Abstract
Sex determination in plants involves a variety of mechanisms. Here, we report the map-based cloning and characterization of the unisexual-flower-controlling gene M. M was identified as a previously characterized putative 1-aminocyclopropane-1-carboxylic acid synthase gene, while the m allele that mutated at a conserved site (Gly33Cys) lost activity in the original enzymatically active allele.
SEX determination in angiosperms, including crop plants, evolves a variety of mechanisms that involve a number of different genetic and epigenetic factors (Tanurdzic and Banks 2004). Due to its diversity in sex types and to the extensive physiological and genetic studies conducted on it, cucumber (Cucumis sativus L.; 2n = 2x = 14) is becoming a model plant for sex-determination research (Atsmon 1968; Tsao 1988; Perl-Treves 1999; Tanurdzic and Banks 2004). In cucumber plants, male and female flowers are generally produced separately in the same individual; however, certain lines also produce bisexual flowers. Preliminary genetic studies have indicated that three major genes are responsible for sex expression and segregation in the cucumber plantF/f, M/m, and A/a. The F gene may promote femaleness, while the m gene regulates the appearance of hermaphroditic flowers on the plant. Furthermore, in combination with the homozygous recessive f gene, the recessive a gene can intensify the androecious nature (Galun 1961; Robinson et al. 1976).
Sex expression in cucumber plants can also be modified by various environmental factors and plant hormones such as ethylene (Atsmon 1968; Takahashi et al. 1983; Takahashi and Jaffe 1984; Perl-Treves 1999; Yamasaki et al. 2005). A series of studies (Kamachi et al. 1997, 2000; Trebitsh et al. 1997; Yamasaki et al. 2003a; Mibus and Tatlioglu 2004; Knopf and Trebitsh 2006) have been conducted to investigate the F/f gene. These studies have shown that CsACS1G, which encodes a key enzyme of the ethylene-synthesis pathway, is the candidate gene for the F/f locus. However, the M/m gene has not been studied in as much detail as the F/f gene. Here, we report the map-based cloning and characterization of the unisexual-flower-controlling M gene.
RESULTS
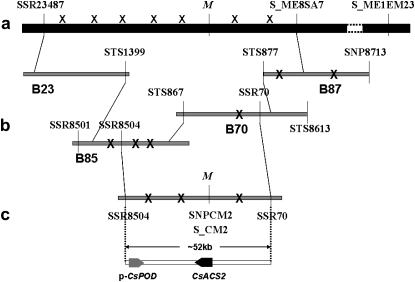
In the previous studies, the M/m locus was independently mapped into a genetic interval of 2.5 cM (Liu et al. 2008) and 6.1 cM (Li et al. 2008). In this study, we developed two larger segregating populations, which included 2830 F2 + BC1 (population 1983) and 2700 F2 (population 5234) individuals and constructed a high-resolution collinear genetic map for the M/m locus (supporting information, Figure S1). After chromosome walking, a bacterial artificial chromosome (BAC) contig formed by two BAC clones (overlapped by an ∼9.2-kb sequence) was found to be anchored to the genetic interval. The sequence of the entire contig covered an ∼52-kb chromosome section, which had two complete candidate genes (Figure 1). One gene sequence showed limited similarity (68%) to a bacterium-induced peroxidase precursor (GenBank accession no. AF155124) found in Gossypium hirsutum. However, the other gene, which was predicted to encode a 445-amino-acid protein, showed 100% sequence identity to the previously characterized CsACS2 gene, which encodes a putative 1-aminocyclopropane-1-carboxylate synthase in C. sativus (Kamachi et al. 1997).
Figure 1.—
M map-based cloning. (a) M genetic map. “X” indicates recombination events identified in population 5234 (the “right side” of the M locus) and population 1983 (the “left side” of the M locus), respectively. (b) Fine mapping M. New marker SNP8713, derived from the B87 end sequence, was mapped beyond the M locus with an additional recombination event. SSR70 was developed from the B70 inner sequence, which was mapped at one recombination event from M between S_ME8SA7 and SSR23487. SSR8501 and SSR8504, derived from the B85, had three and two recombination events, respectively, from the M locus at the other side beyond the SSR70 marker. The final contig was confirmed by an overlap test from STS877, STS867, and STS1399. BAC clones were not drawn to scale. (c) The final contig and the genomic organization for this region. A contig encompassing the M locus, which included three recombination events, was identified by the two markers, SSR8504 and SSR70. Broad arrows indicate the two predicted genes (p-CsPOD and CsACS2) and the predicted transcriptional orientations. Two new markers, S_CM2 and SNPCM2, were developed from the candidate gene CsACS2 and cosegregated with the M locus. The primer sequences are listed in Table S2.
The sequences of the entire genomic region of the putative peroxidase gene (p-CsPOD) and an ∼2.0-kb 5′ upstream region and 1.0-kb 3′ downstream region were identical among the four parent lines. Therefore, we concluded that this putative peroxidase gene in cucumber was not the M gene.
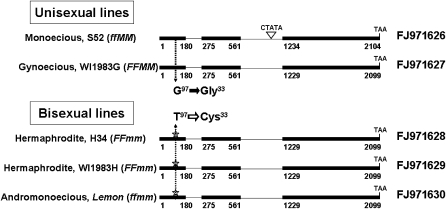
We sequenced the entire genomic section of CsACS2 along with a 1.9-kb 5′ upstream region and a 620-bp 3′ downstream region from all four parental plants. These sequences revealed two types of polymorphism in the four lines used for mapping (Figure 2). First, a 5-bp insertion/deletion difference was found in the second intron between parent plants S52 and H34. A sequence-characterized amplified region marker, S_CM2, was developed to cover this region (Figure 1c). We did not detect any recombination events after mapping in population 5234 (2700 individuals). The second type of polymorphism, a G97T conversion, was identified in the first exon of this gene in all four parent lines (WI1983G and S52 had G, while WI1983H and H34 had T). On the basis of this polymorphism, we developed the SNP marker SNPCM2, which expectedly cosegregated with the M/m locus in the final population of 5530 individuals (Figure 1c). Then we used this SNP marker to analyze a germplasm collection of 65 C. sativus cultivars obtained from different parts of the world (Table S1). All unisexual cultivars (21 monoecious cultivars and 41 gynoecious cultivars) were found to contain G, which was the dominant polymorph. However, a well-known andromonoecious line, Lemon, and the hermaphrodite inbred lines WI1983H and H34 were found to contain the recessive T. The quantitative reverse transcriptase–polymerase chain reaction analysis for CsACS2 indicated significant differences (over ninefold) in the expression levels between the near-isogenic lines (NILs) for the M locus, H34 (FFmm) and M34 (FFMM) (Figure S2). These evidences strongly suggested that CsACS2 was the candidate gene for M.
Figure 2.—
Genomic structures of the M/m alleles. The monoecious plant line S52 had a unique 5-bp (CTATA) insertion/deletion in the second intron. Both of the unisexual parental lines, S52 and WI1983G, had the conserved “G.” The hermaphrodite lines, H34 and WI1983H, with an additional andromonoecious line, Lemon, shared the same framework with WI1983G, except for the nucleotide transition from G to T, which led to the missense mutation of Gly33Cys in the deduced amino acid sequence. Solid boxes represent the full-length mRNA sequence, and thin lines indicate introns. Dashed lines showed the conserved nucleotides in the dominant and recessive (marked with asterisk) alleles.
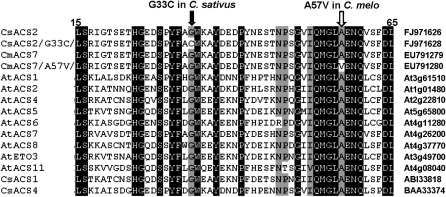
The G97T conversion in the coding region of CsACS2 caused a substitution at position 33 in the protein, thereby producing differences among the proteins produced by the unisexual (gynoecious and monoecious) M (Gly at position 33) and the bisexual m (Cys at position 33) alleles. As shown in Figure 3, Gly was conserved in nearly all the functional 1-aminocyclopropane-1-carboxylic acid (ACC) synthases in plants, while the Gly33Cys isoform may show changed or reduced enzymatic activity.
Figure 3.—
Alignment of the deduced amino acid sequence of ACS isozymes from cucumber, melon, and Arabidopsis thaliana. The conserved-residue conversions, G33C in C. sativus for the M/m gene and A57V in C. melo for the A/a gene, are shown above the alignment. Identical amino acids are against a solid background, and amino acid similarity levels >75% are against a shaded background.
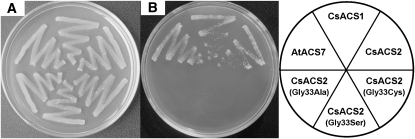
Because of the technical difficulties in cucumber transformation, we used the Escherichia coli system to analyze the activity of the putative ACC synthases encoded by the M and m alleles. Various reports have shown that plant ACC synthase can serve the same function in E. coli (Van Der Straeten et al. 1990; Tarun et al. 1998; Yamagami et al. 2003). Tarun et al. (1998) integrated the ACC deaminase gene into the genome of the mutant strain JHM544, which is an Ile auxotroph, and developed the E. coli strain JAde 6, which is effective for testing putative ACC synthases. To test the potential loss-of-function mutation attributed to the Gly33Cys conversion in CsACS2, we subcloned two full-length coding sequence fragments derived from the cucumber lines M34 (FFMM) and H34 (FFmm) into the expression vector pQE-30. Additionally, we induced artificial mutagenesis at position 33 of the original CsACS2 to produce two mutant proteins (Gly33Ser and Gly33Ala), which were used to study the putative conserved active site of ACC synthase. Furthermore, CsACS1G, encoded by the well-characterized F gene (Knopf and Trebitsh 2006), was used as a positive control in the cucumber plant to test the biochemical activity of the enzyme. After a 3-day incubation at 37° on basic M9 media, CsACS1G and CsACS2 demonstrated ACC synthase activity, while the other three CsACS2 isoforms, namely, Gly33Cys, Gly33Ser, and Gly33Ala, did not show any activity (Figure 4). Therefore, the Gly33Cys conversion may cause a functional alteration resulting in a new phenotype in cucumber plants.
Figure 4.—
Complementation of the E. coli integrative strain JAde 6 with various CsACS and AtACS7 cDNAs. Six plasmids were used to transform E. coli JAde 6 as described in File S1. The streaked sections in A and B correspond to the cDNAs in the pie chart. All the plates were grown at 37° overnight (on LB media) or for 3 days (on minimal media) and then photographed. (A) Growth of all six strains of JAde 6 on LB media for positive control. (B) Growth of all six strains of JAde 6 on minimal media.
DISCUSSION
The conserved ACC synthase gene CsACS2 was first cloned by Kamachi et al. (1997) using a homology-based cloning strategy. This gene was subsequently reported in many studies (Mathooko et al. 1999; Kamachi et al. 2000; Yamasaki et al. 2000, 2001, 2003a,b; Saito et al. 2007). Since the mm genotype was associated with significant expression, CsACS2 was first defined as a downstream product regulated by the M gene. However, despite this aberration, the information from these studies was very useful in clarifying the regulated expression and functional pattern of this gene. Kamachi et al. (1997) reported that the CsACS2 mRNA was detected at the apexes, and the results of the Southern blot analysis suggested that a single copy of the gene was present in the examined cucumber genomes. Furthermore, Kamachi et al. (2000) noted that the expression of CsACS2 at the apexes was localized to the floral buds that developed into female flowers. Yamasaki et al. (2000) found a positive correlation between CsACS2 expression and ethylene evolution in shoot apexes. Later, Yamasaki et al. (2001) showed that ethylene caused substantial increases in the accumulation of CsACS2 mRNA and inhibited stamen development.
Saito et al. (2007) performed comprehensive in situ hybridizations to study the expression pattern of CsACS2. The results revealed that the CsACS2 transcript began to accumulate just beneath the pistil primordia of flower buds at the bisexual stage, and this accumulation coincided with the sex-determination stage in cucumbers (Bai et al. 2004). During the later developmental stages, the accumulation of CsACS2 was correlated with the establishment of female flowers. These results may imply a relationship between the expression of CsACS2 and the continuous arrestment of stamen development.
Although we could not perform the complementation test in cucumber plants, the analysis of the physical location of the gene, the enzymology results obtained from this study, and the results obtained from previous studies indicate that the CsACS2 gene is the M gene in cucumbers.
To our knowledge, the presence of a gene for controlling unisexual expression is unique to the Cucurbitaceae plants, particularly cucumber and melon (Tanurdzic and Banks 2004). Therefore, the unisexuality-determining genes M (in cucumbers) and A (in melons) may represent a homology. Boualem et al. (2008) described an experiment to clone the a gene in melon; homology analysis showed that the two dominant alleles encoded almost identical amino acid sequences, except for eight residues, all of which were in nonconserved active sites (Figure 3). Since the BLASTX analysis did not show any other proteins with such a high degree of similarity (98%), they may be the ortholog gene in the Cucumis plants. Nevertheless, in addition to the different expression patterns of m and a, there were differences between the protein sequences of the natural mutants [Gly33Cys in cucumbers and Ala57Val (Gly19Glu for the targeting-induced local lesions in genomes analysis) in melons]. These results suggest that the evolution of the functional unisexuality-determination genes occurred prior to the differentiation of the two Cucumis species, namely, cucumber and melon. Subsequently, the two species independently evolved two different nonfunctioning recessive mutants.
In cucumber, the F gene and the M gene were shown to encode 1-aminocyclopropane-1-carboxylate synthase, the key regulatory enzyme in the ethylene biosynthetic pathway. Therefore, in cucumber plants, the genes related to the ethylene-signaling pathways might be related to the unique development of sex organs.
Acknowledgments
We thank Rentao Song (Shanghai University, Shanghai, China) for his technical assistance in the BAC library; Athanasios Theologis and Guixia Yu (University of California, Berkeley) for their helpful assistance with E. coli JAde 6; Lingxia Zhao (Shanghai Jiaotong University, Shanghai, China) for the vector pQE-30; and Qiguang Wen (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China) for the chemical reagent 1-aminocyclopropane-1-carboxylic acid. The authors also thank Zhangjun Fei (Cornell University, Ithaca, NY) for his critical reading. This work was supported by the National Natural Science Foundation of China (no. 30871707), National “863” Project (no. 2008AA10Z150), Ministry of Agriculture “948” Program (no. 2008-Z42), Ministry of Science and Technology (no. 2006DFA32140), and the Shanghai Leading Academic Discipline Project (no. B209).
Sequence data have been deposited with the EMBL/GenBank Data Libraries under accession nos. FJ529216 and FJ971626–FJ971630.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.104737/DC1.
References
- Atsmon, D., 1968. The interaction of genetic, environmental, and hormonal factors in stem elongation and floral development of cucumber plants. Ann. Bot. 32 877–882. [Google Scholar]
- Bai, S. L., Y. B. Peng, J. X. Cui, H. T. Gu, L. Y. Xu et al., 2004. Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220 230–240. [DOI] [PubMed] [Google Scholar]
- Boualem, A., M. Fergany, R. Fernandez, C. Troadec, A. Martin et al., 2008. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321 836–838. [DOI] [PubMed] [Google Scholar]
- Galun, E., 1961. Study of the inheritance of sex expression in the cucumber: the interaction of major genes with modifying genetic and non-genetic factors. Genetica 32 134–163. [Google Scholar]
- Kamachi, S., H. Sekimoto, N. Kondo and S. Sakai, 1997. Cloning of a cDNA for a 1-aminocyclopropane-1-carboxylate synthase that is expressed during development of female flowers at the apices of Cucumis sativus L. Plant Cell Physiol. 38 1197–1206. [DOI] [PubMed] [Google Scholar]
- Kamachi, S., H. Mizusawa, S. Matsuura and S. Sakai, 2000. Expression of two 1-aminocyclopropane-1-carboxylate synthase genes, CS-ACS1 and CS-ACS2, correlated with sex phenotypes in Cucumis plants (Cucumis sativus L.). Plant Biotechnol. 17 69–74. [Google Scholar]
- Knopf, R. R., and T. Trebitsh, 2006. The female-specific CS-ACS1G gene of cucumber: a case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol. 47 1217–1228. [DOI] [PubMed] [Google Scholar]
- Li, Z., J. S. Pan, Y. Guan, Q. Y. Tao, H. L. He et al., 2008. Development and fine mapping of three co-dominant SCAR markers linked to the M/m gene in the cucumber plant (Cucumis sativus L.). Theor. Appl. Genet. 117 1253–1260. [DOI] [PubMed] [Google Scholar]
- Liu, S. Q., L. Xu, Z. Q. Jia, Y. Xu, Q. Yang et al., 2008. Genetic association of ETHYLENE-INSENSITIVE3-like sequence with the sex-determining M locus in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 117 927–933. [DOI] [PubMed] [Google Scholar]
- Mathooko, F. M., M. W. Mwaniki, A. Nakatsuka, S. Shiomi, Y. Kubo et al., 1999. Expression characteristics of CS-ACS1, CS-ACS2 and CS-ACS3, three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in cucumber (Cucumis sativus L.) fruit under carbon dioxide stress. Plant Cell Physiol. 40(2): 164–172. [DOI] [PubMed] [Google Scholar]
- Mibus, H., and T. Tatlioglu, 2004. Molecular characterization and isolation of the F/f gene of femaleness in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 109 1669–1676. [DOI] [PubMed] [Google Scholar]
- Perl-Treves, R., 1999. Male to female conversion along the cucumber shoot: approaches to studying sex genes and floral development in Cucumis sativus, pp. 189–286 in Sex Determination in Plants, edited by C. C. Ainsworth. BIOS, Oxford, UK.
- Robinson, R. W., H. M. Munger, T. W. Whitaker and G. M. Bohn, 1976. Genes of Cucubitaceae. Hortscience 11 554–568. [Google Scholar]
- Saito, S., N. Fujii, Y. Miyazawa, S. Yamasaki, S. Matsuura et al., 2007. Correlation between development of female flower buds and expression of the CS-ACS2 gene in cucumber plants. J. Exp. Bot. 58 2897–2907. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., and M. J. Jaffe, 1984. Further studies of auxin and ACC induced feminization in the cucumber plant using ethylene inhibitors. Phyton 44 81–86. [PubMed] [Google Scholar]
- Takahashi, H., T. Saito and H. Suge, 1983. Separation of effects of photoperiod and hormones on sex expression in cucumber. Plant Cell Physiol. 24 142–154. [Google Scholar]
- Tanurdzic, M., and J. A. Banks, 2004. Sex-determining mechanisms in land plants. Plant Cell 16 S61–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, A. S., J. S. Lee and A. Theologis, 1998. Random mutagenesis of 1-aminocyclopropane-1-carboxylate synthase: a key enzyme in ethylene biosynthesis. Proc. Natl. Acad. Sci. USA 95 9796–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh, T., J. E. Staub and S. D. O'Neill, 1997. Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 113 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao, T. H., 1988. Sex expression in flowering. Acta Phytophysiol. Sinica 14 203–207. [Google Scholar]
- Van der Straeten, D., L. Van Wiemeersch, H. M. Goodman and M. Van Montagu, 1990. Cloning and sequence of two different cDNAs encoding 1-aminocyclopropane-1-carboxylate synthase in tomato. Proc. Natl. Acad. Sci. USA 87 4859–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami, T., A. Tsuchisaka, K. Yamada, W. F. Haddon, L. A. Harden et al., 2003. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278 49102–49112. [DOI] [PubMed] [Google Scholar]
- Yamasaki, S., N. Fujii and H. Takahashi, 2000. The ethylene-regulated expression of CS-ETR2 and CS-ERS genes in cucumber plants and their possible involvement with sex expression in flowers. Plant Cell Physiol. 41(5): 608–616. [DOI] [PubMed] [Google Scholar]
- Yamasaki, S., N. Fujii, S. Matsuura, H. Mizusawa and H. Takahashi, 2001. The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant Cell Physiol. 42 608–619. [DOI] [PubMed] [Google Scholar]
- Yamasaki, S., N. Fujii and H. Takahashi, 2003. a Characterization of ethylene effects on sex determination in cucumber plants. Sex Plant Reprod. 16 103–111. [Google Scholar]
- Yamasaki, S., N. Fujii and H. Takahashi, 2003. b Photoperiodic regulation of CS-ACS2, CS-ACS4 and CS-ERS gene expression contributes to the femaleness of cucumber flowers through diurnal ethylene production under short-day conditions. Plant Cell Environ. 26 537–546. [Google Scholar]
- Yamasaki, S., N. Fujii and H. Takahashi, 2005. Hormonal regulation of sex expression in plants. Vitam. Horm. 72 79–110. [DOI] [PubMed] [Google Scholar]