Abstract
Copy number variation (CNV) contributes in phenotypically relevant ways to the genetic variability of many organisms. Cost-effective genomewide methods for identifying copy number variation are necessary to elucidate the contribution that these structural variants make to the genomes of model organisms. We have developed a novel approach for the identification of copy number variation by next generation sequencing. As a proof of concept our method has been applied to map the deletions of three Drosophila deficiency strains. We demonstrate that low sequence coverage is sufficient for identifying and mapping large deletions at kilobase resolution, suggesting that data generated from high-throughput sequencing experiments are sufficient for simultaneously analyzing many strains. Genomic DNA from two Drosophila deficiency stocks was barcoded and sequenced in multiplex, and the breakpoints associated with each deletion were successfully identified. The approach we describe is immediately applicable to the systematic exploration of copy number variation in model organisms and humans.
STRUCTURAL variation is known to contribute extensively to the genetic variability of humans, mammals, and many model organisms. One class of structural variant, termed copy number variation (CNV), includes deletions, duplications, insertions, and genomic rearrangements which affect the number of occurrences of a specific DNA sequence present in the genome (Redon et al. 2006). CNV is known to occur extensively in the Drosophila genome with functionally significant consequences (Bridges 1936; Dopman and Hartl 2007; Tibshirani and Wang 2008; Zhou et al. 2008). In one study of 15 Drosophila strains, as many as 10% of genes were observed to harbor CNVs (Emerson et al. 2008). Cryptic CNVs that affect the phenotype observed in a model organism have the potential to confound research on multiple levels. For example, a recent report indicates that terminal deletions on chromosome (chr) 2L are frequent among deficiency kit stocks with mutations on the second chromosome and that the associated deletion of lgl has distorted the results of several previous studies (Roegiers et al. 2009). Despite widespread existence of CNV, the biological consequences of this phenomenon remain largely unexplored due to the lack of efficient tools for detection and characterization.
Until recently, comparative genomic hybridization with whole-genome tiling arrays (array-CGH) was the primary method for characterizing CNVs (Carter 2007); however, several limitations for this platform reduce its efficacy and efficiency. First, cross-hybridization and reliance on intensity scores lead to data that are difficult to interpret. Second, custom array design and optimization is labor intensive and costly. Third, array-CGH methods can only detect CNV, not other complex rearrangements such as balanced translocations and inversions. Finally, the overall cost of array-CGH methods is relatively high, particularly when high-resolution, whole-genome tiling arrays are employed.
Direct sequencing using next-generation technology has several advantages that make it a potentially powerful alternative to array-CGH for identifying genomic structural variations, including deletions, duplications, and rearrangements (Campbell et al. 2008; Chiang et al. 2009). First, high-throughput sequencing methods overcome the inherent limitations of cross-hybridization and provide a digital count of sequence representation. Second, no prior knowledge or design work is necessary. Third, using paired-end sequencing it is possible to identify complex structural variations. Finally, the current cost of CNV discovery by sequencing is comparable or lower than that of array-CGH and is continuing to decline.
In this report, we describe a sequencing-based strategy for high-throughput, cost-effective, genomewide characterization of structural variation at fine resolution by employing the Illumina sequencing platform. Deletions in three deficiency fly stocks were successfully characterized and the associated breakpoints were accurately determined. As we demonstrate, high-throughput sequencing provides an ideal and cost-effective platform for CNV characterization.
MATERIALS AND METHODS
Fly stocks:
Fly stocks were raised on standard Drosophila media at room temperature (23°–25°). The dac4 deletion mutant was generated by X-ray mutagenesis on the b pr c px sp background and obtained from Graeme Mardon (Mardon et al. 1994). All other stocks used in this report are from the Bloomington Drosophila Stock Center and are described on FlyBase (http://flybase.org/reports/FBst0003779.html). The genotypes of stock no. 3779 and no. 7584 are described as Df(2L)Sd37/SM5 and w1118; Df(3L)Exel6105, P{XP-U}Exel6105/TM6B, Tb1, respectively. Df(2L)Sd37 was generated by X-ray mutagenesis and is cytologically described as a deletion between 37C6-37D1; 38A6-38B2 (Ganetzky 1977; Stathakis et al. 1995). Df(3L)Exel6105 was generated by recombination between two FRT bearing insertions resulting in a molecularly defined deletion 3L: 5359162, 5601375 (Parks et al. 2004). DNA used for genomic sequencing from these strains was obtained from flies heterozygote for the Df over the respective balancer chromosome. Wild type referred to in this manuscript is the w1118 strain obtained from the Bloomington Drosophila Stock Center. DNA used for genomic sequencing was obtained from male adult flies.
Sequencing:
Fly genomic DNA was prepared and sequenced using the Illumina Genome Analyzer according to previously described methodologies (Srivatsan et al. 2008). Sequence reads obtained were mapped to the Drosophila reference genome release 5.1 using the vendor provided Eland pipeline.
Barcoding for multiplex sequencing:
Solexa sequencing primers were modified by the addition of 3 bp (2 of which are unique) for the sequencing in multiplex experiments described in this report. These modified primers were used in library preparations such that the 5′ ends of sequencing products from each sample were standardized with a specific dinucleotide indicating their sample membership. Following multiplex sequencing, reads were separated in silico by a script that identified the leading dinucleotide tag, grouped the sequence products according to sample membership, and trimmed the barcode.
CNV analysis simulation:
Computer simulations were performed in which the dac4 sequencing reads were randomly sampled to generate data sets approximating various levels of sequencing coverage. Data sets were generated for 0.45x, 0.35x, 0.25x, 0.15x, 0.075x, 0.0375x, and 0.01875x with seven replicates each. In simulations CNV was determined by DNA copy, an R implementation of the circular binary segmentation algorithm, which was found to be highly specific and accurate on the basis of self-vs.-self tests and discovery of the dac4 breakpoints (Olshen et al. 2004; Venkatraman and Olshen 2007). To determine the effect of read coverage on the ability of deletion detection and breakpoint mapping, CNV analyses of these data sets were performed at 1-kb and 3-kb average window sizes.
Validation of breakpoints:
PCR primers were designed flanking the breakpoints predicted by CNV analysis (Rozen and Skaletsky 2000). Amplified products were sequenced by traditional Sanger sequencing and subsequently mapped to the Drosophila reference genome to identify the molecular position of breakpoints. For primers used in this study, see Table S2.
Additional methods:
See File S1.
RESULTS
Characterization of the dac4 deletion mutant by direct shotgun sequencing:
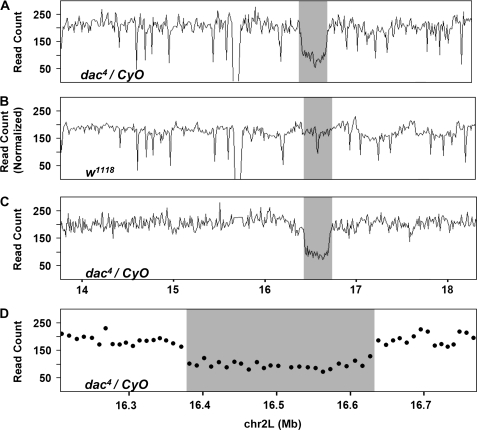
To test the efficiency and accuracy of mapping chromosomal deletions in Drosophila by high-throughput sequencing, we set out to identify the breakpoints for an existing deletion. The dac4 deletion was generated by X-ray mutagenesis and was mapped by analysis of polytene chromosomes to the 35F–36A region that includes the dac gene (Mardon et al. 1994). To molecularly characterize the dac4 deletion, genomic DNA from dac4/CyO flies was sequenced using the Solexa genome analyzer (see materials and methods). A total of 2.4 million 36-bp-long reads were obtained, which could be mapped uniquely to the Drosophila reference genome by the standard eland pipeline for ∼0.48x sequencing coverage on the basis of a genome size of 180 Mb. The average read coverage in nonoverlapping 10-kb windows across the entire 2L chromosome arm is relatively uniform while a drop in the coverage around the dac region is evident (Figure 1A). Oscillation in the coverage likely results from both system biases and variation due to random sampling. Sources of system bias include variable mappability of genomic regions and representation biases from library preparation and sequencing protocols.
Figure 1.—
Sequencing coverage identifies the dac4 deletion region. Distribution of sequence coverage for the 2L chromosome arm of dac4 and wild-type flies. Coverage is measured as the number of reads counted in each bin and plotted along the length of the chromosome by position in megabases from 11 Mb to 18.5 Mb. (A) Sequencing coverage of dac4/CyO with fixed bins of 10 kb in length. (B) Sequencing coverage of w1118 flies with fixed bins of 10 kb in length (read count is normalized to dac4). (C) Sequencing coverage of dac4/CyO heterozygotes with variably sized bins of mean size 10 kb minimizes biases attributable to mappability and sequencing (see materials and methods). (D) Sequencing coverage of dac4/CyO heterozygotes with variably sized bins of mean size 10 kb, a sharp drop in read coverage at the boundaries of the dac4 deletion region is evident.
To estimate the system bias and establish a reference coverage map, deep sequencing of wild-type Drosophila genomic DNA was performed generating 32 million 36-bp-long reads, amounting to 6.4x sequencing coverage. We reasoned that with this depth of sequencing, most of the oscillation in read coverage would be the result of system bias. As expected, read coverage for wild-type DNA is similar to that of the dac4 mutant with the exception of the dac gene region (Figure 1B). To reduce oscillation due to system bias, a set of variably sized bins were determined, which divide the reference genome into pieces of unequal length, each containing a fixed number of wild-type reads (Campbell et al. 2008). Reads from dac4 DNA were partitioned into these variably sized bins and the read coverage calculated. Variation in coverage is significantly reduced by this transformation with the putative deletion region becoming the only significant drop on the dac4 2L chromosome arm (Figure 1C). A significant drop in coverage is observable at both ends of the putative deletion and remains low throughout the dac4 region (Figure 1D).
CNV analysis of dac4 deletion heterozygotes precisely defines deletion breakpoints:
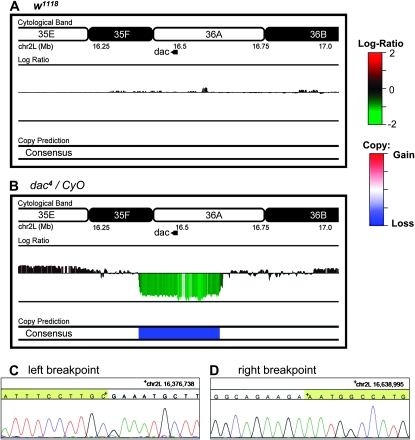
To analyze copy number across the 2L chromosome and identify deletion breakpoints in the dac4 deletion genome, a variety of algorithms that have been developed for CNV discovery with array-CGH data were tested (http://compbio.med.harvard.edu/CGHweb) (Hupe et al. 2004; Olshen et al. 2004; Eilers and de Menezes 2005; Picard et al. 2005; Willenbrock and Fridlyand 2005; Fiegler et al. 2006; Marioni et al. 2006; Carter 2007; Venkatraman and Olshen 2007; Yu et al. 2007; Lai et al. 2008a,b; Tibshirani and Wang 2008). The performance of these algorithms was first assessed with self-vs.-self data sets derived from wild-type sequencing data (see materials and methods). Random samples of ∼2.4 million reads of wild-type sequences were partitioned as described above, in this way no regions of CNV are expected. Interestingly, some of the algorithms demonstrated high levels of sensitivity to oscillations in the wild-type data and identified extensive regions of potential copy change (Figure 2A and see supporting information, Figure S1). All algorithms were then used to analyze the dac4 data for copy change at a resolution of ∼1 kb and the consensus of the algorithms used to determine the final prediction. The most significant prediction was a deletion on chromosome 2L, with breakpoints occurring in the ∼1-kb windows whose midpoints are 16,376,526 and 16,638,211 on chromosome 2L (Figure 2B and see Figure S2). To validate these predictions, PCR primers flanking the predicted breakpoints were designed and a DNA fragment was amplified that spans the junction. The junction fragment was then sequenced and the breakpoints mapped to 16,376,738 and 16,638,995 bp position on chromosome 2L consistent with CNV prediction (Figure 2, C and D). As often occurs with X-ray-generated deletions, the breakpoints of the dac4 deletion are not ligated together, but are separated by a 320-bp sequence. Portions of this 320-bp map in small blocks to multiple chromosomes making it difficult to infer the origin of the inserted sequence.
Figure 2.—
CNV analysis of dac4 reveals copy loss region. Log2 ratio scores generated for the 2L chromosome arm from dac4/CyO and w1118 were analyzed as described in materials and methods. The consensus that identifies losses and gains on the basis of the chosen algorithms is depicted as “consensus.” (A) CNV analysis of wild-type coverage. No regions of copy loss are identified in the wild-type data for dac4 deletion region. (B) CNV analysis of dac4 coverage. A large region of low log2 score is identified on chr 2L from 16.376 to 16.638 Mb by the consensus prediction interpreted as copy loss. Junction PCR using primers flanking the predicted deletion breakpoints amplify a fragment and identify the breakpoints. (C) Trace of left breakpoint identifies position 16,376,738 as the left breakpoint. (D) Trace of right breakpoint identifies position 16,638,995 as the right breakpoint.
Low read coverage is sufficient for CNV detection:
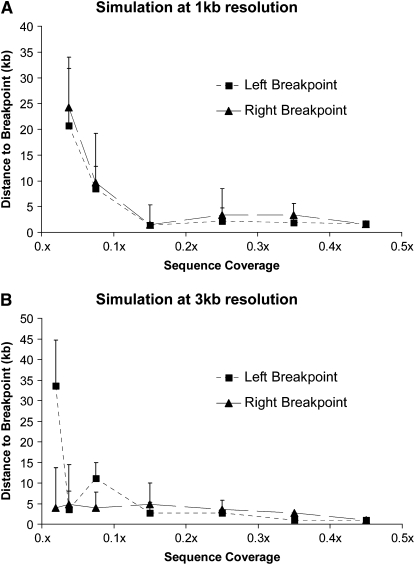
The cost for CNV detection using the high-throughput sequencing platform is proportional to the number of reads required for the analysis. Using the reads obtained from the dac4 deletion flies, a series of computer simulations were performed to determine the effect of read coverage on the ability to detect deletions and map associated breakpoints (see materials and methods). CNV analyses of these data sets were performed at 1-kb and 3-kb resolution. At 1-kb resolution the dac4 deletion was identifiable across all levels of coverage greater than 0.04x or ∼200,000 reads. At 3-kb resolution the dac4 deletion was identified in most replicates at 0.02x sequence coverage or ∼100,000 reads or greater. These results suggest that large CNVs can be detected even with extremely low depth of sequencing coverage. We also observed that decreasing the read coverage has a strong effect on the accuracy of breakpoint detection. For example, when coverage is below 0.1x there is a great deal of error in detection of the dac4 deletion breakpoints (Figure 3, A and B). However, when coverage exceeds 0.1x, the predicted breakpoints are highly consistent and accurate across simulations. From these results we concluded that the reads generated by one lane of Solexa sequencing are more than sufficient to analyze CNVs for multiple genomes at high resolution or that 500,000 reads should be sufficient to identify and accurately characterize the breakpoints of large deletions with high resolution.
Figure 3.—
Simulation studies indicate that low read coverage is sufficient for CNV detection. Analyses of dac4 CNV on chromosome arm 2L were performed on random samples of the dac4 data to simulate different depths of sequencing coverage. Analyses were performed at 1-kb (A) and 3-kb (B) resolution using DNA copy (see materials and methods). The mean observed distance between the midpoint of the predicted window and the empirically derived breakpoint is plotted across seven replicates. Error bars represent the standard deviation across all seven replicates. In both simulations a trend of increasing error is observed as the number of reads sampled decreases.
A barcode system to enable multiplex sequencing:
To allow the simultaneous interrogation of multiple genomes, we developed a barcoding system for multiplex Solexa sequencing. Criteria for an effective barcoding system include the ability to accurately differentiate reads from each sample postsequencing and relatively equal representation of samples among the sequenced reads. By differentially ligating modified primers during genomic library preparation, we enabled the in silico separation of samples postsequencing. We designed primers to test all 16 dinucleotide combinations for their efficiency in library preparation protocols. Barcodes were then further tested by multiplex microbial sequencing. Two microbes, Escherichia coli and Rhodobacter were used to assess the accuracy of the barcoding and multiplex procedures. The DNA of each microbe was differentially labeled with barcoded oligonucleotide adapters and libraries were mixed in equal molar amounts and sequenced simultaneously on one lane of the Illumina genome analyzer. E. coli was labeled with a TT barcode and Rhodobacter with an AC barcode. Of 4.1 million total reads produced in one experiment designed to test these tags, 95% of generated sequences were tagged, and both tags were represented in nearly equal proportions (see Table S1). Furthermore, reads exhibited an extremely low error/cross-contamination rate. About 0.24% of the TT-labeled reads map to Rhodobacter while 0.43% of AC-labeled reads map to E. coli. From these results we concluded that the barcode system is sufficiently specific and can be applied to the simultaneous sequencing of multiple deficiency stocks.
Multiplex sequencing of deficiency stocks:
To test the applicability of multiplex sequencing to the detection of copy number change in Drosophila, two deficiency fly stocks were selected from the Bloomington deficiency kit. Df(3L)Exel6105 (Parks et al. 2004) and Df(2L)Sd37 (Ganetzky 1977) map to different chromosome arms and were multiplex sequenced as a proof of concept. Genomic DNA from male adult flies heterozygote for the deficiency and stock balancer chromosomes was tagged with different barcoded adapters and then mixed and sequenced simultaneously on one lane of the Illumina genome analyzer (see materials and methods). Following sequencing and in silico separation of the samples, 900,000 and 600,000 uniquely mappable reads were obtained for Df(3L)Exel6105 and Df(2L)Sd37, respectively. CNV analysis was then performed on both data sets as described above.
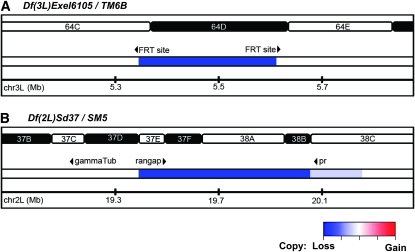
CNV was successfully detected in both deficiency DNAs at the expected regions. Df(3L)Exel6105 was generated by recombination between two distinct FRT bearing insertion sites resulting in a 242-kb deletion with molecularly defined breakpoints of 5,359,162 and 5,601,375 bp (Parks et al. 2004). Consistent with this, CNV analysis identified a deletion on chromosome 3L with breakpoints occurring in ∼3-kb windows whose midpoints were 5,360,035 and 5,600,802 (Figure 4A and see Figure S3). Therefore, not only was the deletion correctly identified but both breakpoints were also mapped within 3 kb. We validated the molecular breakpoints by PCR using primers specific to the insertion element remaining after recombination and the flanking genomic region (see Table S2).
Figure 4.—
CNV analysis of deletion mutants Df(3L)Exel6105 and Df(2L)Sd37 identifies molecular breakpoints. (A) Analysis of chr 3L of Df(3L)Exel6105 identifies a region of low log2 score (blue) consistent with the molecularly defined deletion Df(3L)5,359,162..5,601,375. (B) Analysis of Df(2L)Sd37 identifies a region of low log2 score (blue) consistent with a deletion on chr 2L with breakpoints occurring in 3-kb windows whose midpoints are 19,423,344 and 19,962,150 bp, respectively.
Similar results were obtained for Df(2L)Sd37, a deficiency generated by X-ray mutagenesis (Ganetzky 1977). This deficiency has been mapped cytologically to 37D2–38B2, corresponding to the 19.3–20.7 Mb region. The breakpoints, however, have not been molecularly characterized. CNV analysis identifies a deletion on chromosome 2L with breakpoints occurring in ∼3-kb windows whose midpoints are 19,423,344 and 19,962,150 bp (Figure 4B). This result is consistent with previous genetic complementation data. First, Df(2L)Sd37 complements mutations in the γTub37C gene, which is located on chromosome 2L between 19,183,957 and 19,185,709 bp, and the pr gene, whose location is at chromosome 2L between 20,073,714 and 20,075,479 bp. In addition, Df(2L)Sd37 fails to complement mutations in the RanGap gene, which is located on chromosome 2L between 19,442,041 and 19,447,322 bp (Ganetzky 1977; Pentz et al. 1990; Stathakis et al. 1995). Recovery of the molecular breakpoints by junction PCR with primers flanking the proposed breakpoints was unsuccessful. P-element stocks whose insertions flanked the predicted deletion junction were used to test for the presence or absence of genomic DNA on the deletion chromosome when heterozygote over the insertion. Because the insertion disrupts successful amplification of genomic DNA on the insertion chromosome, failure or success to amplify the PCR product can be interpreted as absence or presence of genomic DNA on the deletion chromosome. Results from these analyses support the breakpoint windows predicted by CNV analysis and indicate that prediction accuracy was within 3 kb (see Figure S5 and Table S2).
DISCUSSION
We have reported a strategy for rapid, cost-effective, high-throughput, genomewide characterization of CNV using next generation sequencing technology. On the basis of our results, large CNVs can be identified and mapped with high resolution. Both computer simulation and experimental studies indicate that low levels of sequence coverage (<0.1x sequencing coverage or ∼458,000 reads) are sufficient for identifying and mapping large CNVs at kilobase resolution. For the characterization of smaller CNVs, such as those in the kilobase range, we estimate that deeper sequencing is required: approximately 4–5 million reads or 1x sequence coverage. Although only tested in Drosophila, the strategy is generally applicable to all organisms.
The accuracy and resolution to which chromosomal deletions and breakpoints can be mapped by our platform is very high. Using the Drosophila deficiency dac4 as a test case, the breakpoints of the deletion were mapped to 1-kb resolution, which were subsequently confirmed by PCR and direct Sanger sequencing. Additionally, paired-end sequencing of size-selected molecules can be used to infer deletion and duplication events and additionally provide information regarding inversions and rearrangements. Due to the advantages of the high-throughput sequencing platform we find it likely this method will become the most commonly used platform for CNV discovery.
The cost and throughput of CNV analysis on the high-throughput sequencing platform can be dramatically reduced by multiplexing, which is enabled by introducing barcodes during sequencing library construction. On the basis of computer simulations using the data obtained from the dac4 deletion, read coverage as low as 0.1x or 458,000 reads is sufficient for breakpoint identification at 3-kb resolution. This result suggests that data generated from a single Solexa lane should be sufficient for simultaneously analyzing as many as five stocks in multiplex. Currently, this puts the cost of characterizing large CNVs at approximately $350 each. In comparison, the cost of an array-CGH experiment with single gene resolution is approximately $350, while a 1-kb resolution would cost approximately $800. As the capacity of high-throughput sequencing continues to increase and costs decline rapidly, the method we describe will be more cost effective than array-CGH. Additionally, the next-generation sequencing approach offers the ability to improve resolution by increasing the depth of sequencing coverage. Thus, CNV discovery by high-throughput sequencing is scalable—the desired coverage-to-resolution balance can be determined and the cost optimized. For CNVs in the subkilobase range, the sequencing platform is likely to be very effective; however, methods of analysis in addition to those described in this report will be required.
One immediate application for CNV discovery in Drosophila by high-throughput sequencing is mapping the deletions of each Bloomington core deficiency stock that have not been molecularly characterized. Two major limitations presently reduce the effectiveness of this important genetic tool. First, the breakpoints of three-quarters of the stocks are mapped cytologically, the size of these deletions remains uncertain, diminishing the utility of these stocks. Second, many stocks may harbor cryptic rearrangements that diminish the reliability of results. Both problems can be largely resolved using the high-throughput sequencing method. The characterization of deficiency and duplication stocks by array-CGH has been described previously (Erickson and Spana 2006). The high-throughput sequencing approach provides a good alternative with greater resolution at a currently comparable and rapidly declining cost.
In addition to identifying the expected deletions and accurately defining the breakpoints for all deficiencies described in this report, our analyses indicated additional copy number variations in each data set (see Figure S2, Figure S3, and Figure S4). From the lack of false positives in the self-vs.-self data sets we find it likely that these variants are legitimate though it is unclear whether they occur on the same chromosome as the expected deletions or are harbored on the balancer chromosome, which was also sequenced. Further inquiry would be required to verify the nature of these CNVs and the chromosome on which they occur; because our interest was in defining the known deficiencies, we did not seek to validate these. These observations do, however, highlight the possibility of cryptic structural variation harbored on the chromosomes of deficiency stocks.
To date CNV studies have been largely limited to humans primarily due to the high cost of the methods used for detection. As described in our report, high-throughput sequencing technology now offers the opportunity for cost-effective characterization of CNV. Taking advantage of this approach, the contribution of CNV to phenotypic variation in model organisms, including Drosophila, can be systematically explored. Such studies are likely to offer important insights regarding the biological consequences of CNV.
Acknowledgments
We thank Graeme Mardon for providing dac4 flies, scientific discussion relevant to the design of experiments, and review of the manuscript. We also thank Hugo Bellen, Shinya Yamamoto, and Kevin Cook for scientific discussion and review of the manuscript. We acknowledge Stephen Richards of the Human Genome Sequencing Center for ongoing collaborations to study CNV in Drosophila. Finally, we thank the staff of the Human Genome Sequencing Center who performed the sequencing of genomic libraries. This work is partially supported by the National Human Genome Research Institute grant U54HG003273 to R.G.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103218/DC1.
References
- Bridges, C. B., 1936. The bar “gene” a duplication. Science 83 210–211. [DOI] [PubMed] [Google Scholar]
- Campbell, P. J., P. J. Stephens, E. D. Pleasance, S. O'Meara, H. Li et al., 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, N. P., 2007. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 39 S16–S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, D. Y., G. Getz, D. B. Jaffe, M. J. O'Kelly, X. Zhao et al., 2009. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods 6 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman, E. B., and D. L. Hartl, 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104 19920–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers, P. H., and R. X. de Menezes, 2005. Quantile smoothing of array CGH data. Bioinformatics 21 1146–1153. [DOI] [PubMed] [Google Scholar]
- Emerson, J. J., M. Cardoso-Moreira, J. O. Borevitz and M. Long, 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320 1629–1631. [DOI] [PubMed] [Google Scholar]
- Erickson, J. N., and E. P. Spana, 2006. Mapping Drosophila genomic aberration breakpoints with comparative genome hybridization on microarrays. Methods Enzymol. 410 377–386. [DOI] [PubMed] [Google Scholar]
- Fiegler, H., R. Redon, D. Andrews, C. Scott, R. Andrews et al., 2006. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 16 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky, B., 1977. On the components of segregation distortion in Drosophila melanogaster. Genetics 86 321–355. [PMC free article] [PubMed] [Google Scholar]
- Hupe, P., N. Stransky, J. P. Thiery, F. Radvanyi and E. Barillot, 2004. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics 20 3413–3422. [DOI] [PubMed] [Google Scholar]
- Lai, T. L., H. Xing and N. Zhang, 2008. a Stochastic segmentation models for array-based comparative genomic hybridization data analysis. Biostatistics 9 290–307. [DOI] [PubMed] [Google Scholar]
- Lai, W., V. Choudhary and P. J. Park, 2008. b CGHweb: a tool for comparing DNA copy number segmentations from multiple algorithms. Bioinformatics 24 1014–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon, G., N. M. Solomon and G. M. Rubin, 1994. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120 3473–3486. [DOI] [PubMed] [Google Scholar]
- Marioni, J. C., N. P. Thorne and S. Tavare, 2006. BioHMM: a heterogeneous hidden Markov model for segmenting array CGH data. Bioinformatics 22 1144–1146. [DOI] [PubMed] [Google Scholar]
- Olshen, A. B., E. S. Venkatraman, R. Lucito and M. Wigler, 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5 557–572. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36 288–292. [DOI] [PubMed] [Google Scholar]
- Pentz, E. S., B. C. Black and T. R. Wright, 1990. Mutations affecting phenol oxidase activity in Drosophila: quicksilver and tyrosinase-1. Biochem. Genet. 28 151–171. [DOI] [PubMed] [Google Scholar]
- Picard, F., S. Robin, M. Lavielle, C. Vaisse and J. J. Daudin, 2005. A statistical approach for array CGH data analysis. BMC Bioinformatics 6 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon, R., S. Ishikawa, K. R. Fitch, L. Feuk, G. H. Perry et al., 2006. Global variation in copy number in the human genome. Nature 444 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegiers, F., J. Kavaler, N. Tolwinski, Y. T. Chou, H. Duan et al., 2009. Frequent unanticipated alleles of lethal giant larvae in Drosophila second chromosome stocks. Genetics 182 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Srivatsan, A., Y. Han, J. Peng, A. K. Tehranchi, R. Gibbs et al., 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4 e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis, D. G., E. S. Pentz, M. E. Freeman, J. Kullman, G. R. Hankins et al., 1995. The genetic and molecular organization of the Dopa decarboxylase gene cluster of Drosophila melanogaster. Genetics 141 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani, R., and P. Wang, 2008. Spatial smoothing and hot spot detection for CGH data using the fused lasso. Biostatistics 9 18–29. [DOI] [PubMed] [Google Scholar]
- Venkatraman, E. S., and A. B. Olshen, 2007. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23 657–663. [DOI] [PubMed] [Google Scholar]
- Willenbrock, H., and J. Fridlyand, 2005. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics 21 4084–4091. [DOI] [PubMed] [Google Scholar]
- Yu, T., H. Ye, W. Sun, K. C. Li, Z. Chen et al., 2007. A forward-backward fragment assembling algorithm for the identification of genomic amplification and deletion breakpoints using high-density single nucleotide polymorphism (SNP) array. BMC Bioinformatics 8 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., G. Zhang, Y. Zhang, S. Xu, R. Zhao et al., 2008. On the origin of new genes in Drosophila. Genome Res. 18 1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]