Abstract
We describe a simple genetic test for assessing the competency of Gal4-based baits prior to a yeast two-hybrid screen, which allows determination of whether a bait protein is expressed appropriately for an interaction to be detected. The novel test, based on interaction with the protein RanBPM, is easier and more predictive than other methods such as Western blotting, allowing identification of ∼80% of incompetent baits prior to screening.
SINCE the development of the yeast two-hybrid assay (Fields and Song 1989), tens of thousands of two-hybrid screens have been carried out. While such screens have been widely successful, revealing new pathways, proteins, and functions for known proteins, individual screens often fail due to lack of interactions, which can be caused by bait-specific or library-specific factors. Bait-specific factors include poor expression, incorrect localization, or degradation of the bait fusion protein (a protein of interest fused to a transcription factor binding domain). Often Western blotting is used to test the competency of a bait fusion before screening, allowing verification that the protein is correctly expressed in the yeast cells. However, this approach is not ideal. First, the Western-blotting procedure is time-consuming and requires antibodies to either the transcription factor binding domain or the bait protein of interest. Second, this method is unable to reveal information about localization within the cell—a protein may be highly expressed but not localized to the nucleus where it is required for the assay. Finally, the bait protein-binding domain fusion may be expressed at a level that is high enough for a successful screen, but too low to detect by Western blotting.
For LexA-based two-hybrid screening, also called the interaction trap method, a bait competency test is available. This test relies on the ability of transcriptionally inactive LexA fusions to repress transcription when bound to specifically positioned LexA operators (Brent and Ptashne 1984). The reporter plasmid uses a galactose-inducible GAL1-LacZ reporter with LexA operators inserted into the GAL1 UAS. Cells carrying the reporter plasmid and bait plasmid are tested for reduced LacZ activity after the addition of galactose. While this method can be useful in bait characterization, it has fallen out of use because the failure of a bait in the repression assay does not necessarily correlate with a poor screen outcome (Golemis et al. 2008). For Gal4-based two-hybrid screens, no genetic test has been developed.
In the interest of improving the efficiency of Gal4-based two-hybrid screening, we reasoned that a protein that interacts with the Gal4-binding domain (Gal4BD) could be used to test the competency of a Gal4BD–bait fusion protein prior to a screen, which would allow the determination of whether the bait is expressed appropriately for an interaction to occur. Such a competency test could be carried out easily with a bait of interest using a mating-based two-hybrid assay. We report here the design and testing of such a bait competency test using a novel Gal4-interacting protein, RanBPM. Comparing the results of the bait competency test with those of Western blotting, we show that the RanBPM competency test is superior in both ease of testing and predictive ability.
RanBPM and CSN5 interact with the Gal4-binding domain:
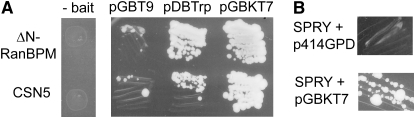
We identified several Gal4 activation domain (Gal4AD) fusion proteins that gave positive two-hybrid interaction results with vectors expressing Gal4BD alone, making them good candidates for competency test proteins. These proteins included CSN5 and an N-terminal truncated version of RanBPM (ΔN-RanBPM, containing amino acids 51–654 of mouse RanBPM). (Figure 1A). CSN5, a part of the COP9 signalosome, was previously found to interact with Gal4BD (Nordgard et al. 2001), but this interaction has not been demonstrated for RanBPM. To assess their interaction capabilities, we tested both proteins for interaction with 34 Gal4-bait fusion proteins that we knew were competently expressed because they had yielded hits in previous two-hybrid screens. CSN5 interacted with 44% of the baits, and ΔN-RanBPM interacted with 91% of the baits tested.
Figure 1.—
Yeast two-hybrid interactions of Gal4BD-interacting proteins. (A) Interaction of CSN5 or ΔN-RanBPM with Gal4BD expressed from vectors pGBT9 (Clontech), pDBTrp [a version of pDBLeu (Invitrogen) with a Trp1 selective marker replacing Leu2)], and pGBKT7 (Clontech). Strains AH109 and Y187 (Clontech) were used, and CSN5 and ΔN-RanBPM Gal4AD fusion proteins were in pGADT7rec (Clontech). Growth shown is on SD medium −Trp/−Leu/−His + 3 mm 3-AT. Also shown (“- bait”, left) are control self-activation tests of yeast expressing Gal4AD-CSN5 or ΔN-RanBPM alone (SD medium −Leu/−His + 3 mm 3-AT). (B) Interaction of the SPRY domain of RanBPM with Gal4BD from vector pGBKT7 or control vector p414GPD (SD medium −Trp/−Leu/−His + 3 mm 3-AT).
RanBPM is a 90-kDa ubiquitously expressed protein of unknown function. It was originally identified in a two-hybrid screen using Ran (Nakamura et al. 1998), where the interacting protein was found to be a truncated 55-kDa version of RanBPM (Nishitani et al. 2001). Full-length RanBPM contains a proline-rich N-terminal region (which is removed in ΔN-RanBPM), a consensus SPRY domain, and LiSH/CTLH motifs (Murrin and Talbot 2007). The SPRY domain has been implicated in mediating protein–protein interactions (Hilton et al. 1998), and previous studies indicated the SPRY domain alone could mediate interactions between RanBPM and some proteins (Rao et al. 2002; Wang et al. 2002; Cheng et al. 2005; Yuan et al. 2006). We found that the SPRY domain alone (amino acids 51–289 of mouse RanBPM) was sufficient for interaction with Gal4BD (Figure 1B), but did not use this construct further since the interaction was not as robust as with ΔN-RanBPM.
ΔN-RanBPM interaction as a predictor of yeast two-hybrid screen outcome:
To examine the ability of ΔN-RanBPM interaction to predict yeast two-hybrid screen outcome, we compared the RanBPM interaction results with results from two-hybrid screens for 75 bait proteins (Table 1). Each protein was expressed in a Gal4-binding domain fusion vector—pGBT9 (low expression), pDBTrp (medium expression), or pGBKT7 (high expression)—and tested for interaction with RanBPM using a mating strategy (see supporting information, File S1). Interactions were scored in three categories: negative (no colonies apparent), weak positive (colonies appearing after 5 days), and strong positive (colonies appearing at or before 5 days). Of the 75 tested baits, 25 failed to pass competency testing, and 46 of the remaining 50 baits (92%) gave hits. When we tested the 25 eliminated baits for interaction in two-hybrid screens, we found that a significant number (21 of the 25, or 84%) failed to give hits. The total number of bait proteins that failed competency testing but gave hits during screening was 4, which gives a false negative rate of 8% (i.e., four “negatives” of 50 screens that gave hits). The false positive rate was also 8% (i.e., four baits did not yield hits, of 50 baits that scored as “competent”).
TABLE 1.
RanBPM interaction correlated with two-hybrid success
Screens were scored as having hits if at least one reproducible interacting protein (as confirmed by individual two-hybrid retests) was identified.
Sixteen baits were in pGBT9, 4 in pDBTrp, and 5 in pGBKT7.
Three baits were in pGBT9 and 6 in pGBKT7.
Fourteen baits were in pGBT9, 6 in pDBTrp, and 21 in pGBKT7.
Comparison of the RanBPM interaction test and Western blotting:
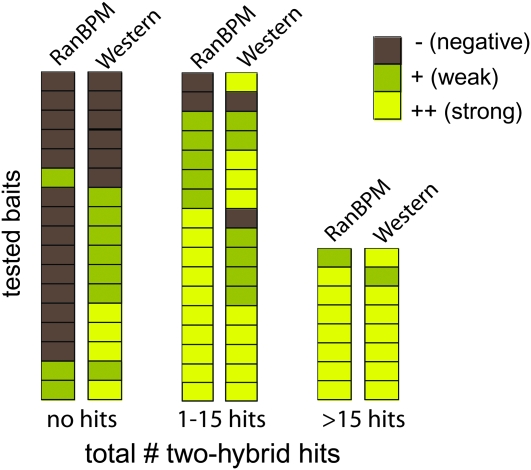
Western blotting, which tests the expression level of a bait fusion, is often used as a test of two-hybrid bait competency. We chose a subset of the proteins examined above, 42 baits in total, to analyze by Western blotting so that we could compare the methods (Figure 2 and Table 2). Because one of our aims was to determine the abilities of the two methods in predicting incompetent baits, about half of the baits, a total of 17, were proteins that had failed to yield hits in two-hybrid screening. Western blot results were scored in three categories: negative (no expression), weak (low-level expression), and strong (high-level expression).
Figure 2.—
Correlation of two-hybrid outcome with bait competency tests. Paired RanBPM and Western blot results are shown for each of the 42 baits tested. Tested baits are grouped into three categories, depending on two-hybrid screen outcome: no hits, 1–15 hits, or >15 hits. Competency test outcome is indicated by a shaded box, with gray indicating no RanBPM interaction or bait protein expression, yellow-gray indicating poor interaction or expression, and yellow indicating strong interaction or expression.
TABLE 2.
Comparison of RanBPM interaction and Western blotting as predictors of two-hybrid success
| No hits | Hits | Total | |
|---|---|---|---|
| RanBPM interaction | |||
| Negative | 14 | 2 | 16 |
| Positive | 3 | 23 | 26 |
| Western blotting | |||
| Negative | 6 | 2 | 8 |
| Positive | 11 | 23 | 34 |
Western blotting and RanBPM interaction gave similar rates of false negatives (8% in both cases): of the 25 baits that yielded hits during screening, only 2 baits failed to interact or failed to yield a positive immunoblot. However, Western blotting had a much higher false positive rate than the RanBPM interaction test: 32% of proteins that showed expression by Western blot failed to give hits, compared with 12% for the RanBPM test with this group of baits. With Western blotting, with the 17 baits that failed to give hits, only 6 scored as “incompetent” (i.e., no expression), while 11 showed some degree of expression. In comparison, with the RanBPM test for the same 17 baits, 14 scored as “incompetent,” with the remaining 3 showing a weak interaction.
Conclusions:
Over 30 different proteins have now been reported to interact with RanBPM. All of these interactions were initially identified by two-hybrid screens, but most were further confirmed by co-immunoprecipitation or pull-down approaches in heterologous systems, suggesting that RanBPM may be an inherently promiscuous or “sticky” protein. As the function of RanBPM remains unknown, it is unclear whether this promiscuity is biologically relevant or rather represents an artifact of assays that use highly overexpressed proteins. In HeLa cells, RanBPM was found in a large protein complex of >670 kDa (Nishitani et al. 2001), which would support the idea that the protein is involved in multiple interactions, protein aggregation, or scaffolding functions.
Independent of its biological role, our results with 75 baits showing that ∼90% of productive Gal4BD-fused baits interact with ΔN-RanBPM and that ∼80% of nonproductive baits fail to interact with ΔN-RanBPM clearly indicate the utility of this truncated protein in assessing bait competency. It should be noted that these data were generated using a specific combination of bait and prey vectors (pGBT9, pDBTrp, pGBKT7, and pGBKT7rec) and strains (AH109 or mated AH109/Y187 diploids) and that the percentages are specific to these vectors and strains. However, preliminary results using other vectors and strains (data not shown) suggest that the method is transferable.
Our studies indicate that the RanBPM interaction test not only is easier than Western blotting, requiring merely a series of streaks on plates, but also is a better predictor of success in two-hybrid screening. Using the RanBPM interaction test, our screen success rate (the percentage of screens yielding hits) was ∼90%, while this rate was only 66% when we used Western blotting as an indicator of bait competency. In particular, we found that a significant number of baits showed abundant expression by Western blotting, but failed to produce hits in two-hybrid screens.
We expect that the RanBPM interaction test will be useful for researchers carrying out high-throughput two-hybrid screens with multiple baits, as well as for investigators testing individual bait proteins (for example, to identify the site of interaction with a known binding partner). Identification of residues and domains important for an interaction often involves mutation or truncation of a bait protein, where loss of interaction due to protein instability or mislocalization can occur. Testing baits for interaction with ΔN-RanBPM, a relatively simple procedure, can provide a valuable control for such interaction studies.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103069/DC1.
References
- Brent, R., and M. Ptashne, 1984. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature 312 612–615. [DOI] [PubMed] [Google Scholar]
- Cheng, L., S. Lemmon and V. Lemmon, 2005. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J. Neurochem. 94 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, S., and O. Song, 1989. A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Golemis, E. A., I. Serebriiskii, R. L. Finley, Jr., M. G. Kolonin, J. Gyuris et al., 2008. Interaction trap/two-hybrid system to identify interacting proteins. Curr. Protoc. Mol. Biol. Chapter 20: Unit 20.1. [DOI] [PubMed]
- Hilton, D. J., R. T. Richardson, W. S. Alexander, E. M. Viney, T. A. Willson et al., 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. USA 95 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrin, L. C., and J. N. Talbot, 2007. RanBPM, a scaffolding protein in the immune and nervous systems. J. Neuroimmune Pharmacol. 2 290–295. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., H. Masuda, J. Horii, K. Kuma, N. Yokoyama et al., 1998. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 143 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani, H., E. Hirose, Y. Uchimura, M. Nakamura, M. Umeda et al., 2001. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene 272 25–33. [DOI] [PubMed] [Google Scholar]
- Nordgard, O., O. Dahle, T. O. Andersen and O. S. Gabrielsen, 2001. JAB1/CSN5 interacts with the GAL4 DNA binding domain: a note of caution about two-hybrid interactions. Biochimie 83 969–971. [DOI] [PubMed] [Google Scholar]
- Rao, M. A., H. Cheng, A. N. Quayle, H. Nishitani, C. C. Nelson et al., 2002. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J. Biol. Chem. 277 48020–48027. [DOI] [PubMed] [Google Scholar]
- Wang, D., Z. Li, E. M. Messing and G. Wu, 2002. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem. 277 36216–36222. [DOI] [PubMed] [Google Scholar]
- Yuan, Y., C. Fu, H. Chen, X. Wang, W. Deng et al., 2006. The Ran binding protein RanBPM interacts with TrkA receptor. Neurosci. Lett. 407 26–31. [DOI] [PubMed] [Google Scholar]