Abstract
DNA sequence analysis and genetic mapping of loci from mating-type-specific chromosomes of the smut fungus Microbotryum violaceum demonstrated that the nonrecombining mating-type-specific region in this species comprises ∼25% (∼1 Mb) of the chromosome length. Divergence between homologous mating-type-linked genes in this region varies between 0 and 8.6%, resembling the evolutionary strata of vertebrate and plant sex chromosomes.
EVOLUTION of mating types or sex-determining systems often involves the suppression of recombination around the primary sex-determining or mating-type-determining locus. In animals and plants, it is often an entire or almost entire chromosome (Y or W in male or female heterogametic species, respectively) that ceases to recombine with its homologous (X or Z) chromosome (Charlesworth and Charlesworth 2000; Charlesworth 2008). Self-incompatibility loci in plants are also thought to be located in regions of suppressed recombination (Charlesworth et al. 2005; Kamau and Charlesworth 2005; Kamau et al. 2007; Li et al. 2007; Yang et al. 2007). Regardless of the phylogenetic position of a species, such nonrecombining regions are known to follow similar evolutionary trajectories. The nonrecombining region on the sex-specific chromosome expands in several steps, forming evolutionary strata—regions of different X/Y (or Z/W) divergence (Lahn and Page 1999; Handley et al. 2004; Sandstedt and Tucker 2004; Nicolas et al. 2005)—and genes in the nonrecombining regions gradually accumulate deleterious mutations that eventually render them dysfunctional (Charlesworth and Charlesworth 2005; Charlesworth 2008).
Fungal mating-type systems are very diverse, with the number of mating types varying from two to several hundred (Casselton 2002). Like sex chromosomes in several animals and plants, suppressed recombination has evolved in regions near fungal mating-type loci, including in Ustilago hordei (Lee et al. 1999), Cryptococcus neoformans (Lengeler et al. 2002), and Neurospora tetrasperma (Menkis et al. 2008). These species have two mating types, but no morphologically distinct sexes. The mating-type locus (the region of suppressed recombination) of C. neoformans is small (∼100 kb) compared with known sex chromosomes and contains only ∼20 genes that, unlike many sex chromosomes (Y or W chromosomes), show no obvious signs of genetic degeneration (Lengeler et al. 2002; Fraser et al. 2004). Judging from the divergence between the homologous genes on the two mating-type-specific chromosomes, C. neoformans started to evolve sex chromosomes a long time ago because silent divergence between the two mating types in the most ancient region exceeds 100% (Fraser et al. 2004). Genes in the younger mating-type-specific region are much less diverged between the two sex chromosomes, suggesting that the evolution of the sex locus in C. neoformans might have proceeded through several steps. The nonrecombining region around the mating-type locus of N. tetrasperma is much larger than in C. neoformans (at least 6.6 Mb), and silent divergence between homologous genes on the mating-type-specific chromosomes ranges from zero to 9%, demonstrating that these mating-type-specific chromosomes evolved recently (Menkis et al. 2008).
M. violaceum, which causes anther smut disease in Silene latifolia and other species in the family Caryophyllaceae, has two mating types, A1 and A2 (reviewed by Giraud et al. 2008), which are determined by the presence of mating-type-specific chromosomes (hereafter A1 and A2 chromosomes, or sex chromosomes) in the haploid stage of the life cycle (Hood 2002; Hood et al. 2004). The A1 and A2 chromosomes are distinguishable by size in pulsed-field electrophoresis, and it is possible to isolate individual chromosomes electrophoretically (Hood et al. 2004). Random fragments of A1 and A2 chromosomes have previously been isolated from mating-type-specific bands of pulsed-field separated chromosomes of M. violaceum (Hood et al. 2004). These fragments were assumed to be linked to mating type. The same method was used to isolate fragments of non-mating-type-specific chromosomes. On the basis of the analysis of their sequences, (Hood et al. 2004) proposed that mating-type-specific chromosomes in M. violaceum might be degenerate because they contained a lower proportion of protein-coding genes than other chromosomes. However, it was not determined whether the sequences isolated from the mating-type chromosomes originated from the mating-type-specific or from the recombining regions (Hood et al. 2004), and the relative sizes of these regions are not known for these M. violaceum chromosomes. We tested the mating-type specificity of 86 of these fragments and demonstrate that fewer than a quarter of these loci are located in the mating-type-specific region, suggesting that the nonrecombining region on the A1 and A2 chromosomes is quite small, while the rest of the chromosome probably recombines (like pseudoautosomal regions of sex chromosomes) and is therefore not expected to undergo genetic degeneration. Genetic mapping confirms the presence of two pseudoautosomal regions in the M. violaceum mating-type-specific chromosomes.
As these chromosomes are mating type specific in the haploid stage of M. violaceum, mating-type-specific loci (or DNA fragments) can be identified by testing whether they are present exclusively in A1 or A2 haploid strains. We therefore prepared haploid A1 and A2 M. violaceum cultures from S. latifolia plants from two geographically remote locations (accessions Sl405 from Sweden and Sl127 from the French Pyrenees). Haploid sporidial cultures were isolated by a standard dilution method (Kaltz and Shykoff 1997; Oudemans and Alexander 1998). Mating types were determined by PCR amplification of each culture with primers designed for A1 and A2 pheromone receptor genes linked to A1 and A2 mating types (Yockteng et al. 2007). The primers were as follows: 5′-TGGCATCCCTCAATGTTTCC-3′ and 5′-CACCTTTTGATGAGAGGCCG-3′ for the A1 pheromone receptor (GenBank accession no. EF584742) and 5′-TGACGAGAGCATTCCTACCG-3′ and 5′-GAAGCGGAACTTGCCTTTCT-3′ for the A2 pheromone receptor (GenBank accession no. EF584741). Cultures with PCR product amplified only from an A1 or A2 pheromone receptor gene were selected for further use. The mating types of the cultures were verified by conjugating them in all combinations.
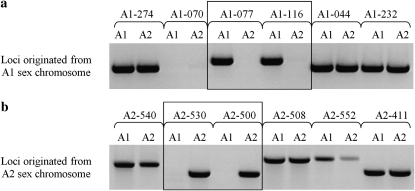
The GenBank nucleotide database was searched using BLAST for sequences similar to those isolated by Hood et al. (2004). Sequences with similarity to transposable elements (TE) and other repeats were excluded. The resulting set of nonredundant sequences was used to design PCR primers for 98 fragments. Half of these were originally isolated from the A1 and half from the A2 chromosomes and are hereafter called A1-NNN or A2-NNN (where NNN is the locus number; supporting information, Table S1), which does not imply that these loci are A1 or A2 specific, but merely indicates that they were originally isolated from the A1 or A2 chromosomes. Amplification of these regions from new A1 and A2 M. violaceum cultures, independently isolated by ourselves, revealed that only 5 of the 49 loci isolated from the A1 chromosome are indeed A1 specific and only 6 of 49 isolated from the A2 chromosome are A2 specific. All other loci amplified from both A1 and A2 cultures. Figure 1 illustrates some of these results from the Swedish sample (Sl405).
Figure 1.—
Testing of mating-type specificity for loci isolated from A1 and A2 chromosomes. (a) PCR amplifications from haploid cultures from Sl405 using primers designed from six A1-originated loci. Loci in which a PCR product could be amplified only from A1 cultures (boxed) were classified as specific to mating type A1. (b) PCR tests of six A2-originated loci on the same set of haploids as in a. Loci in which a PCR product amplified only from A2 cultures (boxed) were classified as specific to mating type A2. Loci amplified from both A1 and A2 cultures are not mating type specific.
The fragments that amplified from both A1 and A2 mating types may be in recombining regions, or they could be present in mating-type-specific regions on both A1 and A2 chromosomes. If they are in recombining regions, the A1- and A2-linked homologs should not be diverged from each other, but if they are in nonrecombining, mating-type-specific regions, the divergence of the A1- and A2-linked homologs should be roughly proportional to the time since recombination stopped in the region. We therefore sequenced and compared PCR fragments amplified from the two mating types of Sl405 or Sl127 cultures (GenBank accession nos. FI855822–FI856001). Sequencing of PCR products showed that 12 (4 A1 and 8 A2) loci have more than one copy, and they were excluded from further analysis. Sequences of 61 loci were identical between the A1 and A2 strains, and four loci demonstrated low total divergence (0.24–0.61%) between the two mating types (Table 1). This level of divergence is similar to the background level of DNA polymorphism in M. violaceum populations, which was measured in 16 mating-type-linked, pseudoautosomal and autosomal genes in a sample of 14 M. violaceum accessions sampled across Europe (A. Votintseva and D. Filatov, unpublished results). Thus, these loci might be located in the recombining part of the mating-type-specific chromosomes. Ten of 75 loci that amplified in both mating types demonstrated multiple polymorphisms fixed between the mating types rather than between the locations. Given that the strains that we used in the analysis originated from two geographically distant locations, it is highly unlikely that multiple polymorphisms distinguishing the A1 and A2 sequences arose purely by chance; thus, these loci are probably located in the nonrecombining mating-type-specific region of the M. violaceum A1 and A2 chromosomes.
TABLE 1.
Loci from mating-type-specific chromosomes of M. violaceum used for PCR analysis and genetic map construction
| With nonzero A1/A2 divergenceb
|
|||||
|---|---|---|---|---|---|
| Loci | Mating type specific | <1% | >1% | With zero A1/A2 divergenceb | Total |
| A1a | 5 | 2 (1) | 3 (3) | 35 (3) | 45 (7) |
| A2a | 6 | 2 (0) | 7 (7) | 26 (3) | 41 (10) |
| Subtotal | 4 (1) | 10 (10) | |||
| Total | 11 | 14 (11) | 61 (6) | 86 (17) | |
A1, loci originated from the A1 sex chromosome; A2, loci originated from the A2 sex chromosome.
The number of loci used for genetic map construction is in parentheses.
To confirm the mating-type-specific or pseudoautosomal locations of the loci with and without A1/A2 divergence, we conducted genetic mapping in a family of 99 individuals, 50 of which were of mating type A1 and 49 of mating type A2. The family was generated by a cross between A1 and A2 M. violaceum strains from S. latifolia accessions Sl405 (Sweden) and Sl127 (France), respectively. The choice of strains from geographically distant locations was motivated by the hope of maximizing the number of DNA sequence differences between them that can be used as molecular genetic markers in segregation analysis. We inoculated S. latifolia seedlings with sporidial cultures of both mating types. For inoculation, petri dishes with 12-day-old seedlings of S. latifolia were flooded with 2.5 ml of inoculum suspension. Inoculum suspension consisted of equal volumes of the A1 and A2 sporidial cultures that were mixed and conjugated overnight at 14° under rotation (Biere and Honders 1996; Van Putten et al. 2003). Seedlings were potted 3 days after inoculation. Two months later, teliospores were collected from the flowers of the infected plant and grown in petri dishes on 3.6% potato dextrose agar medium. Haploid sporidia formed after meiosis were isolated and grown as separate cultures for DNA extraction. The mating types of single sporidia cultures were identified as described above. The loci analyzed in the segregation analysis were sequenced in the two parental haploid strains and in 99 (50 A1 and 49 A2) haploid strains that were generated in the cross. Single nucleotide differences between the parental strains were used as molecular genetic markers for segregation analysis in the progeny. The genetic map was constructed using MAPMAKER/EXP v3.0 (Lincoln et al. 1992) and MapDisto v1.7 (http://mapdisto.free.fr/).
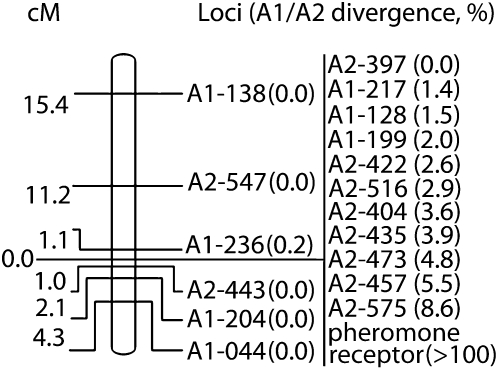
The resulting genetic map is shown in Figure 2. As expected, no recombination was observed between the 10 loci with diverged A1- and A2-linked copies. In addition, one marker with no A1/A2 divergence, A2-397, was also completely linked to the loci with significant A1/A2 divergence. This locus either may be very tightly linked to the nonrecombining mating-type-specific region or may have been added to that region more recently than the loci that had already accumulated some divergence between the alleles in the two mating types. The mating-type-specific pheromone receptor locus (Devier et al. 2009) and 11 mating-type-specific loci are also located in this nonrecombining region (Figure 2). Interestingly, the cluster of nonrecombining markers is flanked on both sides with markers that recombine in meiosis, demonstrating that there are pseudoautosomal regions on both ends of the mating-type-specific chromosomes.
Figure 2.—
Genetic map of the mating-type-determining chromosome in M. violaceum. Genetic distance (in centimorgans) and the relative positions of the markers are shown to the left and the right of the chromosome, respectively. The position of the nonrecombining region corresponds to the cluster of linked markers shown on the right of the figure. Total A1/A2 divergence is shown in parentheses. Eleven mating-type-specific markers (for which sequences are available from only one mating type), located in the nonrecombining mating-type-specific region, are not shown.
Our results demonstrate that although the loci reported by Hood et al. (2004) were isolated from the A1 and A2 chromosomes, most of these loci are not located in the nonrecombining mating-type-specific regions. In fact, the nonrecombining region might be relatively small: of 86 tested fragments, only 21 appeared to be either mating type specific or linked to the mating-type locus. Assuming that these loci represent a random set of DNA fragments isolated from the A1 and A2 chromosomes, it is possible to estimate the size of the nonrecombining region using the binomial distribution: the nonrecombining region is expected to be 24.4% (95% CI: 16.7–33.6%) of the chromosome length. As the sizes of the A1 and A2 chromosomes are ∼3.4 and 4.2 Mb long (Hood 2002; Hood et al. 2004), the nonrecombining region might be ∼1 Mb long.
Interestingly, total A1/A2 divergence for the 11 loci with A1- and A2-linked copies mapped to the nonrecombining region varied from 0% to 8.6% (Table 2 and Figure 2). In addition, 11 loci amplified from only one mating type. These genes could represent degenerated genes, some of which degenerated in A1 strains, and some in A2 strains. Alternatively, they might be highly diverged genes, such that the PCR primers amplify only one allele, and not the other. Variation in divergence may be the result of the stepwise cessation of recombination between the A1 and A2 chromosomes in M. violaceum, resembling the evolutionary strata reported for human, chicken, and white campion sex chromosomes (Lahn and Page 1999; Handley et al. 2004; Bergero et al. 2007). However, only the differences between the most and the least diverged loci are statistically significant (Table 3). Nevertheless, taking into account the much higher A1/A2 divergence of the pheromone receptor locus for which the homologs from the two mating types are barely alignable (Devier et al. 2009), the M. violaceum mating-type region has at least three strata: one oldest stratum, including the pheromone receptor locus; a younger stratum with ∼5–9% A1/A2 divergence; and the youngest stratum with 1–4% divergence between the two mating types. There may also be an additional very recently evolved stratum containing the locus named A2-397, which is also present in all A1 strains tested, with no fixed differences between the A1 and A2 strains (Table 3).
TABLE 2.
Loci with nonzero divergence between the A1 and A2 mating types of M. violaceum
| No. of sites analyzed | Within A1
|
Within A2
|
Fixed differences between A1 and A2 | A1/A2 divergence (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Locia | Sb total | S | π (%)c | S | π (%)c | ||||
| A1/A2 divergence <1% | A1-236 | 456 | 3 | 0 | 0 | 2 | 0.44 | 1 | 0.44 |
| A1-045 | 654 | 4 | 0 | 0 | 0 | 0 | 4 | 0.61 | |
| A2-568 | 413 | 2 | 2 | 0.48 | 2 | 0.48 | 0 | 0.24 | |
| A2-411 | 480 | 2 | 1 | 0.21 | 0 | 0 | 1 | 0.31 | |
| A1/A2 divergence >1% | A1-217 | 667 | 9 | 0 | 0 | 0 | 0 | 9 | 1.35 |
| A1-128 | 569 | 9 | 0 | 0 | 1 | 0.18 | 8 | 1.49 | |
| A1-199 | 618 | 13 | 0 | 0 | 1 | 0.16 | 12 | 2.02 | |
| A2-422 | 344 | 9 | 0 | 0 | 0 | 0 | 9 | 2.62 | |
| A2-516 | 470 | 14 | 0 | 0 | 0 | 0 | 14 | 2.98 | |
| A2-404 | 508 | 20 | 0 | 0 | 3 | 0.59 | 17 | 3.64 | |
| A2-435 | 506 | 22 | 2 | 0.39 | 2 | 0.39 | 18 | 3.95 | |
| A2-473 | 457 | 23 | 1 | 0.22 | 1 | 0.22 | 21 | 4.81 | |
| A2-457 | 303 | 17 | 1 | 0.33 | 0 | 0 | 16 | 5.54 | |
| A2-575 | 503 | 47 | 5 | 0.99 | 3 | 0.59 | 39 | 8.55 | |
A1, loci originated from the A1 sex chromosome; A2, loci originated from the A2 sex chromosome.
S, number of polymorphic sites.
π (%), average number of differences per 100 nucleotides.
TABLE 3.
P-values for the 2 × 2 G-tests for significance of differences in A1/A2 divergence between the loci in the nonrecombining region
| La | Sb | Locus | A2-397 | A1-217 | A1-128 | A1-199 | A2-422 | A2-516 | A2-404 | A2-435 | A2-473 | A2-457 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 519 | 0 | A2-397 | ||||||||||
| 667 | 9 | A1-217 | 0.006 | |||||||||
| 569 | 8 | A1-128 | 0.006 | 0.93 | ||||||||
| 618 | 12 | A1-199 | 0.0007 | 0.41 | 0.48 | |||||||
| 344 | 9 | A2-422 | 0.0003 | 0.17 | 0.21 | 0.51 | ||||||
| 470 | 14 | A2-516 | 0.00003 | 0.06 | 0.086 | 0.28 | 0.76 | |||||
| 508 | 17 | A2-404 | 0 | 0.025 | 0.038 | 0.15 | 0.55 | 0.75 | ||||
| 506 | 18 | A2-435 | 0 | 0.015 | 0.024 | 0.104 | 0.45 | 0.62 | 0.86 | |||
| 457 | 21 | A2-473 | 0 | 0.001 | 0.003 | 0.0163 | 0.15 | 0.21 | 0.34 | 0.43 | ||
| 303 | 16 | A2-457 | 0 | 0.0009 | 0.0017 | 0.0097 | 0.09 | 0.13 | 0.203 | 0.26 | 0.69 | |
| 503 | 39 | A2-575 | 0 | 0 | 0 | 0.00001 | 0.002 | 0.002 | 0.003 | 0.006 | 0.055 | 0.199 |
P-values <0.05 are in boldface type.
L, the length of the region compared.
S, the number of nucleotide differences observed.
As most of the loci isolated from the A1 and A2 chromosomes recombine in meiosis, they are not expected to degenerate. Thus, the observation of a higher proportion of TEs in these loci, compared to other chromosomes (Hood et al. 2004), is unlikely to reflect genetic degeneration attributable to a lack of recombination in these loci. A higher abundance of TEs in the sequences isolated from the A1 and A2 chromosomes, as reported by Hood et al. (2004), may simply reflect variation in the TE density across the genome. Thus, it remains to be seen whether M. violaceum mating-type-specific regions degenerate, similar to vertebrate Y (or W) chromosomes, or remain largely intact, as in C. neoformans (Lengeler et al. 2002). If the latter were the case, it may suggest that nonrecombining regions in fungi do not necessarily follow the same degenerative path as animal Y and W chromosomes. The analysis of sequences from the M. violaceum genome (and perhaps other fungal genomes) will hopefully provide the answer to this question.
The lack of degeneration of mating-type-specific regions in C. neoformans may be due to the relatively small size of the nonrecombining regions. The 20 genes present in this region may not be sufficient for the operation of such detrimental population genetic processes as background selection or Muller's ratchet because the speed of these processes depends critically on the number of active genes linked together (Charlesworth 2008). Larger mating-type-specific regions in M. violaceum might contain more genes; thus, more active genetic degeneration may be expected in this species. Indeed, many strains of M. violaceum show haplolethality linked to one of the mating types (Hood and Antonivics 2000; Thomas et al. 2003; Tellier et al. 2005), which may reflect the accumulation of deleterious mutations in the nonrecombining regions around the mating-type loci. Mating-type specificity of the markers that amplified in only A1 or A2 strains in this study may also reflect genetic degeneration.
Another factor that may potentially prevent degeneration of genes linked to mating-type loci in fungi is the haploid expression of genes in these regions. In animals, many Y-linked genes have functional homologs on the X chromosome, and loss of the Y-linked gene may be compensated for by expression of the X-linked homologs. The haploid stage in an animal's life cycle is very short, and very few genes are actively expressed in animal gametes (Schultz et al. 2003). In plants, on the other hand, a significant proportion of the genome is expressed in pollen (da Costa-Nunes and Grossniklaus 2003), and so the loss of Y-linked genes expressed in gametes may be more detrimental than in animals. Indeed, most genes isolated from the white campion X chromosome have intact Y-linked copies (Filatov 2005; Bergero et al. 2007), but due to the small number of genes available, it is still unclear whether genetic degeneration of Y-linked genes is indeed slower in this species (and in plants generally) compared to animal Y chromosomes. Haploid expression could be an even more powerful force in fungi and other organisms with haploid sexes, such as bryophytes, as most genes are expressed in the haploid stage. Further analysis of genetic degeneration in nonrecombining sex- or mating-type-specific regions in fungi and bryophytes will help to shed light on this question.
Acknowledgments
We thank Lorna Casselton, Deborah Charlesworth, and two anonymous reviewers for critical reading of the manuscript and helpful comments and Christopher Dixon for assistance in teliospore collection and correction of the manuscript. This project was funded by the University of Oxford John Fell Fund and by the Biotechnology and Biological Sciences Research Council.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103192/DC1.
References
- Bergero, R., A. Forrest, E. Kamau and D. Charlesworth, 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biere, A., and S. Honders, 1996. Host adaptation in the anther smut fungus Ustilago violacea (Microbotryum violaceum): infection success, spore production and alteration of floral traits on two host species and their F1-hybrid. Oecologia 107 307–320. [DOI] [PubMed] [Google Scholar]
- Casselton, L. A., 2002. Mate recognition in fungi. Heredity 88 142–147. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., 2008. Sex chromosome origins and evolution, pp. 207–240 in Evolutionary Genomics and Proteomics, edited by P. A. Pagel. Sinauer Associates, Sunderland, MA.
- Charlesworth, D., and B. Charlesworth, 2005. Sex chromosomes: evolution of the weird and wonderful. Curr. Biol. 15 R129–R131. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., X. Vekemans, V. Castric and S. Glemin, 2005. Plant self-incompatibility systems: a molecular evolutionary perspective. New Phytol. 168 61–69. [DOI] [PubMed] [Google Scholar]
- da Costa-Nunes, J. A., and U. Grossniklaus, 2003. Unveiling the gene-expression profile of pollen. Genome Biol. 5 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devier, B., G. Aguileta, M. E. Hood and T. Giraud, 2009. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics 181 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D. A., 2005. Isolation of genes from plant Y chromosomes. Methods Enzymol. 395 418–442. [DOI] [PubMed] [Google Scholar]
- Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler et al., 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2 e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud, T., R. Yockteng, M. Lopez-Villavicencio, G. Refregier and M. E. Hood, 2008. Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryot. Cell 7 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley, L. J., H. Ceplitis and H. Ellegren, 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E., 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E., and J. Antonivics, 2000. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea). Heredity 85 231–241. [DOI] [PubMed] [Google Scholar]
- Hood, M. E., J. Antonovics and B. Koskella, 2004. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics 168 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltz, O., and J. A. Shykoff, 1997. Sporidial mating-type ratios of teliospores from natural populations of the anther smut fungus Microbotryum (=Ustilago) violaceum. Int. J. Plant. Sci. 158 575–584. [Google Scholar]
- Kamau, E., and D. Charlesworth, 2005. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr. Biol. 15 1773–1778. [DOI] [PubMed] [Google Scholar]
- Kamau, E., B. Charlesworth and D. Charlesworth, 2007. Linkage disequilibrium and recombination rate estimates in the self-incompatibility region of Arabidopsis lyrata. Genetics 176 2357–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. Four evolutionary strata on the human X chromosome. Science 286 964–967. [DOI] [PubMed] [Google Scholar]
- Lee, N., G. Bakkeren, K. Wong, J. E. Sherwood and J. W. Kronstad, 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 96 15026–15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester et al., 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1 704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., M. Webster, M. Furuya and P. M. Gilmartin, 2007. Identification and characterization of pin and thrum alleles of two genes that co-segregate with the Primula S locus. Plant J. 51 18–31. [DOI] [PubMed] [Google Scholar]
- Lincoln, S., M. Daly and E. Lander, 1992. Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report. Whitehead Institute, Cambridge, MA.
- Menkis, A., D. J. Jacobson, T. Gustafsson and H. Johannesson, 2008. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 4 e1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, M., G. Marais, V. Hykelova, B. Janousek, V. Laporte et al., 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudemans, P. V., and H. M. Alexander, 1998. The distribution of mating-type bias in natural populations of the anther-smut Ustilago violacea on Silene alba in Virginia. Mycologia 90 372–381. [Google Scholar]
- Sandstedt, S. A., and P. K. Tucker, 2004. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 14 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, N., F. K. Hamra and D. L. Garbers, 2003. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. USA 100 12201–12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier, A., L. M. Villareal and T. Giraud, 2005. Maintenance of sex-linked deleterious alleles by selfing and group selection in metapopulations of the phytopathogenic fungus Microbotryum violaceum. Am. Nat. 165 577–589. [DOI] [PubMed] [Google Scholar]
- Thomas, A. J., J. A. Shykoff, O. Jonot and T. Giraud, 2003. Mating-type ratio bias in populations of the phytopathogenic fungus Microbotryum violaceum from several host species. Int. J. Plant Sci. 164 641–647. [Google Scholar]
- Van Putten, W. F., A. Biere and J. M. Van Damme, 2003. Intraspecific competition and mating between fungal strains of the anther smut Microbotryum violaceum from the host plants Silene latifolia and S. dioica. Evolution 57 766–776. [PubMed] [Google Scholar]
- Yang, Q., D. Zhang, Q. Li, Z. Cheng and Y. Xue, 2007. Heterochromatic and genetic features are consistent with recombination suppression of the self-incompatibility locus in Antirrhinum. Plant J. 51 140–151. [DOI] [PubMed] [Google Scholar]
- Yockteng, R., S. Marthey, H. Chiapello, A. Gendrault, M. E. Hood et al., 2007. Expressed sequences tags of the anther smut fungus, Microbotryum violaceum, identify mating and pathology genes. BMC Genomics 8 272. [DOI] [PMC free article] [PubMed] [Google Scholar]