Abstract
Lateral gene transfer (LGT) and gene rearrangement are essential for shaping bacterial genomes during evolution. Separate attention has been focused on understanding the process of lateral gene transfer and the process of gene translocation. However, little is known about how gene translocation affects laterally transferred genes. Here we have examined gene translocations and lateral gene transfers in closely related genome pairs. The results reveal that translocated genes undergo elevated rates of evolution and gene translocation tends to take place preferentially in recently acquired genes. Translocated genes have a high probability to be truncated, suggesting that translocation followed by truncation/deletion might play an important role in the fast turnover of laterally transferred genes. Furthermore, more recently acquired genes have a higher proportion of genes on the leading strand, suggesting a strong strand bias of lateral gene transfer.
GENE insertions and deletions, together with gene translocations play important roles in bacterial genome evolution (Garcia-Vallvé et al. 2000; Ochman and Jones 2000; Tillier and Collins 2000a; Fraser-Liggett 2005). Gene insertions and deletions, as the essential driving forces in influencing gene content (Kunin and Ouzounis 2003), have received a great deal of attention. Various methods have been employed to study gene insertions and deletions previously; for instance, there are studies of population dynamics (Nielsen and Townsend 2004), such as a birth-and-death model of evolution (Berg and Kurland 2002; Novozhilov et al. 2005), phylogeny-dependent studies including parsimony methods (Daubin et al. 2003a,b; Mirkin et al. 2003; Hao and Golding 2004), and maximum-likelihood methods (Hao and Golding 2006b, 2008b). It has been shown that recently laterally transferred genes have high evolutionary rates and high rates of gene turnover (Daubin et al. 2003b; Hao and Golding 2004, 2006b).
Gene rearrangement has also been commonly studied as another important driving force that shapes bacterial genomes (for a review, see Rocha 2004). Gene order changes in genomes are history dependent; for instance, fewer gene rearrangements are expected among more closely related species. Gene order within genomes has therefore been used to reconstruct phylogeny (Sankoff et al. 2000; Tamames 2001; Rogozin et al. 2004; Belda et al. 2005). Previous studies have focused mainly on lateral gene transfer (LGT) and gene rearrangement individually, but little is known about any association between laterally transferred genes and gene rearrangements. The study of gene order of laterally acquired genes might shed some light on the understanding of the LGT process.
In this study, we have examined gene translocations and lateral gene transfers in closely related genome pairs. It is shown that the proportion of translocated genes among recently acquired genes is always high, while the proportion of translocated genes is always low in ancient genes, suggesting that gene translocation tends to take place in recently transferred genes. The results also reveal that translocated genes have elevated rates of evolution compared with positionally conserved genes and gene truncation is more prevalent in translocated genes. These findings suggest that gene translocation might accelerate the gene turnover of recently transferred genes and/or that genes likely to undergo translocation are those genes more likely to be laterally transferred and dispensable for the genome. Furthermore, the proportion of recently acquired genes is higher on the leading strand, suggesting that laterally transferred genes are biased toward being on the leading strand. After lateral transfer, some genes could be translocated to the lagging strand and some translocated genes are likely to be eliminated during evolution.
METHODS
The Bacillaceae group was chosen in this study due to the abundance of completely sequenced congeneric species. Complete genome sequences (Table 1 and Figure 1) were downloaded from the NCBI database (ftp://ftp.ncbi.nlm.nih.gov/). Annotated protein sequences were extracted from each complete genome. Four genome pairs (BlBp, BamBs, BwBc4, and Bc2Bc3) were examined for gene translocation because of the variation in gene content and the absence of large-scale genome rearrangement between each genome pair (Figure 2). The reciprocal best hit procedure has been commonly used for identifying orthologous pairs (Eisen 2000; Hirsh and Fraser 2001); in this study, orthologs were inferred from reciprocal best hits via a BLASTP search (Altschul et al. 1997). Significant matches are required to have an E-value <10−5. To avoid the confounding effects of duplication during evolution (Gu et al. 2002; Zhang et al. 2003), all paralogs in the analyzed genomes were excluded from further analysis. To do this, a TBLASTN search was conducted to search against both the query and the subject genomes with an E-value <10−5. If there was more than one significant hit in either genome, the query sequence was removed from further analysis. To avoid the potential effect of nonorthologous matches, a series of different cutoff thresholds on protein identity (from 30 to 80%) were employed in addition to the existing criteria for identifying orthologs.
TABLE 1.
Strain information in the Bacillaceae group
| Taxa | Abbreviation | Accession No. |
|---|---|---|
| Bacillus anthracis str. “Ames Ancestor” | Ba1 | NC_007530 |
| B. anthracis str. Ames | Ba2 | NC_003997 |
| B. anthracis str. Sterne | Ba3 | NC_005945 |
| B. amyloliquefaciens | Bam | NC_009725 |
| B. cereus E33L | Bc1 | NC_006274 |
| B. cereus ATCC 10987 | Bc2 | NC_003909 |
| B. cereus ATCC 14579 | Bc3 | NC_004722 |
| B. cereus subsp. cytotoxis | Bc4 | NC_009674 |
| B. clausii | Bcl | NC_006582 |
| B. halodurans | Bh | NC_002570 |
| B. licheniformis ATCC 14580 | Bl | NC_006322 |
| B. subtilis | Bs | NC_000964 |
| B. thuringiensis serovar konkukian | Bt1 | NC_005957 |
| B. thuringiensis str. Al Hakam | Bt2 | NC_008600 |
| B. pumilus | Bp | NC_009848 |
| B. weihenstephanensis | Bw | NC_010184 |
| Geobacillus kaustophilus | Gk | NC_006510 |
| G. thermodenitrificans | Gt | NC_009328 |
| Lysinibacillus sphaericus | Ls | NC_010382 |
| Oceanobacillus iheyensis | Oi | NC_004193 |
| Listeria innocua | Outgroup | NC_003212 |
| L. monocytogenes | Outgroup | NC_003210 |
Figure 1.—
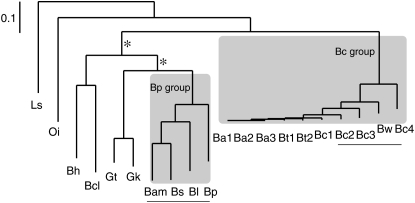
Phylogeny of the Bacillaceae group. Maximum-likelihood phylogeny was obtained from concatenated DNA sequences of 325 universally present nonduplicated genes in the Bacillaceae group. The topology is identical to the consensus of 325 individual gene trees, and the major part of phylogeny except two internal branches (labeled as *) is consistent with the neighbor-joining tree of the concatenated sequences. Abbreviations of strain names are listed in Table 1. The genomes used in genome pair analysis are underlined. Two clades are within shaded boxes. The Bc group is commonly used for the clade of B. anthracis (Ba), B. cereus (Bc), and B. thuringiensis (Bt), and the Bp group (including Bam, Bs, Bl, and Bp) is for description purposes in this study.
Figure 2.—
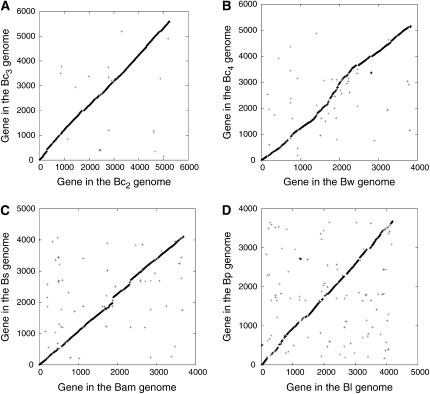
Genome synteny. (A) Bc2 vs. Bc3; (B) Bw vs. Bc4; (C) Bam vs. Bs; (D) Bl vs. Bp. Homologous matches are taken to have an expected value <10−5 between nonduplicated genes in a BLASTP search.
Genes were further categorized into group-specific genes and nonspecific genes. For instance, Bc group-specific (see Figure 1 for group definition) genes are present only in the Bc group but absent (with an E-value >10−5) from any other Bacillaceae genomes. Similarly, Bp-specific genes are present only in the Bp group but absent from any other Bacillaceae genomes. Members of orthologous genes were sorted according to their physical location on the chromosomes in each genome. The pairs that do not show conserved location on the chromosomes were deemed as translocated genes. Gene truncation was also identified in each genome pair. Annotated gene sequences in one genome were used as query sequences to BLAST against another genome. Significant hits are required to have an E-value <10−5. The match length of each hit was shown and the fraction of imperfect matches was used as an indicator for the degree of gene truncation as in Hao and Golding (2008a). To avoid the potential effect of nonorthologous matches, a series of more restrictive cutoff thresholds on E-values were examined (10−5, 10−10, 10−15, and 10−20).
No large-scale genome rearrangement was observed in the four genome pairs (Figure 2), which makes it easier to study individual gene translocation. Among the four genome pairs, BlBp is the most diverse pair, and Bc2Bc3 is the least diverse pair (see supporting information, Figure S1). The lower ends of the 95% confidence interval on protein identity for BlBp, BamBs, BwBc4, and Bc2Bc3 are 38.1, 52.1, 58.5, and 80.1, respectively. When different cutoff thresholds were not used, the lower ends of the 95% confidence interval were used to avoid the potential effect of nonorthologous matches.
Regions associated with insertion sequences (ISs) and prophages were identified. ISs were identified by the IScan program (Wagner et al. 2007), using query sequences of 20 reference sequences from Wagner et al. (2007) and 82 additional IS sequences that have been discovered in Bacillus species (names are given in Table S1). The sequences of all 102 ISs were obtained from the ISfinder website (Siguier et al. 2006b). Genes present in the IS regions were deemed to be IS associated. Prophages in each genome were identified by the Prophinder web server (Lima-Mendez et al. 2008). Genes present in the prophage regions were deemed to be prophage associated.
The origins and termini of replication for all genomes were identified by GC skew as done in previous studies (Lobry 1996; Morton and Morton 2007). GC skew was computed from the function (G − C)/(G + C) on 1000-bp windows across each genome. Gene location together with its orientation was used to determine whether the gene is on the leading strand or not. The number of genes on the leading strand was counted (see Table S2). The proportion of genes on the leading strand was further analyzed at different phylogenetic depths in both the Bc group and the Bp group. In the Bc group, group-specific genes in the Ba1 genome were examined and classified according to their depth in the phylogeny. In brief, genes present in Bc4 were categorized as n0, genes present in Bw but not present in Bc4 were categorized as n1, genes present in Bc3 but not present in Bw or Bc4 were categorized as n2, genes present in either Bc1 or Bc2 but not present in Bc4, Bw, or Bc3 were categorized as n3, genes present in Bt genomes but not present in Bc4, Bw, Bc3, Bc2, or Bc1 were categorized as n4, and genes present only in the Ba strains were categorized as n5.
Alignments of homologous sequences were constructed using the MUSCLE program (Edgar 2004). Three hundred twenty-five nonduplicated genes that are universally present in all Bacillaceae genomes were used for phylogeny reconstruction. A maximum-likelihood tree and a neighbor-joining tree were generated on concatenated sequences of the 325 genes (335,380 characters), using the PHYLIP package (Felsenstein 1989) version 3.67, and the rate variation parameter alpha was estimated using the PUZZLE program (Strimmer and von Haeseler 1996). The ratio of nonsynonymous changes to synonymous changes (Ka/Ks ratio) was measured by the Yang and Nielsen (2000) method, using yn00 in the PAML package (Yang 2007) based on nucleotide sequence alignments that were created from the corresponding protein alignments. To obtain a more reliable measurement of Ka/Ks, we excluded protein pairs that have protein identity <50%, since in this case synonymous changes might be greatly saturated. Statistical analyses were conducted using the R package (R Development Core Team 2008).
RESULTS
Molecular evolution of translocated genes:
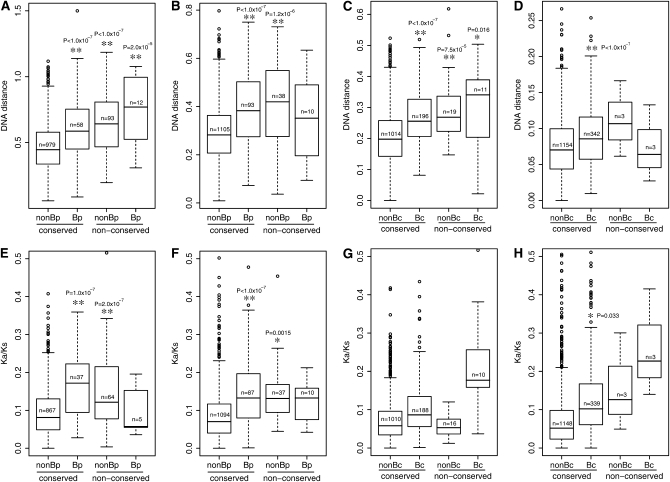
Evolutionary distance of different genes was examined separately in each genome pair (Figure 3, A–D). Strikingly, conserved specific genes have greater evolutionary distance than conserved nonspecific genes in all genome pairs. Ka/Ks values, as an indicator for the degree of functional constraints, were also examined for different gene groups (Figure 3, E–H). Conserved specific genes have greater Ka/Ks values than conserved nonspecific genes in all genome pairs. This is consistent with previous findings that recently transferred genes have faster rates of evolution (Hao and Golding 2006b). In nonspecific genes, translocated genes have faster rates of evolution over positionally conserved genes in the BlBp, BamBs, and BwBc4 genome pairs, suggesting that translocated genes tend to have greater rates of evolution over positionally conserved genes. Translocated nonspecific genes also show significantly higher Ka/Ks values over positionally conserved genes in the BlBp and BamBs pairs. A MANOVA test (see Table S3) also supports that both LGT and gene translocation contribute to the elevated substitution rates and Ka/Ks values.
Figure 3.—
DNA distance [(A) BlBp; (B) BamBs; (C) BwBc4; (D) Bc2Bc3] and Ka/Ks values [(E) BlBp; (F) BamBs; (G) BwBc4; (H) Bc2Bc3] in each genome pair. Abbreviations: Bc (Bp), Bc (Bp) group-specific genes; nonBc (nonBp), gene not specific to the Bc (Bp) group. The size of each class is shown. In Ka/Ks estimation, gene pairs that have protein identity <50% were excluded. Difference among classes was tested in a Tukey's honestly significant differences test. All observed significant comparisons are associated with the conserved nonspecific genes (the left box plot in each panel), and levels of significance (**P < 0.001 or *P < 0.05) together with P-values are shown.
Translocation in recently acquired genes:
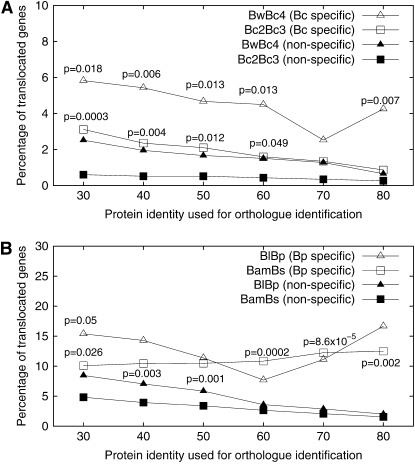
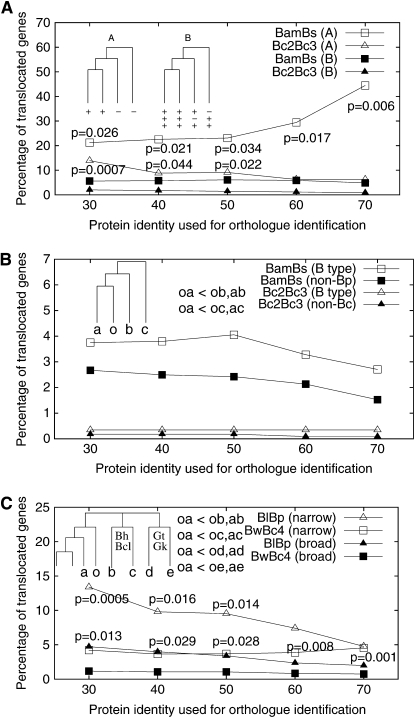
The proportion of translocated genes was calculated and is shown in Figure 4. The results reveal that recently transferred genes have a high proportion of translocated genes in all four genome pairs, while a high proportion of translocated genes was not observed in ancient genes (nonspecific genes). In fact, the proportion of translocated genes in genes that are present in all Bacillaceae genomes is even lower than that in nonspecific genes (data not shown). Together, the data show that gene translocation tends to take place in recently transferred genes. If gene translocation is a constant process throughout bacterial genome evolution, the results suggest that many translocated genes are deleted rapidly during evolution. These results are robust when different cutoff thresholds are used (protein identity from 30 to 80%). In other words, the high proportion of translocated genes in recently transferred genes is not an artifact of relaxed cutoff thresholds used to identify orthologs.
Figure 4.—
Higher proportion of translocation in taxa-specific genes than in nonspecific genes. A variety of protein identity cutoffs were used for ortholog identification, and genes associated with ISs and prophages were excluded. (A) The BwBc4 and Bc2Bc3 genome pairs; (B) the BlBp and BamBs genome pairs. In each genome pair, the proportion of translocation in taxa-specific genes is higher than that in nonspecific genes. In each panel, open triangles are higher than solid triangles and open squares are higher than solid squares (P-values of a χ2-test are shown).
This trend holds true in genes acquired at different evolutionary depths. Group-specific genes were further divided and analyzed in two types (“A” and “B,” Figure 5). The A type of genes is present in a narrower spectrum of genomes than the B type of genes, and, very likely, the A type of genes is more recently acquired than the B type of genes. Figure 5A shows that the A type of genes yields a higher percentage of translocated genes than the B type of genes in both BamBs and Bc2Bc3 genome pairs. To minimize the effect of xenologous gene displacement (with the original copy missing), we excluded genes with exceptionally large phylogenetic distance in Figure 5, B and C. In brief, for a gene, if the DNA distance from a closely related strain is larger than the distance from a slightly more distantly related strain (e.g., Bam-Bs > Bs-Bl or Bam-Bs > Bs-Bp), the gene is excluded from further analysis. Figure 5B shows that the B type of genes has a higher proportion of translocated genes than nonspecific genes in both BamBs and Bc2Bc3 genome pairs. We then expanded the same analysis on nonspecific genes in Figure 5C. It shows that genes present in a broader spectrum have a higher proportion of translocated genes than those present in a narrower spectrum. In other words, the proportion of translocated genes is nicely associated with the phylogenetic depths. It is worth mentioning that the inverse relationship between the proportion of translocated genes and their gene age is not likely an artifact of different degrees of divergence among gene categories. Genes within some genome pairs are highly similar in terms of their protein identity. For instance, >95% of gene pairs between Bc2Bc3 have protein identity >80.1% (see Figure S1). Divergent orthologs in these closely related genomes would still be able to be detected using low protein identities as cutoffs. In fact, the inverse relationship between the proportion of translocated genes and their gene age is robust in closely related pairs (and distantly related pairs) regardless of cutoff thresholds (Figures 4 and 5). These data support that more recently acquired genes are more likely to be translocated.
Figure 5.—
Translocated genes in recently acquired genes. (A) Translocated genes at different phylogenetic depths. Group-specific genes (labeled as “specific” in Figure 4) in two genome pairs BamBs and Bc2Bc3 were further examined at different phylogenetic depths using reference genomes. Bp and Bl are a reference for the BamBs pair; Bw and Bc4 are a reference for the Bc2Bc3 pair. The “A” types of genes are present in the analyzed genome pair but not in the reference genomes, while the “B” types of genes are present in the analyzed genome pair and in at least one of the reference genomes. Note that both A and B types of genes are group specific. (B) Comparison between the B type of genes and nonspecific genes (as in Figure 4). Genes with exceptionally large phylogenetic distance were excluded. (C) Comparison within “nonspecific” genes. Genes were designated as “broad” if they have at least one homolog in Ls, Oi, or Listeria and otherwise designated as “narrow.” Genes with exceptionally large phylogenetic distance were excluded using Bh, Bcl, Gt, and Gk as a reference. P-values of a χ2-test are shown.
Truncation in translocated genes:
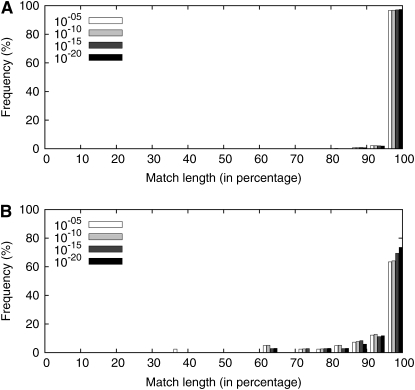
If gene truncation, as an imperfect form of gene deletion, takes place constantly as does gene deletion, different numbers of truncated genes might reflect different levels of gene deletions (Hao and Golding 2006a). Figure 6 shows the fraction of imperfect match length in a TBLASTN search after excluding genes associated with ISs and prophages. The results reveal that translocated genes have a higher proportion of truncated genes over positionally conserved ones in the BwBc4 pair. This trend is robust after more restrictive cutoff thresholds on E-values were used in identifying orthologs (from 10−5 to 10−20). To avoid the potential effect of frameshift mutation, a BLASTN search was conducted using the DNA sequences of annotated genes as query sequences. The result is consistent that translocated genes have a higher proportion of truncated genes over positionally conserved ones (see Figure S2). Comparison was also conducted reciprocally within the BwBc4 pair and within each of the other three genome pairs, the trend holds true for all of them (data not shown). This suggests that the high proportion of gene truncation in translocated genes is not an artifact of the particular analyzed genome, but rather it is a general phenomenon in bacterial genome evolution.
Figure 6.—
Fraction of homologous sequences that do not have a perfect match length in a TBLASTN search using different cutoffs for E-values. Annotated genes from the Bc4 genome were used as query sequences to search against the Bw genome. Genes associated with ISs and prophages were excluded. (A) Positionally conserved genes; (B) translocated genes.
Dynamic strand bias:
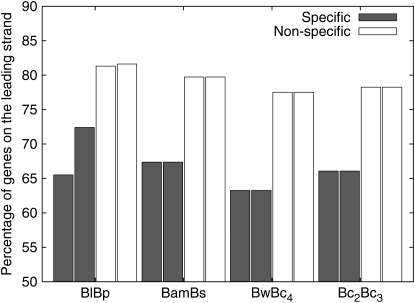
Among positionally conserved genes, group-specific genes have a lower proportion on the leading strand than nonspecific genes (Figure 7). Since it has been shown that essential genes tend to be more conserved on the leading strand (Rocha and Danchin 2003; Fang et al. 2005), one should expect that functionally important genes are more likely on the leading strand. Genes on leading/lagging strands were counted according to their COG classification (Tatusov et al. 2000). Poorly characterized genes and genes not included in COG classification have a lower percentage of genes on the leading strand compared with other genes (data not shown). Genes could also be translocated to a different strand during evolution. Among the translocated genes, ∼30% of them have been translocated to a different strand (see Table S2).
Figure 7.—
Proportion of positionally conserved genes on the leading strand. Two genomes are shown for each genome pair; group-specific and nonspecific genes are shown separately. Note that there is a slight difference between the Bl and Bp genomes due to individual gene inversion.
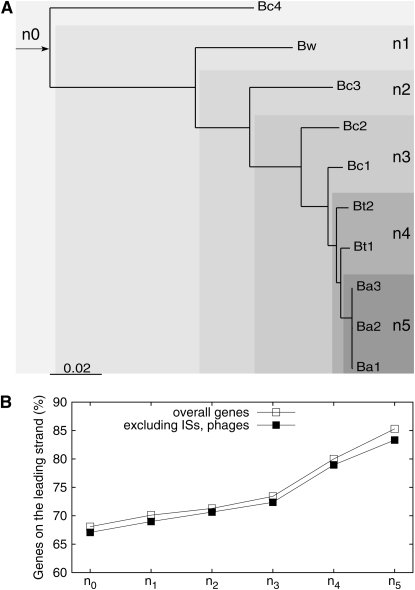
The proportion of genes on the leading strand was further examined at different phylogenetic depths in the Bc group (Figure 8). It is clear that more recently acquired genes have a higher proportion of genes on the leading strand. This trend is the opposite of the result that ancient genes have a higher proportion of genes on the leading strand than overall group specific genes (Figure 7). The result of more recently acquired genes on the leading strand could not be explained by the essentiality of recently acquired genes, since previous analyses have shown faster evolutionary rates and higher Ka/Ks ratios in more recently acquired genes (Hao and Golding 2006b). Genes and the ISs and prophage regions in the Ba1 genome were mapped on the chromosome (see Figure S3). The most recently acquired class n5 has a significantly higher proportion of genes associated with prophages (see Table S4). This might explain a small part of the higher proportion of genes on the leading strand in more recently acquired genes, since phages tend to integrate in such a way that most of their genes are coded on the leading strand (Campbell 2002). The negative association between phylogenetic depth and proportion of genes on the leading strand, however, still holds after excluding genes associated with ISs and prophages (Figure 8).
Figure 8.—
Proportion of genes on the leading strand associated with phylogenetic depths. (A) Distribution of clade-specific genes at different phylogenetic depths in the Bc group (n0 = 426, n1 = 368, n2 = 101, n3 = 79, n4 = 20, and n5 = 68); (B) proportion of genes on the leading strand.
DISCUSSION
Robustness:
Inferring gene translocation relies heavily on the identification of orthologous pairs. Any single threshold for ortholog identification might be problematic. We therefore made use of a series of cutoff thresholds to detect orthologs. Different threshold values caused some variation of the number of orthologous pairs, such as a decrease in the numbers of orthologous pairs when using restrictive cutoffs and an increase in the numbers of orthologous pairs when using relaxed cutoffs. Importantly, the proportion of translocated genes in recently transferred genes is always higher than that in ancient genes when using different cutoff thresholds. The high frequency of gene translocation in recently acquired genes, therefore, is not likely an artifact of the methodology used in this study.
Gene duplication is very common during genome evolution and substitution rates are often accelerated following gene duplication (Zhang et al. 2003). After gene duplication, duplicates may be retained and undergo neofunctionalization or subfunctionalization (Lynch and Force 2000; Lynch et al. 2001). There is a possibility that some orthologs inferred in this study were involved in differential loss after duplication. It has been shown that differential loss and gene conversion might happen after ancient duplication (Lathe and Bork 2001), and gene duplication followed by differential loss can always be invoked as an alternative to lateral gene transfer and vice versa (Gogarten and Townsend 2005). Differential loss will result in a relatively high level of divergence at the sequence level. In this study, the high proportion of translocated genes in recently acquired genes holds true even when the cutoff threshold for ortholog identification is very restrictive (up to 80% of protein identity). This supports the robustness of the concept that translocation tends to take place in recently acquired genes.
It is possible that some orthologous pairs detected in this study might be due to gene replacement via LGT. First, a distantly related gene copy could be introduced into a different location of the genome (lineage) and the original copy in the genome is deleted during evolution. This is the case of xenologous gene displacement. Second, it is also possible that the distantly related gene copy could be introduced to the same location of a genome and replace the original copy. This is known as gene displacement in situ (Omelchenko et al. 2003). Third, it is possible that a distinct gene is introduced into one lineage and then laterally transferred to another lineage. The first scenario is similar to the case of differential loss after duplication. In Figure 5, B and C, we have excluded genes with exceptionally large DNA distance by comparing slightly more distantly related strains. The trends are consistent in both cases, even though no significant P-values were obtained in Figure 5B, which might be due to the small number of genes in comparison. The second scenario does not result in gene translocation since the diverged copy just replaced the original copy in situ. In fact, some genes were found to have conserved gene order but have significant levels of sequence divergence (data not shown). The third scenario is difficult to distinguish from gene translocation. The likelihood of two successive transfers of one gene should be low, since closely related genomes are usually diverged due to niche separation but gene transfers are likely to take place among organisms that live in similar niches (Jain et al. 2003). Furthermore, the B types of genes in Figure 5 are not very likely subject to successive transfers because of their presence in a broader spectrum of genomes, and they also show a higher proportion of translocated genes than nonspecific genes (Figure 5B). Therefore, successive transfer events, if they happen, would not alter the conclusion that recently acquired genes tend to be translocated.
The evolution of translocated genes:
Besides the high frequency of gene translocation in recently transferred genes, this study reveals that translocated genes undergo faster rates of evolution compared with positionally conserved genes (Figure 3). Since translocated genes are under faster rates of evolution than positionally conserved genes, when more restrictive cutoff thresholds are used in identifying orthologs, the number of identified translocated genes might decrease more dramatically than that of positionally conserved genes, which results in a decrease in the proportion of translocated genes with more restrictive cutoffs (Figures 4 and 5). Indeed, a fast rate of evolution has been reported to result in a failure to detect homologs in similarity searches (Hao and Golding 2006a).
Previous studies have suggested that many recently transferred genes tend to be deleted rapidly (Hao and Golding 2004, 2006b). Gene translocations tend to take place in recently transferred genes that tend to be deleted rapidly; as a consequence, gene translocation should be considered as a local phenomenon. Indeed, relatively high rates of gene rearrangements have been found in closely related Salmonella strains (Liu and Sanderson 1998; Liu et al. 2003; Kothapalli et al. 2005), whereas the genome structures between Escherichia coli and Salmonella remain highly similar (Krawiec and Riley 1990; Liu et al. 1993). Furthermore, most truncated genes were found in translocated genes and the proportion of truncated genes is much higher in translocated genes than in positionally conserved genes (Figure 5). This holds true in all four genome pairs (data not shown). In other words, after being translocated, many genes tend to be deleted rapidly.
Compared with ancient genes, recently transferred genes were shown to be under relaxed functional constraints and translocated genes might be under more relaxed functional constraints (Ka/Ks ratios, Figure 3). It is plausible that genes under relaxed constraints are more likely to be translocated and tend to change more freely or even be deleted due to these relaxed functional constraints. On the other hand, some gene translocations might be considered as adaptive. The host with translocated genes might be able to adapt to a new niche faster than if it depended solely on substitution. Indeed, it has been shown that large-scale genome rearrangements, such as gene inversion and gene translocation, alter gene expression (Brinig et al. 2006) and might play roles in niche adaptation (Colson et al. 2004; Kuwahara et al. 2004; Burgetz et al. 2006; Coleman et al. 2006; Liu et al. 2006).
The occurrence of gene translocation seems to be influenced by gene function. The distribution of COG classification was compared between translocated genes and positionally conserved genes (see Figure S4). A significant difference in distribution was observed in BlBp, BamBs, and BwBc4. Gene translocation is generally rare in genes involved in translation, ribosomal structure, and biogenesis (“J” class), while gene translocation is more common in genes involved in carbohydrate transport and metabolism (“G” class) and amino acid transport and metabolism (“E” class) and in genes not included in COG (“−” class in Figure S4). In other words, besides the elevated evolutionary rates, translocated genes have a biased distribution of functional classification. This finding is a snapshot of the evolutionary process with the presence of selection. Gene translocation has deleterious effects on genes, and translocation that occurred in ancient genes or functionally essential genes is likely strongly deleterious, while translocation that has occurred in recently acquired genes is likely less deleterious or might be adaptive. Adaptive translocations are likely to be retained and slightly deleterious translocations could be retained in a population for some period of time, while strongly deleterious translocations should be extremely rare. The fate of many translocated genes in recently acquired genes is to be eliminated during evolution. Therefore, gene translocation serves as a factor that speeds up the turnover of laterally transferred genes.
Genes distributed on the leading strand:
Genes on the leading strand were examined but different pictures were obtained at different levels of comparison. A large-scale comparison shows that ancient genes are more likely on the leading strand than group-specific genes (Figure 7). The proportion of genes on the leading strand is higher in genes universally present in all Bacillaceae genomes and further inflated in the universal genes in Bacillaceae also present in Listeria genomes (data not shown). A similar pattern has been found by Fang et al. (2005). The high proportion of genes on the leading strand in ancient genes is likely due to their functional essentiality, since essential genes tend to be on the leading strand (Rocha and Danchin 2003).
In a fine-scale comparison it is found that more recently acquired genes have an even higher proportion of genes on the leading strand (Figure 8). We examined the effect of prophage genes, since lambdoid phages tend to integrate in such a way that most of their genes are coded on the leading strand (Campbell 2002). Genes associated with prophages do show a higher proportion of being on the leading strand (Table S5) and the most recently acquired class n5 has a significantly higher proportion of genes associated with prophages (Table S4). However, the removal of genes associated with ISs and prophages resulted in little change of the trend. One possible explanation is that recently acquired genes are of phage origin but have become difficult to identify. Indeed, most of the recently acquired genes have features similar to genes in lambdoid phages (Daubin et al. 2003a). Another possibility is that some foreign genes are from some nonphage sources, but like lambdoid phages, they also tend to be inserted into the leading strand of the host genome. The high proportion of newly transferred genes on the leading strand could also be explained by the fact that transfers to the lagging strand are likely less successful. The substantial difference between the large-scale comparison and the fine-scale comparison is that, in the short term, gene translocation is likely neutral or nearly neutral, whereas, in the long term, gene translocation could be deleterious and selected against.
It has been shown that genes evolve faster after shifting from one replicating strand to the other due to mutational biases (Tillier and Collins 2000b; Rocha and Danchin 2001). We have examined the translocated genes that shifted strand, but no significant difference in DNA distance was found between genes that shifted strand and those that did not shift strand (see Figure S5). The trend, though not significant, that translocated genes that shifted strand evolve faster than those that did not shift strand was observed in BlBp and BamBs. It is possible that the test lacks statistical power due to the small number of translocated genes. Importantly, translocated genes that did not shift strand have shown a significantly larger distance than positionally conserved genes. This suggests that the elevated rate of evolution in translocated genes is not mainly due to mutational bias after shifting strand.
Gene translocation mechanisms:
Genome rearrangement can be the result of a number of specific molecular mechanisms (Arber 2003), initiated or aided by prophage, IS elements, and site-specific recombination. Prophages have been well documented to play an important role in large-scale genome rearrangements (Canchaya et al. 2004), and quite often prophages are associated with insertions of a number of novel sequences (Ivanova et al. 2003). Translocated genes identified in this study tend to be spatially dispersed rather than clustered together (Figure 2, Figure S6, and Figure S7). Therefore, bacterial phages might play a role in translocation of several genes in a cluster, but it is not likely the main driving force for individual gene translocation during evolution.
Mobile elements (IS elements) have been known to play an important role in extensive genome rearrangement, such as in Bordetella (Brinig et al. 2006). In this study, the results are robust even after excluding genes associated with ISs and prophages. However, the possibility that IS elements are involved in gene translocation cannot be ruled out since most of the IS elements in genomes are evolutionarily young and under fast rates of turnover (Siguier et al. 2006a; Wagner 2006a,b; Touchon and Rocha 2007). It has been shown that elements involved in gene transfer have undergone a decay process (Sirand-Pugnet et al. 2007). Similarly, it might be possible that IS elements involved in gene translocation in this study have been deleted during evolution.
Site-specific recombination has also been reported to be involved in lateral gene transfer and deletion in bacterial genome evolution (Gillings et al. 2005; MacDonald et al. 2006). Furthermore, short palindromic sequences (Lewis et al. 1999; Tobes and Pareja 2006) or short signature sequences (Robins et al. 2005) have been suggested to serve as a source of recombination sites for gene movement. However, detection of recombination sites requires more experimental evidence.
Conclusion:
We have uncovered significant associations between gene translocation and lateral gene transfer. Translocated genes have accelerated rates of evolution and gene translocation tends to be observed in recently acquired genes. Many translocated genes undergo gene truncation and will ultimately be deleted from the genome. Furthermore, there is a strong leading strand bias of lateral gene transfer and in the course of evolution the strand bias of the laterally transferred genes will be influenced by gene translocation and many other factors. In conclusion, gene translocation plays an important role in shaping the evolution of laterally transferred genes.
Acknowledgments
The authors thank the reviewers for many useful suggestions. This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to G.B.G.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.104216/DC1.
References
- Altschul, S. F., T. L. Madden, A. A. Schffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, W., 2003. Elements for a theory of molecular evolution. Gene 317 3–11. [DOI] [PubMed] [Google Scholar]
- Belda, E., A. Moya and F. J. Silva, 2005. Genome rearrangement distances and gene order phylogeny in gamma-proteobacteria. Mol. Biol. Evol. 22 1456–1467. [DOI] [PubMed] [Google Scholar]
- Berg, O. G., and C. G. Kurland, 2002. Evolution of microbial genomes: sequence acquisition and loss. Mol. Biol. Evol. 19 2265–2276. [DOI] [PubMed] [Google Scholar]
- Brinig, M. M., C. A. Cummings, G. N. Sanden, P. Stefanelli, A. Lawrence et al., 2006. Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J. Bacteriol. 188 2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgetz, I. J., S. Shariff, A. Pang and E. R. M. Tillier, 2006. Positional homology in bacterial genomes. Evol. Bioinform. 2 42–55. [PMC free article] [PubMed] [Google Scholar]
- Campbell, A. M., 2002. Preferential orientation of natural lambdoid prophages and bacterial chromosome organization. Theor. Popul. Biol. 61 503–507. [DOI] [PubMed] [Google Scholar]
- Canchaya, C., G. Fournous and H. Brussow, 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53 9–18. [DOI] [PubMed] [Google Scholar]
- Coleman, M. L., M. B. Sullivan, A. C. Martiny, C. Steglich, K. Barry et al., 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311 1768–1770. [DOI] [PubMed] [Google Scholar]
- Colson, I., D. Delneri and S. G. Oliver, 2004. Effects of reciprocal chromosomal translocations on the fitness of Saccharomyces cerevisiae. EMBO Rep. 5 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin, V., E. Lerat and G. Perriere, 2003. a The source of laterally transferred genes in bacterial genomes. Genome Biol. 4 R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin, V., N. A. Moran and H. Ochman, 2003. b Phylogenetics and the cohesion of bacterial genomes. Science 301 829–832. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J. A., 2000. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr. Opin. Microbiol. 3 475–480. [DOI] [PubMed] [Google Scholar]
- Fang, G., E. Rocha and A. Danchin, 2005. How essential are nonessential genes? Mol. Biol. Evol. 22 2147–2156. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1989. PHYLIP (phylogeny inference package). Version 3.2. Cladistics 5 164–166. [Google Scholar]
- Fraser-Liggett, C. M., 2005. Insights on biology and evolution from microbial genome sequencing. Genome Res. 15 1603–1610. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallvé, S., A. Romeu and J. Palau, 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings, M. R., M. P. Holley, H. W. Stokes and A. J. Holmes, 2005. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. USA 102 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten, J. P., and J. P. Townsend, 2005. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3 679–687. [DOI] [PubMed] [Google Scholar]
- Gu, Z., D. Nicolae, H. H. Lu and W. H. Li, 2002. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18 609–613. [DOI] [PubMed] [Google Scholar]
- Hao, W., and G. B. Golding, 2004. Patterns of bacterial gene movement. Mol. Biol. Evol. 21 1294–1307. [DOI] [PubMed] [Google Scholar]
- Hao, W., and G. B. Golding, 2006. a Asymmetrical evolution of cytochrome bd subunits. J. Mol. Evol. 62 132–142. [DOI] [PubMed] [Google Scholar]
- Hao, W., and G. B. Golding, 2006. b The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res. 16 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, W., and G. B. Golding, 2008. a High rates of lateral gene transfer are not due to false diagnosis of gene absence. Gene 421 27–31. [DOI] [PubMed] [Google Scholar]
- Hao, W., and G. B. Golding, 2008. b Uncovering rate variation of lateral gene transfer during bacterial genome evolution. BMC Genomics 9 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh, A. E., and H. B. Fraser, 2001. Protein dispensability and rate of evolution. Nature 411 1046–1049. [DOI] [PubMed] [Google Scholar]
- Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon et al., 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423 87–91. [DOI] [PubMed] [Google Scholar]
- Jain, R., M. C. Rivera, J. E. Moore and J. A. Lake, 2003. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20 1598–1602. [DOI] [PubMed] [Google Scholar]
- Kothapalli, S., S. Nair, S. Alokam, T. Pang, R. Khakhria et al., 2005. Diversity of genome structure in Salmonella enterica serovar Typhi populations. J. Bacteriol. 187 2638–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawiec, S., and M. Riley, 1990. Organization of the bacterial chromosome. Microbiol. Rev. 54 502–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin, V., and C. A. Ouzounis, 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh et al., 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101 14919–14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe, 3rd, W. C., and P. Bork, 2001. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS Lett. 502 113–116. [DOI] [PubMed] [Google Scholar]
- Lewis, S., E. Akgun and M. Jasin, 1999. Palindromic DNA and genome stability. Further studies. Ann. N Y Acad. Sci. 870 45–57. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez, G., J. Van Helden, A. Toussaint and R. Leplae, 2008. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24 863–865. [DOI] [PubMed] [Google Scholar]
- Liu, G. R., K. Edwards, A. Eisenstark, Y. M. Fu, W. Q. Liu et al., 2003. Genomic diversification among archival strains of Salmonella enterica serovar typhimurium LT7. J. Bacteriol. 185 2131–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. R., W. Q. Liu, R. N. Johnston, K. E. Sanderson, S. X. Li et al., 2006. Genome plasticity and ori-ter rebalancing in Salmonella typhi. Mol. Biol. Evol. 23 365–371. [DOI] [PubMed] [Google Scholar]
- Liu, S. L., and K. E. Sanderson, 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol. Lett. 164 275–281. [DOI] [PubMed] [Google Scholar]
- Liu, S. L., A. Hessel and K. E. Sanderson, 1993. Genomic mapping with I–Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90 6874–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry, J. R., 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13 660–665. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., M. O'Hely, B. Walsh and A. Force, 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, D., G. Demarre, M. Bouvier, D. Mazel and D. N. Gopaul, 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440 1157–1162. [DOI] [PubMed] [Google Scholar]
- Mirkin, B. G., T. I. Fenner, M. Y. Galperin and E. V. Koonin, 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 3 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, R. A., and B. R. Morton, 2007. Separating the effects of mutation and selection in producing DNA skew in bacterial chromosomes. BMC Genomics 8 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K. M., and J. P. Townsend, 2004. Monitoring and modeling horizontal gene transfer. Nat. Biotechnol. 22 1110–1114. [DOI] [PubMed] [Google Scholar]
- Novozhilov, A. S., G. P. Karev and E. V. Koonin, 2005. Mathematical modeling of evolution of horizontally transferred genes. Mol. Biol. Evol. 22 1721–1732. [DOI] [PubMed] [Google Scholar]
- Ochman, H., and I. B. Jones, 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19 6637–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko, M. V., K. S. Makarova, Y. I. Wolf, I. B. Rogozin and E. V. Koonin, 2003. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 4 R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Robins, H., M. Krasnitz, H. Barak and A. J. Levine, 2005. A relative-entropy algorithm for genomic fingerprinting captures host-phage similarities. J. Bacteriol. 187 8370–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E. P., 2004. Order and disorder in bacterial genomes. Curr. Opin. Microbiol. 7 519–527. [DOI] [PubMed] [Google Scholar]
- Rocha, E. P., and A. Danchin, 2001. Ongoing evolution of strand composition in bacterial genomes. Mol. Biol. Evol. 18 1789–1799. [DOI] [PubMed] [Google Scholar]
- Rocha, E. P., and A. Danchin, 2003. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 31 6570–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I. B., K. S. Makarova, Y. I. Wolf and E. V. Koonin, 2004. Computational approaches for the analysis of gene neighbourhoods in prokaryotic genomes. Brief. Bioinform. 5 131–149. [DOI] [PubMed] [Google Scholar]
- Sankoff, D., D. Bryant, M. Deneault, B. F. Lang and G. Burger, 2000. Early eukaryote evolution based on mitochondrial gene order breakpoints. J. Comput. Biol. 7 521–535. [DOI] [PubMed] [Google Scholar]
- Siguier, P., J. Filee and M. Chandler, 2006. a Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9 526–531. [DOI] [PubMed] [Google Scholar]
- Siguier, P., J. Perochon, L. Lestrade, J. Mahillon and M. Chandler, 2006. b ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirand-Pugnet, P., C. Lartigue, M. Marenda, D. Jacob, A. Barre et al., 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 3 e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer, K., and A. von Haeseler, 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13 964–969. [Google Scholar]
- Tamames, J., 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2 RESEARCH0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R. L., M. Y. Galperin, D. A. Natale and E. V. Koonin, 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier, E. R., and R. A. Collins, 2000. a Genome rearrangement by replication-directed translocation. Nat. Genet. 26 195–197. [DOI] [PubMed] [Google Scholar]
- Tillier, E. R., and R. A. Collins, 2000. b Replication orientation affects the rate and direction of bacterial gene evolution. J. Mol. Evol. 51 459–463. [DOI] [PubMed] [Google Scholar]
- Tobes, R., and E. Pareja, 2006. Bacterial repetitive extragenic palindromic sequences are DNA targets for insertion sequence elements. BMC Genomics 7 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon, M., and E. P. Rocha, 2007. Causes of insertion sequences abundance in prokaryotic genomes. Mol. Biol. Evol. 24 969–981. [DOI] [PubMed] [Google Scholar]
- Wagner, A., 2006. a Cooperation is fleeting in the world of transposable elements. PLoS Comput. Biol. 2 e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A., 2006. b Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol. Biol. Evol. 23 723–733. [DOI] [PubMed] [Google Scholar]
- Wagner, A., C. Lewis and M. Bichsel, 2007. A survey of bacterial insertion sequences using IScan. Nucleic Acids Res. 35 5284–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17 32–43. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Z. Gu and W. H. Li, 2003. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 4 R56. [DOI] [PMC free article] [PubMed] [Google Scholar]