Abstract
In yeast, assembly of the septins at the cell cortex is required for a series of key cell cycle events: bud-site selection, the morphogenesis and mitotic exit checkpoints, and cytokinesis. Here we establish that the Ccr4-Pop2-NOT mRNA deadenylase contributes to septin organization. mRNAs encoding regulators of septin assembly (Ccd42, Cdc24, Rga1, Rga2, Bem3, Gin4, Cla4, and Elm1) presented with short poly(A) tails at steady state in wild-type (wt) cells, suggesting their translation could be restricted by deadenylation. Deadenylation of septin regulators was dependent on the major cellular mRNA deadenylase Ccr4-Pop2-NOT, whereas the alternative deadenylase Pan2 played a minor role. Consistent with deadenylation of septin regulators being important for function, deletion of deadenylase subunits CCR4 or POP2, but not PAN2, resulted in septin morphology defects (e.g., ectopic bud-localized septin rings), particularly upon activation of the Cdc28-inhibitory kinase Swe1. Aberrant septin staining was also observed in the deadenylase-dead ccr4-1 mutant, demonstrating the deadenylase activity of Ccr4-Pop2 is required. Moreover, ccr4Δ, pop2Δ, and ccr4-1 mutants showed aberrant cell morphology previously observed in septin assembly mutants and exhibited genetic interactions with mutations that compromise septin assembly (shs1Δ, cla4Δ, elm1Δ, and gin4Δ). Mutations in the Not subunits of Ccr4-Pop2-NOT, which are thought to predominantly function in transcriptional control, also resulted in septin organization defects. Therefore, both mRNA deadenylase and transcriptional functions of Ccr4-Pop2-NOT contribute to septin organization in yeast.
THE septins are evolutionarily conserved filament-forming proteins, whose functions and assembly are best understood in the model yeast Saccharomyces cerevisiae. The mitotic septins Cdc3, Cdc10, Cdc11, Cdc12, and Shs1 assemble a ring at the mother-bud neck just before bud emergence, which expands into a collar as the cell cycle progresses and is split between the mother and daughter cells at cytokinesis (Ford and Pringle 1991; Kim et al. 1991; reviewed in Douglas et al. 2005). The septins are thought to function as scaffolds to recruit proteins to specific parts of the cell cortex and as membrane diffusion barriers that enable asymmetric distribution of proteins between the mother and daughter cells (Douglas et al. 2005). The cellular functions of the septins include cytokinesis, bud-site selection, cell wall deposition, and mitotic checkpoints related to bud emergence and exit from mitosis (reviewed in Douglas et al. 2005; Park and Bi 2007).
Septin assembly is controlled at the post-translational level in a pathway coordinated by the small GTPase Cdc42 (reviewed in Douglas et al. 2005; Park and Bi 2007). Proposed roles for Cdc42 include a direct role in septin recruitment to the cortex for assembly into the ring (Gladfelter et al. 2002; Caviston et al. 2003), as well as regulation of effector proteins (Douglas et al. 2005; Park and Bi 2007). Cdc42 regulators and/or effectors are also required for proper septin assembly. Mutations in the Cdc42 GTP exchange factor (GEF) Cdc24 and the GTPase activating proteins (GAPs) Rga1, Rga2, and Bem3 cause defects in septin assembly, as do mutations in Cdc42 effectors Gic1 and Gic2 and the kinases Cla4, Gin4, and Elm1 (Cvrckova et al. 1995; Longtine et al. 1998; Caviston et al. 2003; Versele and Thorner 2004; Iwase et al. 2006; reviewed in Douglas et al. 2005; Park and Bi 2007). Cla4 and Gin4 phosphorylate the septins to stabilize their association (Dobbelaere et al. 2003; Versele and Thorner 2004), whereas Elm1 phosphorylates Gin4 to activate it and cause Gin4-dependent phosphorylation of the septin Shs1 (Asano et al. 2006).
Post-transcriptional shortening of mRNA poly(A) tails (deadenylation) is an important mechanism in regulation of gene expression (Parker and Song 2004; Goldstrohm and Wickens 2008). mRNA poly(A) tail length can affect both transcript stability and translation; long tails positively correlate with translation efficiency (Preiss et al. 1998), while tail shortening by mRNA deadenylation serves as an entry point into mRNA decay (Parker and Song 2004; Goldstrohm and Wickens 2008). During mRNA maturation in yeast, poly(A) length is set by the Pan2 mRNA deadenylase, which trims the tails following nuclear polyadenylation. The cytoplasmic mRNA deadenylase Ccr4-Pop2-NOT shortens the tail further to a length of ∼(A)10, and the mRNA then becomes the substrate for decapping and 5′→3′ exonucleolytic decay. These generic processes of mRNA ageing and decay are thought to operate on most cellular mRNAs (Parker and Song 2004; Goldstrohm and Wickens 2008). Nevertheless, different mRNA species can display distinct spectra of poly(A) tail lengths at steady state that reflect mRNA-specific control of deadenylation and decay, and such post-transcriptional control is typically driven by elements in the mRNA 3′-untranslated region (3′-UTR).
Microarray-based analyses of mRNA polyadenylation state uncovered widespread use of mRNA-specific poly(A) tail length control in the S. cerevisiae and Schizosaccharomyces pombe transcriptomes (Beilharz and Preiss 2007; Lackner et al. 2007). Of note here, many mRNAs encoding cell cycle, cell polarity, and morphogenesis-related functions including septin assembly, were found to be present with mostly short oligo(A) tails in nonsynchronized, exponentially growing wild-type (wt) cell cultures. These data suggested that rapid deadenylation by Ccr4-Pop2-NOT of such “short-tailed” mRNAs serves as a mechanism to restrict their translation to a tight time window during the cell cycle (Beilharz and Preiss 2007). Thus, even though the Ccr4-Pop2-NOT deadenylase shortens the poly(A) tails of virtually all mRNAs (Parker and Song 2004; Woolstencroft et al. 2006; Beilharz and Preiss 2007; Goldstrohm and Wickens 2008), its effects on the group of “short-tailed” mRNAs are particularly critical for cell function, and their deregulation in deadenylase mutant cells is therefore likely to contribute to observable phenotypes.
Our transcriptome-wide analysis had indicated that mRNAs that preferentially possess short poly(A) tails are enriched in gene ontology (GO) terms such as bud neck, cell cortex, establishment and maintenance of cell polarity, and cellular morphogenesis, and deadenylase mutant strains showed morphology defects (e.g., wide necks and elongated cells) that could result from problems in septin organization (Beilharz and Preiss 2007). We therefore decided to examine the role of the Ccr4-Pop2-NOT mRNA deadenylase in septin organization. We show that a short poly(A) tail is a property of all tested mRNAs coding for major septin assembly factors and that Ccr4-Pop2-NOT contributes to septin organization in yeast.
MATERIALS AND METHODS
Yeast strains and growth conditions:
The strains used in this study are listed in Table 1. All strains are isogenic to KY803 (MATa trp1Δ1 ura3-52 gcn4 leu2∷PET56). Deletion mutants were created by standard gene replacement methods. GFP was fused to the C terminus of Cdc3 at the endogenous locus using the GFP-KANMX6 cassette. Standard growth conditions were YPD (2% glucose, 2% peptone, 1% yeast extract) media at 30°. For assaying growth at 37°, cells were pregrown in YPD at 30° and then 2.5 μl of 10-fold serial dilution starting from OD600 = 0.5 were plated on YPD plates and incubated at 37° for 2–3 days. For assaying suppression by sorbitol, sorbitol was added to the YPD plates at 1 m concentration.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| Y136 (KY803) | MATatrp1Δ1 ura3-52 gcn4 leu2∷PET56 | Deniset al. (2001) |
| Y181 (KY804) | AS Y136 but MATα | Deniset al. (2001) |
| Y215 | AS Y136 but not1-1 | Deniset al. (2001) |
| Y214 | AS Y136 but not4Δ∷URA3 | Deniset al. (2001) |
| Y294 | AS Y136 but ccr4Δ∷klURA | Traven et al. (2005) |
| Y297 | AS Y136 but pop2Δ∷klURA | Traven et al. (2005) |
| Y298 | AS Y136 but not2Δ∷klURA | Traven et al. (2005) |
| Y299 | AS Y136 but not5Δ∷klURA | Traven et al. (2005) |
| Y310 | AS Y136 but not3Δ∷klURA | Traven et al. (2005) |
| Y369 | (ccr4-1) AS Y136 but ccr4-E556A | Traven et al. (2005) |
| Y763 | AS Y136 but swe1Δ∷KAN | This study |
| Y816 | AS Y136 but pop2Δ∷klURA swe1Δ∷KAN | This study |
| Y956 | AS Y136 but cla4Δ∷KAN | This study |
| Y960 | AS Y181 but cla4Δ∷KAN | This study |
| Y961 | AS Y136 pop2Δ∷klURA cla4Δ∷KAN | This study |
| Y968 | AS Y181 but elm1Δ∷KAN | This study |
| Y972 | AS Y136 but shs1Δ∷KAN | This study |
| Y976 | AS Y136 but pop2Δ∷klURA shs1Δ∷KAN | This study |
| Y980 | AS Y136 but ccr4-E556A shs1Δ∷KAN | This study |
| Y982 | AS Y136 but ccr4Δ∷klURA shs1Δ∷KAN | This study |
| Y989 | AS Y136 but CDC3-GFP-KAN | This study |
| Y992 | AS Y136 but ccr4Δ∷klURA CDC-GFP-KAN | This study |
| Y993 | AS Y136 but pop2Δ∷klURA CDC3-GFP-KAN | This study |
| Y1013 | AS Y136 ccr4Δ∷klURA cla4Δ∷KAN | This study |
| Y1126 | AS Y136 but ccr4Δ∷klURA swe1Δ∷KAN | This study |
| Y1128 | AS Y136 but ccr4-E556A swe1Δ∷KAN | This study |
| Y1172 | AS Y136 but pan2Δ | Beilharz et al. (2007) |
| Y1173 | AS Y136 but ccr4-E556A pan2Δ | Beilharz et al. (2007) |
| YAT164 | AS Y136 but gin4Δ∷KAN | This study |
| YAT165 | AS Y136 but ccr4Δ∷klURA gin4Δ∷KAN | This study |
| YAT170 | AS Y136 but pop2Δ∷klURA gin4Δ∷KAN | This study |
klURA-Kluyveromyces lactis URA
Double ccr4Δ/pop2Δ and cla4Δ or elm1Δ mutants were obtained from dissection of tetrads after sporulation of the respective heterozygote diploid strains. Diploid strains were sporulated using standard methods and spores were dissected onto YPD plates using a Zeiss dissecting microscope. The spores were genotyped by plating on selective plates for the markers used to delete the relevant genes (see Table 1).
Cell morphology assays and microscopy:
To assay cell morphologies in the presence or absence of hydroxyurea (HU), cells were grown to early-to-mid logarithmic phase in YPD at 30° and then treated with or without 0.2 m HU for 16 hr. We observed that the morphology defects of the mutants (particularly ccr4Δ) decreased with chronological age (when cultures were inoculated from plates kept at 4° for >2 weeks), and therefore plates from strains freshly streaked from stocks were used in all experiments. For microscopy, cells were fixed with 70% ethanol for 30 min–1 hr. After fixation, cells were rehydrated in 1× PBS (phosphate-buffered saline) and samples were kept at 4° until viewing. Cells were viewed with a Zeiss Axiovert 25 microscope using a 100× magnification objective. Photographs were taken on Kodak film at an original magnification of 250× or with an AxioCam MRc camera using the Axiovision Rel. 4.6 software. Typically between 100 and 300 cells were scored for each strain, for each independent culture. Averages are from at least three independent cultures and the experimental error represents the standard deviation.
For immunofluorescence microscopy, cells were fixed for 1 hr in 3.7% formaldehyde at 30°. Cell walls were digested by treatment with zymolyase 20T (250 μg/ml) for 10–20 min and cells were applied to polylysine-coated slides. All antibody incubations and blocking were done in 3% BSA (bovine serum albumin) in PBS. The anti-Cdc11 antibody (Santa Cruz Biotechnologies) was used at 1:100 dilution, followed by 1:1000 dilution of secondary anti-rabbit antibody conjugated to Alexa488. Cover slips were sealed with clear nail polish and slides were stored at −20°. The slides were viewed and photographed with the Zeiss Axiovert 25 microscope using the Axiocam MRc camera, and the Axiovision Rel. 4.6 software. For Figures 3B and 4B, cells were viewed with an Olympus BX51 microscope and photographs were taken with the DP Controller software.
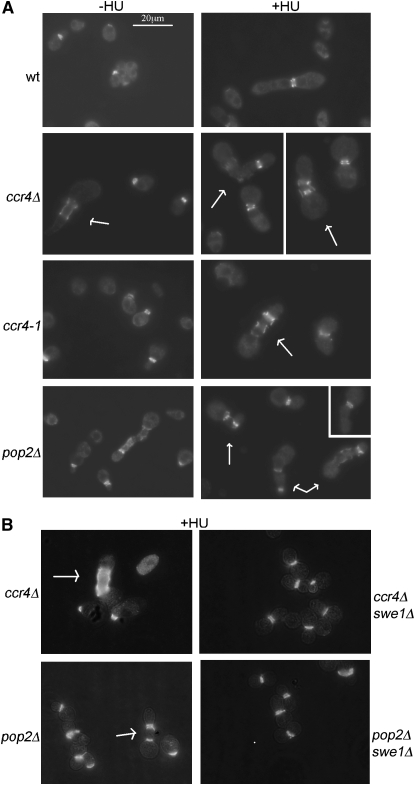
Figure 3.—
Ccr4 and Pop2 are required for proper septin organization. Septins were stained in HU-treated and untreated cultures by immunofluorescence with the anti-Cdc11 antibody. The arrows represent cells with ectopic septin rings in the buds.
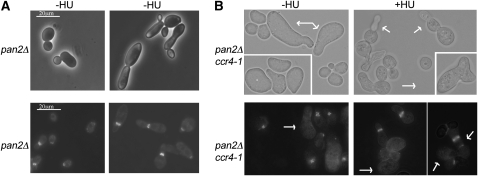
Figure 4.—
The Pan2 mRNA deadenylase is dispensable for septin organization. (A) Upper panel: Cells were treated with or without 0.2 m HU for 16 hr and processed for microscopy as in Figure 2A. Lower panel: Immunofluorescence with the anti-Cdc11 antibody in pan2Δ cultures, with or without treatment with 0.2 m HU for 16 hr. (B) Double pan2Δ ccr4-1 mutants were either from log phase cultures (−HU) or after treatment with 0.2 m HU for 16 hr. Cells were processed for microscopy and immunofluorescence as in Figures 2 and 3. The arrows indicate the big elongated cells that did not stain for septins, as well as cells displaying bud chains and ectopic septin rings.
Western and Northern blotting:
For Western blots, cells were grown overnight, diluted to OD600 = 0.2–0.3, grown for 3 hr at 30°, and then treated with 0.2 m HU for the times indicated in the figures. Protein extracts were prepared by precipitation of proteins with trichloro acetic acid (TCA), followed by resolubilization in Laemli buffer. The antibodies were used at the following dilutions: anti-actin 1:4000 (Chemicon) and anti-Swe1 1:200 (Santa Cruz Biotechnologies). The secondary antibodies were conjugated to horseradish peroxidase and detection was performed with the ECL reagent (Amersham).
Northern analysis of gene expression (supporting information, Figure S1) was performed from wild type, ccr4Δ, ccr4-1, and pop2Δ cultures not treated by HU. RNA was isolated by the hot phenol method and processed for Northern blot analysis as described (Hammet et al. 2002). Probes were obtained by PCR with ORF-specific primers from genomic DNA of wild-type KY803 yeast and were labeled with 32P. The signals were quantified with the ImageQuant software after exposure on Phosphorimager screens. The expression levels of the relevant genes were normalized to ACT1 levels and are expressed as fold change compared to expression in the wild type (with wild-type levels set to 1).
Ligation-mediated polyadenytation test:
Ligation-mediated polyadenytation test (LM-PAT) was performed as described previously (Beilharz and Preiss 2007). The TVN-PAT samples (see Figure 1) were obtained from wt cDNA using a PAT-T12-VN reverse transcription primer (5′ GCGAGCTCCGCGGCCGCGTTTTTTTTTTTTVN; where V is any nucleotide except T and N is any nucleotide). The PAT-T12-VN binds at the 3′-UTR and poly(A) junction, and therefore the TVN-PAT samples show the shortest PCR product detected by the assay. The shared 5′ sequence between the regular LM-PAT reverse primer and the TVN-PAT primer allows parallel PCR amplification from these control samples and the PAT cDNAs that report on the poly(A) length.
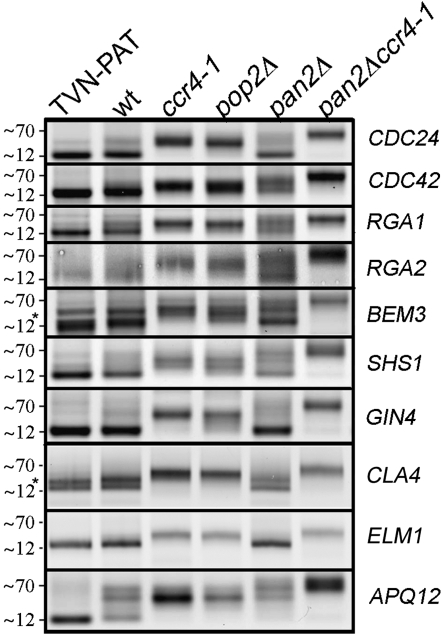
Figure 1.—
Transcripts encoding septin regulators have short mRNA poly(A) tails and their deadenylation is controlled by Ccr4-Pop2. LM-PAT was performed on cDNAs from the indicated strains using gene-specific primers. The TVN-PAT samples represent the shortest products detected by the assay. The lengths, ∼12 and ∼70, of the poly(A) tail are approximate lengths, and the * indicates alternate 3′-UTR usage in the BEM3 and CLA4 transcripts.
RESULTS
mRNAs encoding septin assembly factors are short-tailed and their deadenylation is controlled by Ccr4-Pop2:
Gene ontology analysis of microarray data (Beilharz and Preiss 2007) suggested that terms such as “establishment and maintenance of cell polarity,” “cellular morphogenesis,” “bud neck,” and “cell cortex” are prevalent in the group of short-tailed mRNAs. Among the identified short-tailed genes were several with known roles in septin assembly, such as the septin SHS1, the kinase GIN4, the Cdc42 GAPs RGA1 and RGA2, as well as other genes implicated in septin organization (eg., BNI1, BNI4, and BUD3; Gladfelter et al. 2005).
We wanted to address whether a short poly(A) tail was a general property of mRNAs encoding proteins required for septin assembly. To that end we analyzed poly(A) tail length distribution on transcripts encoding the central regulator of septin assembly, the small GTPase Cdc42, as well as its regulators and effectors: the Cdc42 GEF Cdc24, the Cdc42 GAPs Rga1, Rga2, and Bem3, and the kinases Gin4, Cla4, and Elm1. GIN4, RGA1, and RGA2 were assigned to the short-tailed group in our whole transcriptome analysis and here we verified their poly(A) tail length distribution by direct assessment on the individual mRNAs. CDC42, CDC24, BEM3, CLA4, and ELM1 had not been assigned to a tail length category by microarray analysis (Beilharz and Preiss 2007). We also looked at the mRNA encoding the septin Shs1, as it was found to be short-tailed by microarray analysis (Beilharz and Preiss 2007).
The lengths of the mRNA poly(A) tails were assessed by ligation-mediated poly(A) test (LM-PAT). LM-PAT is a reverse-transcription/PCR assay where product sizes reflect the distribution of poly(A) tail lengths on the tested mRNAs (Sallés and Strickland 1995; Beilharz and Preiss 2007). As a control for the shortest possible tail lengths in each case, we performed PCRs from cDNAs generated with a primer that anneals to 3′-UTR and poly(A) junction (TVN-PAT lanes in Figure 1, see materials and methods). Whether the mRNA poly(A) tail on the tested mRNAs was short or long in wild-type cells was determined by comparison to the TVN-PAT samples. The mRNA poly(A) tail lengths were analyzed in the wild type and in mutants in the subunits of Ccr4-Pop2-NOT that are essential for mRNA deadenylase activity: the exonuclease-inactive ccr4-1 and pop2Δ mutants (Tucker et al. 2001, 2002; Chen et al. 2002). We also analyzed mutants deleted for the alternative mRNA deadenylase PAN2, and the pan2Δ ccr4-1 double mutant was included to determine the maximal poly(A) tail length (Figure 1).
All septin assembly factors, as well as the septin SHS1, were predominantly short-tailed mRNAs in the wild-type cells at steady state, barely differing from the minimally short TVN-PAT samples that reflect the shortest detectable product in these assays (Figure 1). In the mRNA deadenylase mutants ccr4-1 and pop2Δ the tails on the tested mRNAs increased dramatically in size, confirming that their deadenylation is dependent on Ccr4-Pop2 (Figure 1). As a “long-tailed” control we used the APQ12 mRNA because, in addition to some short-tailed species, this transcript presents with long poly(A) tails at steady state. Although there was less APQ12 length heterogeneity in the ccr4-pop2 mutants with an apparent loss of the short-tailed fraction observed in the wild type, the majority of the transcripts were not altered in length by loss of Ccr4-Pop2 activity.
Longer poly(A) tails on mRNAs for septins and septin assembly factors could lead to stabilization and higher mRNA levels and/or increased translation of the transcripts. mRNA levels for the septins and septin assembly factors that we tested were not elevated in ccr4 and pop2 mutants (Figure S1), suggesting that, as it has been previously suggested for the Ccr4 target gene CRT1 (Woolstencroft et al. 2006), translational rates, rather than mRNA stability is affected by longer poly(A) tails. We tested protein levels of some proteins for which antibodies were commercially available (Cdc42 and the septin Cdc11); however we did not observe higher levels in ccr4 and pop2 mutants (data not shown), indicating that perhaps some of the other mRNAs are the relevant targets.
In contrast to the tightly stabilized longer mRNA poly(A) tails (which were devoid of short-tailed molecules and were of a uniform longer size) observed in the ccr4-1 and pop2Δ mutants, the alternative mRNA deadenylase Pan2 had a less dramatic effect on poly(A) tail lengths for transcripts encoding septin assembly factors (see pan2Δ mutants in Figure 1). This result is consistent with previous transcriptome data that showed the short-tailed mRNAs strongly depended on Ccr4 to establish appropriate tail length control (Beilharz and Preiss 2007). Some transcripts (eg., CDC24, GIN4, CLA4, and ELM1) were not affected by loss of Pan2. In other cases (for example RGA1, BEM3, and SHS1), the mRNAs did have longer tails in pan2Δ mutants. However, only a subset of the molecules for each affected mRNA had a longer tail in the pan2Δ cells, and shorter tails could also be observed, including tails as short as in the wild type (Figure 1).
Ccr4 and Pop2 are required for normal morphogenesis and septin ring organization:
If Ccr4-Pop2-dependent deadenylation of septin assembly factors (ie., their physiologically short mRNA poly(A) tails) is functionally relevant, ccr4 and pop2 mutants should display aberrant septin morphology and/or localization and, consequently, also cell morphology defects (for examples see Barral et al. 1999; Longtine et al. 2000; Gladfelter et al. 2004, 2005).
The ccr4Δ mutant indeed displayed various morphology defects, such as bigger and misshapen cells and elongated and/or misshapen buds (the −HU panel in Figure 2A and data not shown), whereas the pop2Δ strain and the deadenylase-dead ccr4-1 mutant had a milder phenotype with occasionally bigger, multi-budded cells or cells with wide necks and misshapen and/or elongated buds (see the −HU left panel in Figure 2A and data not shown).
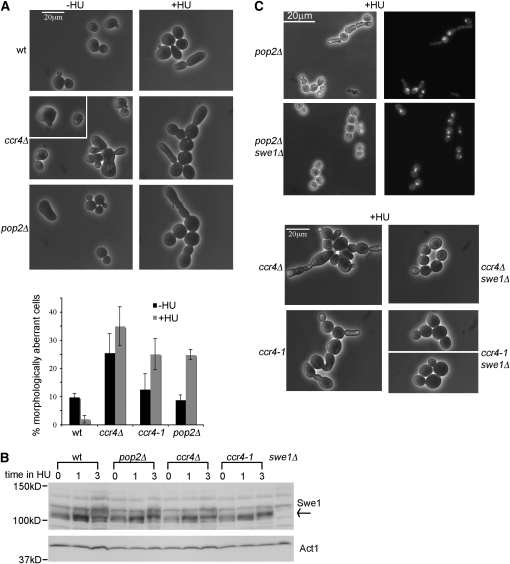
Figure 2.—
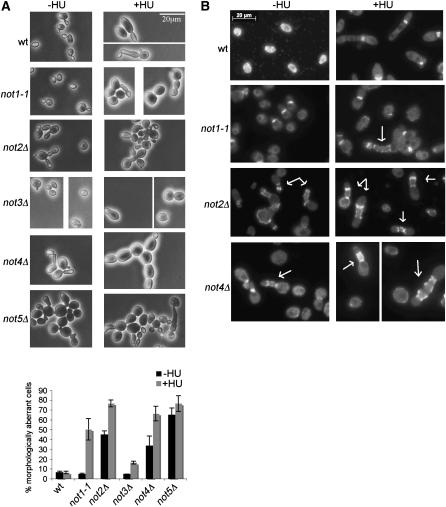
ccr4 and pop2 mutants show Swe1-dependent aberrant cell morphology. (A) Cells were treated with or without 0.2 m HU for 16 hr. The graph represents averages from at least three independent cultures for each strain. Error bars depict the standard deviation. (B) Cell extracts were from cells treated with 0.2 m HU for the indicated times (hours). Membranes were probed with the antibody against Swe1, followed by the antibody against actin (loading control). (C) Cells from the indicated mutants were treated with 0.2 m HU for 16 hr. Nuclear DNA was stained with DAPI.
It has been previously shown that activation of the Cdc28-inhibitory kinase Swe1 increases the morphology defects of septin assembly mutants (Gladfelter et al. 2005; Enserink et al. 2006; Keaton and Lew 2006; Liu and Wang 2006; Smolka et al. 2006). Therefore, we activated Swe1 by treatment with the DNA replication inhibitor HU (Liu and Wang 2006), and then observed the morphology of the deadenylase mutant strains. Figure 2B shows that Swe1 was activated by HU in the wild type and ccr4 and pop2 mutants, as evidenced by HU-induced Swe1 stabilization and change in mobility toward slower migrating forms by immunoblot analyses.
Consistent with the hypothesis that Ccr4-Pop2 contributes to septin organization, the morphology defects of the ccr4Δ, pop2Δ, and ccr4-1 mutants became more pronounced after HU treatment (Figure 2A, +HU panel on the right; the ccr4-1 mutant is shown in Figure 2C). Similar to previous observations for septin assembly mutants (Gladfelter et al. 2005), HU treatment caused a bud-chain phenotype in the ccr4Δ, pop2Δ, and ccr4-1 strains: the cells displayed elongated and misshapen buds, which often had constrictions along the length of the bud (see the +HU samples in Figure 2, A and C). DAPI staining of HU-treated pop2Δ mutants showed one nucleus in the cells with bud chains (Figure 2C), suggesting uncoupling of budding from nuclear division. In addition to bud chains, HU-treated ccr4Δ mutants (and to a lesser extent the pop2Δ and ccr4-1 mutants) also displayed formation of cell aggregates (clumps), which could be dispersed by brief sonication, as well as some cell chains.
To confirm that the effects of HU on the morphology of the ccr4 and pop2 mutants were due to Swe1 activation, we deleted SWE1 in the ccr4Δ, pop2Δ, and ccr4-1 mutants and asked whether that could suppress the morphology defects (Figure 2C). Deletion of SWE1 suppressed the elongated bud-chain morphology defect of HU-treated pop2Δ cells by approximately threefold (38.1 ± 6.5% reduced to 11.8 ± 4.5%, averages ± standard deviation from three independent cultures), resulting in predominantly large budded cells (Figure 2C). swe1Δ also suppressed the elongation and bud morphology defects of the ccr4Δ and ccr4-1 mutants (Figure 2C, note that the double mutants still formed clumps and chains). Therefore, Swe1-dependent mechanisms are at least in part responsible for the morphology defects of Ccr4-Pop2 deadenylase mutants, particularly for formation of elongated buds and bud chains.
The Swe1-dependent morphology defect of the ccr4 and pop2 mutants (in particular formation of bud chains) was a strong indication that the mutants have problems with septin organization (Gladfelter et al. 2005). To test this directly, we stained the septins in the wild type and ccr4Δ, pop2Δ, and ccr4-1 mutants, using an antibody against the Cdc11 septin. As shown in Figure 3A, wild-type cells mostly displayed normal septin staining at the neck after HU treatment, whereas ccr4Δ, pop2Δ, and ccr4-1 showed aberrant septin staining. The mutants formed additional septin rings within the elongated buds, and absent or diffuse septin staining could also be observed, particularly in bigger and misshapen cells and cells with wide necks (4.8% of HU-treated wild-type cells showed aberrant septin staining and this was increased to 23, 30.3, and 18.8% in the ccr4Δ, pop2Δ, and ccr4-1 mutants, respectively; quantification is from single cultures, n ≥ 200). Consistent with what we observed in the morphology assays, Swe1 was required for formation of ectopic bud-localized rings in ccr4Δ and pop2Δ mutants in HU-treated cells (Figure 3B). In those experiments, 21 and 17.6% of cells from ccr4Δ and pop2Δ cultures, respectively, showed aberrant septin staining (predominantly ectopic septin rings in the bud), whereas in the double ccr4Δ swe1Δ and pop2Δ swe1Δ mutants ≤1.5% cells showed ectopic septin rings and most cells were large budded with wild-type septin staining at the mother-bud neck (n ≥ 200 for all strains, the wild-type and swe1Δ mutants displayed ≤2% aberrant septin staining).
Roles of the alternative mRNA deadenylase Pan2 in septin assembly:
Pan2 had a less pronounced role in deadenylation of septin assembly factors than Ccr4-Pop2 (Figure 1), and we next tested whether Pan2 was required for septin assembly. We found that pan2Δ mutants did not show bud chains in response to Swe1 activation by HU (even though the cells did display an elongated cell morphology/pseudohyphal phenotype, see upper panel in Figure 4A; compare with ccr4 and pop2 mutants in Figure 1). Furthermore, the mutants displayed normal septin staining at the neck, in the presence or absence of HU treatment (Figure 4A), indicating that, unlike Ccr4-Pop2, Pan2 is not required for septin organization.
The role of Pan2 in deadenylation of transcripts encoding septin assembly regulators becomes more pronounced in the absence of Ccr4 activity (double pan2Δ ccr4-1 mutants displayed longer poly(A) tails on mRNAs for septin assembly factors than ccr4-1 single mutants, Figure 1). Therefore, we also tested the morphology and septin organization in pan2Δ ccr4-1 vs. ccr4-1 mutant strains. As shown in Figure 4B, cultures from strains in which both mRNA deadenylases are inactivated displayed a proportion of very large, sometimes elongated cells, which were not observed in cultures from single ccr4-1 or pan2Δ mutants. These cells did not show any detectable septin staining with the anti-Cdc11 antibody (Figure 4B): 23.63 ± 8.8% of cells in pan2Δ ccr4-1 did not stain with the anti-Cdc11 antibody, compared to 7.4 ± 3.3% in ccr4-1 single mutants (the cells that showed absence of septin staining were bigger and/or elongated or had wide mother-bud necks), showing the number of cells with no septin staining increased in the absence of both Ccr4 and Pan2 activity compared to cells in which only Ccr4 is inactivated. Upon treatment with 0.2 m HU, a proportion of cells from pan2Δ ccr4-1 mutant cultures displayed bud chains, and these cells showed formation of ectopic septin rings in immunofluorescence experiments with the anti-Cdc11 antibody (Figure 4B). Counting of cells with septin staining defects (cells forming ectopic bud-localized septin rings, as well as big cells and cells with wide necks displaying no septin staining) showed that HU-treated pan2Δ ccr4-1 mutants have a comparable number of cells with aberrant septin staining to ccr4-1 single mutants (17.45 ± 2.5% in double mutants, 14.8% in single ccr4-1 mutants, n ≥ 200 per individual culture).
Because the morphology defects in the pan2Δ ccr4-1 mutants are somewhat different than ccr4-1 cells, it is hard to directly compare the defects in septin assembly. For example, it is not clear what the etiology of the big cells in the double pan2Δ ccr4-1 mutants is and whether the absence of normal septin organization is a cause or consequence of the aberrant morphology in those cells. However, overall the data suggest that, even though per se Pan2 does not play a detectable role in septin organization, in the absence of Ccr4 activity, the function of Pan2 in determination of morphology/cell size and possibly also septin organization becomes more important.
CCR4 and POP2 genetically interact with genes encoding the septins and septin assembly kinases Cla4, Elm1, and Gin4:
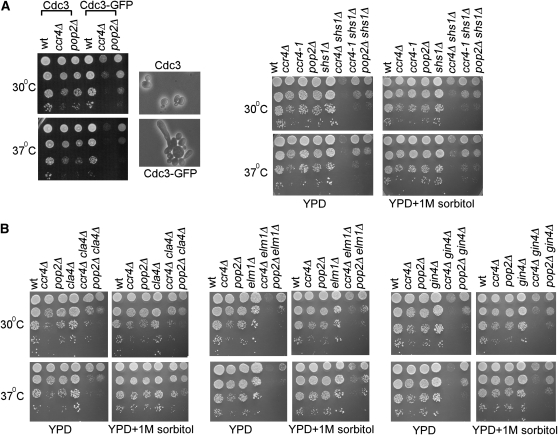
To further evaluate the role for the Ccr4-Pop2 deadenylase in septin organization, we tested if ccr4Δ, ccr4-1, and pop2Δ mutations genetically interacted with mutations that compromised septin ring structure. All mitotic septins except Shs1 are essential in S. cerevisiae, but we took advantage of the fact that fusion of GFP to the C terminus of septins can affect their function (for an example see Enserink et al. 2006) to test the genetic interaction with an essential septin gene. We used a CDC3-GFP allele to test the effects of lower Cdc3 function in the deadenylase mutant strains. In agreement with diminished Cdc3 function, we observed an increase in aberrant morphology of wild-type cells upon fusion of GFP to Cdc3 (∼20% of cells were aberrant, compared to 5% in the absence of GFP and the cells were also more prone to lysis during ethanol fixation, Figure 5A and data not shown). We found that fusion of GFP to the C terminus of Cdc3 resulted in slower growth of ccr4Δ and pop2Δ cells (ccr4Δ mutants were considerably more affected) and the synthetic growth defect was stronger at 37° (Figure 5A). ccr4Δ, pop2Δ, and the catalytically inactive ccr4-1 allele also showed a synthetic slow growth phenotype with deletion of the only nonessential mitotic septin SHS1. Again, ccr4Δ showed significantly stronger slow growth phenotype with shs1Δ than pop2Δ or the catalytically inactive ccr4-1 mutant and the synthetic growth defects of double ccr4Δ shs1Δ mutants were slightly more pronounced at 37° (Figure 5A).
Figure 5.—
CCR4 and POP2 show genetic interactions with mutations affecting septin assembly. (A) Tenfold serial dilutions of wild-type or mutant cultures were spotted on YPD plates with or without 1 m sorbitol and incubated for 2–3 days at 30° or 37°. The micrographs are from wild-type cultures at 30°, with or without GFP fused C terminally to Cdc3. (B) Tenfold serial dilutions of cells from the indicated strains were plated on YPD plates with or without 1 m sorbitol and incubated for 2 days at 30° or 37°.
Next we tested the genetic interactions of ccr4Δ and pop2Δ with deletions of the genes encoding the septin assembly kinases CLA4, ELM1, and GIN4 (Douglas et al. 2005). With cla4Δ, pop2Δ, but not ccr4Δ, showed a slight synthetic growth defect already at 30°, whereas at 37° both ccr4Δ cla4Δ and pop2Δ cla4Δ mutants showed a synthetic growth defect (Figure 5B). The synthetic growth defects of ccr4Δ cla4Δ and pop2Δ cla4Δ were the weakest of all the double mutants analyzed. CLA4 is a short-tailed transcript and its deadenylation depends on Ccr4-Pop2 (Figure 1). The weaker synthetic interactions between CCR4/POP2 and CLA4 compared to the other tested genes, indicates that CLA4 could be a relevant target of the deadenylase for septin organization.
The ccr4Δ elm1Δ and the pop2Δ elm1Δ double mutants also displayed a synthetic growth defect (the strongest growth defect of all double mutants tested) and the defects were again stronger at 37° (Figure 5B). ccr4Δ and pop2Δ were also synthetic sick with gin4Δ and again the slow growth phenotype of the double mutants was more pronounced at 37°.
Mutants in the Ccr4-Pop2-NOT complex have phenotypes consistent with cell wall defects (see Chen et al. 2002) and the septins are also required for wall biogenesis: septins are needed for deposition of chitin in the cell wall (Demarini et al. 1997) and mutants in the septins and septin assembly regulators have cell wall defects in S. cerevisiae and Candida albicans (Gonzales-Novo et al. 2006; Schmidt et al. 2008). Therefore, we wondered if the synthetic slow growth phenotype of cells in which both the Ccr4-Pop2 deadenylase and septin assembly are compromised is due to increased cell wall defects. To address this, we tested whether the synthetic growth defects of the double mutants could be suppressed by addition of the osmostabilizer sorbitol to the media (Figure 5B).
The synthetic growth defects of double ccr4Δ elm1Δ and pop2Δ elm1Δ, as well as ccr4Δ gin4Δ and pop2Δ gin4Δ could be partially suppressed by sorbitol (Figure 5B). However, the double mutants still grew slower than the single mutants, suggesting the synthetic growth defects are due to cell wall-dependent and -independent functions. This was also true in the case of double ccr4/pop2 mutants with deletion of the SHS1 septin: the growth of ccr4Δ shs1Δ mutants was moderately improved by addition of sorbitol (but not completely rescued), whereas the modest growth defect of double ccr4-1 shs1Δ and pop2Δ shs1Δ was not affected by sorbitol (Figure 5A).
Supression of growth defects by sorbitol was most pronounced for double ccr4Δ cla4Δ and pop2Δ cla4Δ mutants, for which sorbitol could restore growth almost completely to levels of single ccr4Δ and pop2Δ mutants at 37° (Figure 5B). This indicates that the slow growth of the ccr4Δ cla4Δ and pop2Δ cla4Δ mutants at 37° is largely due to increased cell wall defects. Of note, the slower growth of pop2Δ cla4Δ at 30° could not be suppressed by sorbitol, suggesting it is not a consequence of augmented cell wall biogenesis defects.
In conclusion, the synthetic sick genetic interactions between CCR4/POP2 and SHS1, CDC3, CLA4, ELM1, and GIN4 support a role for Ccr4-Pop2 in septin assembly. Addition of sorbitol can partially suppress the synthetic growth defect of the double mutants, indicating that in part the synthetic genetic interactions are due to the roles of septins and Ccr4-Pop2 in cell wall biogenesis. Other cell wall-unrelated functions of Ccr4-Pop2 in septin organization also contribute to the observed genetic interactions.
The Not subunits of Ccr4-Pop2-NOT also contribute to morphogenesis and septin organization:
In addition to the mRNA deadenylase catalytic subunits Ccr4 and Pop2, the Ccr4-Pop2-NOT complex also contains five NOT proteins (Not1–Not5). The Not proteins have overlapping, but also separate functional roles from Ccr4-Pop2 and they are thought to predominantly function in transcriptional control (Tucker et al. 2002; Collart 2003; Traven et al. 2005; Laribee et al. 2007; Mulder et al. 2007). We wanted to address if the NOT subunits of Ccr4-Pop2-NOT were also required for septin organization and cellular morphogenesis.
As shown in Figure 6A, mutations in the NOT genes resulted in aberrant cell morphology and the defects were again exacerbated by Swe1 activation in response to HU treatment. The morphological defects in the not mutants were similar to the defects of ccr4 and pop2 strains and were characterized by the presence of long, often irregularly shaped buds and formation of bud chains. not2Δ and particularly not5Δ mutants had the strongest morphological defect, consistent with the very slow growth of cells deleted for NOT2 or NOT5.
Figure 6.—
The Not subunits of Ccr4-NOT perform roles in septin assembly and cell morphology. (A) Cells were treated with or without 0.2 m HU for 16 hr and processed for microscopy after fixation in 70% ethanol. The graph shows averages from three independent cultures for each of the strains. The error bar is the standard deviation. (B) Immunofluorescence with the anti-Cdc11 antibody in HU-treated and -untreated strains. Arrows indicate cells that formed ectopic septin rings in the elongated buds.
Immunofluorescence with anti-Cdc11 demonstrated that the not1-1, not2Δ, and not4Δ mutants also mislocalized the septins (Figure 6B; not5Δ were very sensitive to zymolyase treatment and were therefore excluded from immunofluorescence analysis of septin organization). Similarly to what we observed in ccr4 and pop2 strains, the not mutants also displayed formation of additional, bud-localized septin rings in the elongated buds (Figure 6B). Other septin morphology defects in the mutants included diffused or absent staining particularly on cells with wide necks. In conclusion, our data shows that the NOT subunits of Ccr4-Pop2-NOT are also required for septin assembly and establishment of wild-type cell morphology.
DISCUSSION
mRNA deadenylation affects both transcript stability and translation and is therefore a crucial mechanism in the control of gene expression. Because each mRNA deadenylase can in principle regulate a large number of genes, it remains poorly understood which cellular processes are particularly dependent on proper deadenylase activity.
In the present report, we documented short poly(A) tails on all tested mRNAs encoding septins (Shs1) and septin assembly factors (Cdc42, Cdc24, Rga1, Rga2, Bem3, Gin4, Cla4, and Elm1) and we therefore conclude that a short poly(A) tail is very likely a general property of mRNA encoding proteins required for septin assembly. The tails were subject to Ccr4-Pop2-dependent deadenylation, as the tested mRNAs displayed tightly stabilized longer tails in deadenylase mutant strains ccr4-1 and pop2Δ.
In agreement with mRNA deadenylation being important for function of the septin regulators, the Ccr4-Pop2 mRNA deadenylase contributes to septin organization as follows: (i) ccr4Δ and pop2Δ mutants display morphogenesis defects (eg., bud chains) particularly in response to activation of the kinase Swe1, a phenotype previously reported for mutants with compromised septin structure (Gladfelter et al. 2005); (ii) ccr4Δ and pop2Δ mutants showed synthetic sick genetic interaction with mutations that compromise septin assembly (shs1Δ, cla4Δ, elm1Δ, gin4Δ, and a fusion of GFP to Cdc3); and (iii) septin staining demonstrated septin organization defects in the ccr4Δ and pop2Δ mutant strains, such as formation of ectopic bud-localized septin rings. The ectopic septin rings are thought to form in the presence of an unstable initial ring at the mother-bud neck and therefore their formation is indicative of defects in primary septin ring organization (Gladfelter et al. 2005). The exonuclease inactive ccr4-1 mutant also showed Swe1-dependent morphogenesis defects and aberrant septin organization, demonstrating that the exonuclease activity of Ccr4-Pop2-NOT contributes to proper septin assembly.
Mutants in the alternative yeast mRNA deadenylase Pan2 displayed wild-type septin staining at the mother-bud neck, even after activation of Swe1 by HU. This is in contrast to similar phenotypes for mutants in the two deadenylases for other cellular functions such as the DNA damage response (Hammet et al. 2002; Traven et al. 2005). A possible explanation for the phenotypic difference in septin organization could lie in the differential effects on mRNA poly(A) tail lengths seen for the two deadenylase mutants. The length of the poly(A) tail on mRNAs for some of the tested transcripts (eg., GIN4, CLA4, and ELM1) was not affected at all by loss of Pan2. Even for the mRNAs that did accumulate some “longer-tailed” forms in pan2Δ cells (for example RGA1, RGA2, BEM3, and SHS1), the length of the tail was less tightly stabilized than in ccr4-1 and pop2Δ cells, and tails as short as in the wild type still predominated. However, Pan2 could compensate for the absence of Ccr4: the poly(A) tails on the short-tailed septin assembly regulators were longer in pan2Δ ccr4-1 than in ccr4-1 mutants, and pan2Δ ccr4-1 double mutants displayed bigger and elongated cells (not seen in pan2Δ or ccr4-1 single mutants), which showed no septin staining, but it remains to be determined if this is a cause or a consequence of the cell size/morphology defect.
How does Ccr4-Pop2 contribute to septin assembly? We suggest that post-transcriptional control of the expression of septins and septin assembly factors by Ccr4-Pop2-NOT-dependent deadenylation contributes to proper septin organization. Longer mRNA poly(A) tails on transcripts encoding septin assembly factors likely lead to higher mRNA levels and/or more efficient translation. Alternatively timely expression during the cell cycle might be compromised (Beilharz and Preiss 2007). The process of septin assembly is sensitive to protein levels, for example overexpression of the septins compromises cell morphology and septin organization (Sopko et al. 2006; Iwase et al. 2007) and therefore higher levels of septins and/or septin assembly factors could lead to septin organization defects.
It remains to be determined how longer mRNA poly(A) tails on septins and septin assembly factors control their expression. Our data suggest that translation rather than mRNA stability is affected, a result consistent with a previous report for the Ccr4 target gene CRT1 (Woolstencroft et al. 2006). We tested protein levels for a few of the septins and septin assembly regulators for which antibodies were available to us (Cdc42 and Cdc11), but did not observe higher levels in ccr4 and pop2 mutants, indicating that perhaps these are not the relevant targets. However, the experiments addressing mRNA and protein levels of the septins and septin assembly factors in cultures of ccr4 and pop2 mutant are complicated by the fact that only a proportion of the cells in the deadenylase mutant cultures display septin defects (generally around 15–30% in HU-treated samples, see Figure 3). Therefore the change in expression levels in the affected cells could be buffered by the remaining cells that display normal septin morphology, making it difficult to address the effects of Ccr4-Pop2-NOT. The heterogeneity of deadenylase mutant cultures suggests the presence of alternative or redundant pathways that compensate for absence of Ccr4-Pop2 in regulating the expression of genes that contribute to septin assembly.
Because Ccr4-Pop2-NOT affects a large number of genes, it is likely its effects on septin assembly are complex and, in addition to regulation of expression of septins and septin assembly factors, they include effects on other cellular processes, such as the cell cycle (Westmoreland et al. 2004; Traven et al. 2005; Woolstencroft et al. 2006; Manukyan et al. 2008). An obvious connection to test was between Ccr4 and G1 cyclin expression: G1 cyclin activity is required for proper septin ring assembly (Moffat and Andrews 2004; Gladfelter et al. 2005) and a recent report demonstrated that timely expression of CLN1 and CLN2 in the cell cycle is compromised in ccr4Δ mutants, leading to increased cell size (Manukyan et al. 2008). However, even though overexpression of Cln2 was enough to suppress the cell size defects of ccr4Δ (Manukyan et al. 2008), we found that it did not suppress the morphology defects (data not shown), suggesting perturbations in G1 cyclins activity are unlikely to be the reason for the observed septin assembly defects in ccr4Δ mutants.
Our data also indicate that, in addition to the mRNA deadenylase catalytic activity, other functions of Ccr4-Pop2-NOT are required for regulation of septin organization and morphogenesis: (i) the ccr4Δ mutant showed a more pronounced morphology defect and stronger synthetic interactions with mutations in the septins than the exonuclease-deficient ccr4-1 strain, even though ccr4Δ and ccr4-1 mutants show no difference in their deadenylation defects for all of the transcripts tested so far (T. H. Beilharz and T. Preiss, unpublished data) and (ii) mutants in the NOT genes, which have minor deadenylation defects (Tucker et al. 2002), also displayed morphogenesis and septin defects. In particular, the morphology defects seen in not2Δ and not5Δ mutants were more pronounced than the defects seen in the absence of Ccr4-Pop2 deadenylase activity.
What is the deadenylase-independent function of Ccr4-Pop2-NOT? Ccr4 might have roles in post-transcriptional control that are additional to its catalytic role as an exonuclease; for example, it might facilitate recruitment of factors to target mRNAs by protein–protein interactions. A prime candidate is the translational repressor and decapping activator Dhh1, which makes direct physical contact with Ccr4-Pop2-NOT complex via Not1, Not4, and Caf40 (Tarassov et al. 2008). Interestingly, it has been reported that deletion of the RRP6 and RRP44 exonuclease subunits of the exosome (a nuclear and cytoplasmic RNA degradation complex) also leads to a stronger phenotype than inactivation of their nuclease activity, possibly because of requirements for complex stability and/or scaffold functions for interactions with cofactors (Schmid and Jensen 2008).
The NOT subunits of Ccr4-Pop2-NOT are thought to predominantly function in transcriptional control, affecting transcription initiation and chromatin modification (Collart 2003; Laribee et al. 2007; Mulder et al. 2007). Therefore, the morphogenesis and septin defects in the not mutants suggest that transcriptional functions of Ccr4-Pop2-NOT also contribute to septin assembly.
The septins perform diverse and essential cellular roles in lower and higher eukaryotes including humans, and thus how their organization and function is controlled is an important issue. We have identified that the Ccr4-Pop2-NOT complex contributes to septin organization in yeast. The mRNA deadenylase subunits Ccr4 and Pop2 contribute to septin organization, as do the NOT subunits, which have additional functions in transcriptional regulation. Future studies will be aimed at identifying the Ccr4-Pop2-NOT-targeted mRNAs relevant for septin organization.
Acknowledgments
We thank Andrew Hammet, Trevor Lithgow, and Amy Gladfelter for comments on the manuscript. This work was supported by a Peter Doherty fellowship from the Australian National Health and Medical Research Council (NHMRC), and an establishment grant from the Ramaciotti Foundation Australia (to A.T), as well as an NHMRC project grant (to J.H.). T.B. is the recipient of an Australian Research Council (ARC) ARF fellowship. T.P. acknowledges funding from the NHMRC, ARC, and the Sylvia & Charles Viertel Charitable Foundation.
Supporting information is available online at: http://www.genetics.org/cgi/content/full/genetics.109.104414/DC1.
References
- Asano, S., J. E. Park, L. R. Yu, M. Zhou, K. Sakchaisri et al., 2006. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J. Biol. Chem. 281 27090–27098. [DOI] [PubMed] [Google Scholar]
- Barral, Y., M. Parra, S. Bidlingmaier and M. Snyder, 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz, T. H., and T. Preiss, 2007. Widespread use of polyA tail length control to accentuate expression of the yeast transcriptome. RNA 13 982–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J. P., M. Longtine, J. R. Pringle and E. Bi, 2003. The role of Cdc42 GTPase activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14 4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Y. C. Chiang and C. L. Denis, 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart, M., 2003. Global control of gene expression in yeast by the Ccr4-NOT complex. Gene 313 1–16. [DOI] [PubMed] [Google Scholar]
- Cvrckova, F., C. de Virgilio, C. E. Manser, J. R. Pringle and K. Nasmyth, 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9 1817–1830. [DOI] [PubMed] [Google Scholar]
- DeMarini, D. J., A. E. Adams, H. Fares, C. de Virgilio, G. Valle et al., 1997. A septin-based hierarchy of proteins required for a localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere, J., M. S. Gentry, R. L. Hallberg and Y. Barral, 2003. Phosphorylation-dependent regulaton of septin dynamics during the cell cycle. Dev. Cell 4 345–357. [DOI] [PubMed] [Google Scholar]
- Douglas, L. M., F. J. Alvarez, C. McCreary and J. B. Konopka, 2005. Septin function in yeast model systems and pathogenic fungi. Eukaryot. Cell 4 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink, J. M., M. B. Smolka, H. Zhou and R. D. Kolodner, 2006. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J. Cell Biol. 175 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, S. K., and J. R. Pringle, 1991. Cellular morphogenesis in Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 12 281–292. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A. S., I. Bose, T. R. Zyla, E. S. Bardes and D. J. Lew, 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 21 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A. S., L. Kozubowski, T. R. Zyla and D. J. Lew, 2005. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 118 1617–1628. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A. S., T. R. Zyla and D. J. Lew, 2004. Genetic interactions among regulators of septin organization. Eukaryot. Cell 3 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm, A. C., and M. Wickens, 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell. Biol. 9 337–344. [DOI] [PubMed] [Google Scholar]
- Gonzales-Novo, A., L. Labrado, A. Jimenez, M. Sanchez-Perez and J. Jimenez, 2006. Role of the septin Cdc10 in virulence of Candida albicans. Microbiol. Immunol. 50 499–511. [DOI] [PubMed] [Google Scholar]
- Hammet, A., B. L. Pike and J. Heierhorst, 2002. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A) nuclease complex contributes to survival of replication blocks. J. Biol. Chem. 277 22469–22474. [DOI] [PubMed] [Google Scholar]
- Iwase, M., J. Luo, E. Bi and A. Toh-e, 2007. Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae. Genetics 177 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M., L. Luo, S. Nagaraj, M. Longtine, H. B. Kim et al., 2006. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Cell Biol. 17 1110–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton, M. A., and D. J. Lew, 2006. Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint. Curr. Opin. Microbiol. 9 540–546. [DOI] [PubMed] [Google Scholar]
- Kim, H. B., B. K. Haarer and J. R. Pringle, 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, D. H, T. H. Beilharz, S. Marguerat, J. Matta, S. Watt et al., 2007. A network of multiple regulatory layers shape gene expression in fission yeast. Mol. Cell 26 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee, R. N., Y. Shibata, D. P. Mersman, S. R. Collins, P. Kemmeren, et al., 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. USA 104 5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., and Y. Wang, 2006. The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol. Biol. Cell 17 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., H. Fares and J. R. Pringle, 1998. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle et al., 2000. Septin-dependent assembly of a cell-cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell Biol. 20 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manukyan, A., J. Zhang, U. Thippeswamy, J. Yang, N. Zavala et al., 2008. Ccr4 alters cell size in yeast by modulating the timing of CLN1 and CLN2 expression. Genetics 179 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, J., and B. Andrews, 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 6 59–66. [DOI] [PubMed] [Google Scholar]
- Mulder, K. W., A. B. Brenkman, A. Inagaki, N. J. van der Broek and H. T. Timmers, 2007. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 35 2428–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H. O., and E. Bi, 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 7 48–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and H. Song, 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11 121–127. [DOI] [PubMed] [Google Scholar]
- Preiss, T., M. Muckenthaler and M. W. Hentze, 1998. Poly(A)-tail promoted translation in yeast: implications for translational control. RNA 4 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallés, F. J, and S. Strickland, 1995. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl. 4 317–321. [DOI] [PubMed] [Google Scholar]
- Schmid, M., and T. H. Jensen, 2008. The exosome, a multipurpose RNA decay machine. Trends Biochem. Sci. 33 501–510. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., T. Drgon, B. Bowers and E. Cabib, 2008. Hyperpolarized growth of Saccharomyces cerevisiae cak1P212S and cla4 mutants weakens cell walls and renders cells dependent on chitin synthase 3. FEMS Yeast Res. 8 362–373. [DOI] [PubMed] [Google Scholar]
- Smolka, M. B., S. H. Chen, P. S. Maddox, J. M. Enserink, C. P. Albuquerque et al., 2006. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J. Cell Biol. 175 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko, R., D. Huang, N. Preston, G. Chua, B. Papp et al., 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 3 319–330. [DOI] [PubMed] [Google Scholar]
- Tarassov, K., V. Messier, C. R. Landry, S. Radinovic, M. M. Serna Molina et al., 2008. An in vivo map of the yeast protein interactome. Science 320 1465–1470. [DOI] [PubMed] [Google Scholar]
- Traven, A., A. Hammet, N. Tenis, C. L. Denis and J. Heierhorst, 2005. Ccr4-NOT complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 169 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis et al., 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104 377–386. [DOI] [PubMed] [Google Scholar]
- Tucker, M., R. R. Staple, M. A. Valencia-Sanchez, D. Mulhard and R. Parker, 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M., and J. Thorner, 2004. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland, T. J., J. R. Marks, J. A. Olson, Jr., E. M. Thompson, M. A. Resnick et al., 2004. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell 3 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstencroft, R. N., T. H. Beilharz, M. A. Cook, T. Preiss, D. Durocher et al., 2006. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 119 5178–5192. [DOI] [PubMed] [Google Scholar]