRK7 gene, was unexpected. The Capsella and Arabidopsis genera share a large number of trans-species polymorphisms (Paetsch et al. 2006). Because the CgSRK7 gene is expressed and the eSRK7:AlSRKb fusion is functional, the nonfunctionality of CgSRK7 reveals significant divergence between the two taxa either in processing of the SRK protein or in downstream targets of the receptor. Additional experiments are required to determine if the ineffectiveness of the CgSRK7 gene in A. thaliana is a function of its transmembrane or kinase domains, and whether all Capsella SRK alleles fail to function in A. thaliana.
Despite successful expression of six distinct SI specificities, however, one SRK/SCR chimeric gene pair tested, AleSRK37:AlSRKb and AlSCRb:AlSCR37, failed to confer an incompatibility response in A. thaliana. This failure does not seem to be a function of overall sequence divergence, because AleSRK37 and AlSCR37 are not significantly more diverged from AleSRKb and AlSCRb than the variants that conferred an incompatibility response. At present, it is not known if failure to express incompatibility is due to the nonfunctionality of AleSRK37 or of AlSCR37. However, an earlier report had indicated that SRK variants bear polymorphisms that influence not only their ligand specificity, but also their ability to form homodimers and heterodimers (Naithani et al. 2007). The region that influences specificity in dimerization is located within the PAN_APPLE domain of eSRK (Naithani et al. 2007) at the junction of the SRK fusion constructs used in our study [C-terminal variable region (CVR) in Figure 3]. It is possible that receptor dimerization is negatively affected in the AleSRK37:AlSRKb fusion but not in the eSRK:AlSRKb fusions that conferred SI. Alternatively, the AleSRK37:AlSRKb protein might not assume a functional conformation and thus might fail to accumulate to appropriate levels or to be correctly targeted to the plasma membrane in A. thaliana stigmas.
Nevertheless, the chimeric protein approach is clearly a valuable strategy for analysis of SRK and SCR function. Not only has it allowed the transfer of several distinct SI specificities into A. thaliana, but it has also allowed us to determine that the eSRKs and SCRs of the A. thaliana ΨSB haplotype have accumulated function-altering substitutions in addition to the previously noted obvious open reading frame disrupting mutations or rearrangements. At present, it is not possible to infer which mutations in these pseudogenes are primary mutations that might have caused loss of SI in the A. thaliana lineage and which mutations are secondary mutations that occurred after the switch to self-fertility due to relaxation of selective pressure on the S locus.
The chimeric protein approach also provides a facile means for testing the functional equivalence or nonequivalence of S haplotypes from different species, as shown by our demonstration that AlSa and CgS7 determine distinct SI specificities despite high similarity in the sequences and arrangement of their SRK and SCR genes. More generally, the chimeric gene expression system might provide a means for future transfer of SI specificities between various crucifer species. SI is a valuable trait in breeding schemes for hybrid seed production, and a reliable and efficient means of transferring SI specificities to agronomically important crucifers is desirable. Therefore, understanding the factors required for proper function of SRK and SCR in heterologous crucifer species, as may be achieved using the protein fusion expression system described here, is a critical goal of future research. In the short term, further in planta analysis of the various SI specificities already introduced into A. thaliana promises to provide a mechanistic understanding of several aspects of SRK and SCR function. In particular, this analysis may allow us to elucidate the molecular basis of dominance relationships between SRK alleles (Hatakeyama et al. 2001; Schierup et al. 2001; Mable 2003; Prigoda et al. 2005) and to delineate the residues in the eSRK and SCR that are required for specificity and functionality of these highly specific receptor-ligand pairs.
Acknowledgments
A. thaliana seed was obtained from the Arabidopsis Biological Resource Center in Columbus, Ohio. This article is based upon work supported by National Science Foundation grant IOS-0744579.
Supporting information is available online at: http://www.genetics.org/cgi/content/full/genetics.109.102442/DC1.
References
- Bechsgaard, J. S., V. Castric, D. Charlesworth, X. Vekemans and M. H. Schierup, 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23 1741–1750. [DOI] [PubMed] [Google Scholar]
- Bergwitz, C., T. J. Gardella, M. R. Flannery, J. T. Potts, Jr., H. M. Kronenberg et al., 1996. Full activation of chimeric receptors by hybrids between parathyroid hormone and calcitonin. Evidence for a common pattern of ligand-receptor interaction. J. Biol. Chem. 271 26469–26472. [DOI] [PubMed] [Google Scholar]
- Bi, Y. M., N. Brugiere, Y. Cui, D. R. Goring and S. J. Rothstein, 2000. Transformation of Arabidopsis with a Brassica SLG/SRK region and ARC1 gene is not sufficient to transfer the self-incompatibility phenotype. Mol. Gen. Genet. 263 648–654. [DOI] [PubMed] [Google Scholar]
- Boggs, N. A., J. B. Nasrallah and M. E. Nasrallah, 2009. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5 e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D. C., M. E. Nasrallah, J. Vrebalov and J. B. Nasrallah, 1997. The self-incompatibility (S) haplotypes of Brassica contain highly divergent and rearranged sequences of ancient origin. Plant Cell 9 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A. J., and L. A. Casselton, 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17 393–400. [DOI] [PubMed] [Google Scholar]
- Casselman, A. L., J. Vrebalov, J. A. Conner, A. Singhal, J. Giovannoni et al., 2000. Determining the physical limits of the Brassica S locus by recombinational analysis. Plant Cell 12 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., C. Bartolome, M. H. Schierup and B. K. Mable, 2003. Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol. Biol. Evol. 20 1741–1753. [DOI] [PubMed] [Google Scholar]
- Chookajorn, T., A. Kachroo, D. R. Ripoll, A. G. Clark and J. B. Nasrallah, 2004. Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc. Natl. Acad. Sci. USA 101 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer, K. G., B. A. Lalonde, J. B. Nasrallah and M. E. Nasrallah, 1992. Structure and expression of AtS1, an Arabidopsis thaliana gene homologous to the S-locus related genes of Brassica. Mol. Gen. Genet. 231 442–448. [DOI] [PubMed] [Google Scholar]
- Gleave, A. P., 1992. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into plants genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, K., T. Takasaki, G. Suzuki, T. Nishio, M. Watanabe et al., 2001. The S receptor kinase gene determines dominance relationships in stigma expression of self-incompatibility in Brassica. Plant J. 26 69–76. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., C. R. Schopfer, M. E. Nasrallah and J. B. Nasrallah, 2001. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kho, Y. O., and J. Baer, 1968. Observing pollen tubes by means of fluorescence. Euphytica 17 298–302. [Google Scholar]
- Kimura, R., K. Sato, R. Fujimoto and T. Nishio, 2002. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant J. 29 215–223. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and J. Schell, 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kusaba, M., K. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13 627–643. [PMC free article] [PubMed] [Google Scholar]
- Mable, B. K., 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90 422–431. [DOI] [PubMed] [Google Scholar]
- Mable, B. K., M. H. Schierup and D. Charlesworth, 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata(Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90 422–431. [DOI] [PubMed] [Google Scholar]
- Naithani, S., T. Chookajorn, D. R. Ripoll and J. B. Nasrallah, 2007. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 104 12211–12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, J. B., 2000. Cell-cell signaling in the self-incompatibility response. Curr. Opin. Plant Biol. 3 368–373. [DOI] [PubMed] [Google Scholar]
- Nasrallah, J. B., 2005. Recognition and rejection of self in plant self-incompatibility: comparisons to animal histocompatibility. Trends Immunol. 26 412–418. [DOI] [PubMed] [Google Scholar]
- Nasrallah, J. B., P. Liu, S. Sherman-Broyles, R. Schmidt and M. E. Nasrallah, 2007. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics 175 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, M., P. Liu, S. Sherman-Broyles, N. Boggs and J. Nasrallah, 2004. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101 16070–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, M. E., P. Liu and J. B. Nasrallah, 2002. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297 247–249. [DOI] [PubMed] [Google Scholar]
- Paetsch, M., S. Mayland-Quellhorst and B. Neuffer, 2006. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97 283–290. [DOI] [PubMed] [Google Scholar]
- Prigoda, N. L., A. Nassuth and B. K. Mable, 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22 1609–1620. [DOI] [PubMed] [Google Scholar]
- Sato, Y., R. Fujimoto, K. Toriyama and T. Nishio, 2003. Commonality of self-recognition specificity of S haplotypes between Brassica oleracea and Brassica rapa. Plant Mol. Biol. 52 617–626. [DOI] [PubMed] [Google Scholar]
- Sato, Y., S. Okamoto and T. Nishio, 2004. Diversification and alteration of recognition specificity of the pollen ligand SP11/SCR in self-incompatibility of Brassica and Raphanus. Plant Cell 16 3230–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., K. Sato and T. Nishio, 2006. Interspecific pairs of class II S haplotypes having different recognition specificities between Brassica oleracea and Brassica rapa. Plant Cell Physiol. 47 340–345. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., B. K. Mable, P. Awadalla and D. Charlesworth, 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C. R., M. E. Nasrallah and J. B. Nasrallah, 1999. The male determinant of self-incompatibility in Brassica. Science 286 1697–1700. [DOI] [PubMed] [Google Scholar]
- Shimizu, K. K., J. M. Cork, A. L. Caicedo, C. A. Mays, R. C. Moore et al., 2004. Darwinian selection on a selfing locus. Science 306 2081–2084. [DOI] [PubMed] [Google Scholar]
- Shimizu, K. K., R. Shimizu-Inatsugi, T. Tsuchimatsu and M. D. Purugganan, 2008. Independent origins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17 704–714. [DOI] [PubMed] [Google Scholar]
- Shimosato, H., N. Yokota, H. Shiba, M. Iwano, T. Entani et al., 2007. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in brassica self-incompatibility. Plant Cell 19 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J. C., B. Howlett, D. C. Boyes, M. E. Nasrallah and J. B. Nasrallah, 1991. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S., H. Shimosato, H. Shiba, M. Funato, F. S. Che et al., 2001. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413 534–538. [DOI] [PubMed] [Google Scholar]
- van Engelen, F. A., J. W. Molthoff, A. J. Conner, J. P. Nap, A. Pereira et al., 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4 288–290. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., A. Ito, Y. Takada, C. Ninomiya, T. Kakizaki et al., 2000. Highly divergent sequences of the pollen self-incompatibility (S) gene in class-I S haplotypes of Brassica campestris (syn. rapa) L. FEBS Lett. 473 139–144. [DOI] [PubMed] [Google Scholar]
- Yamazaki, K., and G. K. Beauchamp, 2007. Genetic basis for MHC-dependent mate choice. Adv. Genet. 59 129–145. [DOI] [PubMed] [Google Scholar]
- Zhang, X., R. Henriques, S. S. Lin, Q. W. Niu and N. H. Chua, 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1 641–646. [DOI] [PubMed] [Google Scholar]