Abstract
The Su(z)2 complex contains Posterior sex combs (Psc) and Suppressor 2 of zeste [Su(z)2], two paralogous genes that likely arose by gene duplication. Psc encodes a Polycomb group protein that functions as a central component of the PRC1 complex, which maintains transcriptional repression of a wide array of genes. Although much is known about Psc, very little is known about Su(z)2, the analysis of which has been hampered by a dearth of alleles. We have generated new alleles of Su(z)2 and analyzed them at the genetic and molecular levels. Some of these alleles display negative complementation in that they cause lethality when heterozygous with the gain-of-function Su(z)21 allele but are hemizygous and, in some cases, homozygous viable. Interestingly, alleles of this class identify protein domains within Su(z)2 that are highly conserved in Psc and the mammalian Bmi-1 and Mel-18 proteins. We also find several domains of intrinsic disorder in the C-terminal regions of both Psc and Su(z)2 and suggest that these domains may contribute to the essential functions of both proteins.
THE Su(z)2 complex of Drosophila spans ∼100 kb and contains two divergently transcribed genes, Posterior sex combs (Psc) and Suppressor 2 of zeste [Su(z)2] (Adler et al. 1989; Wu et al. 1989; Wu and Howe 1995). Of the two, Su(z)2 is the lesser known. It stands in stark contrast to Psc, which has been the focus of extensive genetic, molecular, and biochemical analyses for many years. Psc is a member of the Polycomb group (PcG) of genes, many of which function at the level of chromatin as part of at least two PcG repressive complexes, called PRC1 and PRC2 (reviewed by Brock and Fisher 2005; Breiling et al. 2007; Schuettengruber et al. 2007; Schwartz and Pirrotta 2007, 2008; Mateos-Langerak and Cavalli 2008). PRC2 contains the Enhancer of zeste [E(z)] protein, which provides a histone methyltransferase activity that methylates histone H3 on lysine 27 (reviewed by Cao and Zhang 2004). This epigenetic chromatin mark is believed to recruit PRC1 (Fischle et al. 2003; Min et al. 2003; but see also Kahn et al. 2006), which then functions to maintain target gene silencing. PRC1 contains >15 subunits (Saurin et al. 2001) and blocks both transcription and chromatin remodeling in vitro (Shao et al. 1999). These inhibitory activities can be reproduced by a minimal complex, called the PRC1 core complex (PCC), consisting of four proteins, including Psc, Polycomb (Pc), Polyhomeotic (Ph), and Sex combs extra (Sce) (Francis et al. 2001; Sce is also known as dRing1; Fritsch et al. 2003; Gorfinkiel et al. 2004). Psc can reproduce the inhibitory activities by itself, suggesting that it is a central component of PCC (Francis et al. 2001).
Many lines of evidence suggest that Su(z)2 is functionally related to, and even partially redundant with, Psc. For example, overexpression of either gene leads to bristle defects (Brunk et al. 1991b; Sharp et al. 1994; Wu and Howe 1995) and, as detailed below, certain alleles of either gene can act as suppressors or enhancers of an allele of the zeste (z) gene (Wu and Howe 1995). In addition, embryos homozygous for a deficiency that removes both genes, Su(z)21.b8, display cuticle defects that are more severe than those of embryos lacking either Psc or Su(z)2 alone (Adler et al. 1991; Soto et al. 1995; Wu and Howe 1995). Similarly, somatic clones homozygous for Su(z)21.b8 in wing imaginal discs show derepression of homeotic genes and cellular overgrowth, whereas clones homozygous for loss-of-function (l-o-f) alleles of either Psc or Su(z)2 do not (Beuchle et al. 2001). Su(z)2 also colocalizes with Psc and Pc at many sites on polytene chromosomes (Rastelli et al. 1993; Platero et al. 1996; Sharp et al. 1997) and, very recently, co-immunoprecipitation experiments using Drosophila and cell-line extracts suggest that Su(z)2 exits in a complex that also contains Pc, Ph, and Sce/dRing1, which are the three non-Psc members of PCC (Lo et al. 2009).
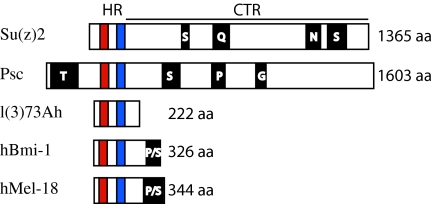
Psc and Su(z)2 also resemble each other at the structural level. First, both Psc and Su(z)2 are large proteins, consisting of 1603 and 1365 amino acids, respectively. Second, they are homologous over a 200-amino-acid interval located in their N-terminal regions. This interval, called the homology region (HR), contains a ring-finger (RF) domain and a helix-turn-helix (HTH) domain and is 37% identical between the two proteins (Figure 1) (Brunk et al. 1991a; van Lohuizen et al. 1991). RF and HTH domains have been implicated in mediating protein interactions. Finally, the two proteins are similar in the amino acid content of the ∼1000 amino acids of their C-terminal regions (CTRs). While the CTRs are not conserved at the level of the primary amino acid sequence, both show a high level of flexibility, are enriched in proline and serine, and contain runs of one or more of the following amino acids: asparagine, glutamine, glycine, proline, serine, and threonine (Figure 1) (Brunk et al. 1991a). Functional studies of Psc have confirmed that both the HR and the CTR contribute to the activity of the protein. In particular, genetic and molecular analyses indicate that the RF is required for Psc function in vivo, the HR is necessary for assembly of the PCC in vitro, and the CTR, which is functionally separable from the RF, is essential for wild-type Psc activity in vivo as well as for the inhibition of transcription and chromatin remodeling in vitro (King et al. 2005). Importantly, Su(z)2 behaves similarly to Psc in in vitro assays; it can replace Psc in a complex with Pc, Ph, and Sce/dRing1, its HR is essential for formation of the complex, and its CTR inhibits chromatin remodeling (Lo et al. 2009). This latter finding is consistent with studies in mammalian cells showing that Su(z)2, either full length or lacking the majority of its HR, can repress activator function (Bunker and Kingston 1994).
Figure 1.—
Comparison of Drosophila Su(z)2, Psc, and l(3)73Ah with human Bmi-1 and Mel-18. The HR and CTR are labeled. The RF is in red and the HTH is in blue. Regions enriched for specific amino acids are in black and labeled with the relevant residue. The figure is drawn to scale.
The mammalian orthologs of Psc and Su(z)2 are Bmi-1, which is involved in stem cell maintenance and cancer (for example, see Pietersen et al. 2008; Sangiorgi and Capecchi 2008; reviewed in Sparmann and van Lohuizen 2006; Pietersen and van Lohuizen 2008), and Mel-18 (also known as PCGF2), which has been implicated in tumor suppression and the regulation of c-myc and bmi-1 (for example, see Guo et al. 2007a,b; Wiederschain et al. 2007; Lee et al. 2008). Bmi-1 and Mel-18 are homologous to Psc and Su(z)2 throughout the HR (Brunk et al. 1991a; van Lohuizen et al. 1991; Alkema et al. 1993; Ishida et al. 1993) and, although the CTRs of these mammalian proteins are relatively short, they resemble the long CTRs of Psc and Su(z)2 in that they are enriched in proline and serine.
Despite the many similarities between Psc and Su(z)2, there are also differences between the two. For example, Psc and Su(z)2 alleles differ with respect to their lethal phases and cuticular phenotypes (Jürgens 1985; Adler et al. 1989, 1991; Wu et al. 1989; Wu and Howe 1995) as well as with respect to their interactions with trithorax group genes, which act in opposition to PcG genes (reviewed by Brock and Fisher 2005; Breiling et al. 2007; Schuettengruber et al. 2007; Schwartz and Pirrotta 2007, 2008). Given the structural similarities and partial functional redundancy between Psc and Su(z)2, these differences suggest that Su(z)2 has roles beyond those associated with prototypical PcG genes.
Su(z)2 was first identified by the Su(z)21 allele (Kalisch and Rasmuson 1974). This allele was isolated as a gain-of-function (g-o-f) dominant suppressor of an allele of the X-linked zeste gene, called z1, which represses expression from the white+ (w+) eye-color gene in a manner that is sensitive to whether the w+ gene is paired with another w+ gene (Jack and Judd 1979; reviewed by Wu and Goldberg 1989; Pirrotta 1991; Kassis 2002). For example, the eye color of z1 w+/z1 w+ females is yellow instead of wild-type red because the somatic homolog pairing that occurs in Drosophila (Stevens 1907, 1908; Metz 1916; Lewis 1954; reviewed by McKee 2004) brings the two w+ genes together, making them subject to silencing by z1. Strikingly, Su(z)21 suppresses the z1 eye-color phenotype in a dominant antimorphic fashion, such that z1 w+/z1 w+; Su(z)21/+ females have eyes that are red, rather than yellow (Kalisch and Rasmuson 1974; Wu and Howe 1995). A dominant allele of Psc, called Psc1, is also a strong suppressor of the z1 phenotype. Interestingly, Psc1 displays second site noncomplementation (SSNC) (reviewed by Hawley and Gilliland 2006) with Su(z)21 such that Psc1 +/+ Su(z)21 heterozygotes are not viable (Wu 1984; Adler et al. 1989; Wu et al. 1989; Wu and Howe 1995). This SSNC suggests that the gene products of Su(z)2 and Psc may interact, which is consistent with their colocalization at many sites on polytene chromosomes (Rastelli et al. 1993; Platero et al. 1996; Sharp et al. 1997).
How Su(z)21 and Psc1 suppress z1 is unclear, although much has been learned about Zeste, which is found in PRC1 (Saurin et al. 2001; Mulholland et al. 2003) and is known to bind DNA (reviewed by Pirrotta 1991; also see Mohrmann et al. 2002), self-associate or aggregate (reviewed by Pirrotta 1991; also see Chen and Pirrotta 1993; Rosen et al. 1998), and participate in both gene activation and gene repression (Biggin et al. 1988; Laney and Biggin 1992; Kal et al. 2000; Hur et al. 2002; Mulholland et al. 2003; Dejardin and Cavalli 2004). Of the mechanisms being considered, several suggest that Su(z)2 and Psc interact with Zeste directly or within the context of a larger complex, the form or occurrence of such interactions being contingent on the mutant or wild-type state of the proteins (Mansukhani et al. 1988; Wu and Goldberg 1989; Wu et al. 1989; Chen and Pirrotta 1993; Rastelli et al. 1993; Rosen et al. 1998; Saurin et al. 2001; Mulholland et al. 2003). For example, the z1 protein may silence w+ by drawing Su(z)2 and/or Psc to the locus or, if Su(z)2 and/or Psc are normally present at the target, may induce them to silence w+ to an abnormal degree. If so, Su(z)21 and Psc1 may suppress z1 by antagonizing that silencing. Alternatively, as Zeste has been implicated in silencing, it is possible that Zeste1 is hypermorphic for that activity and that Su(z)21 and Psc1 suppress z1 by antagonizing Zeste1 directly.
Our studies have focused on extant and newly isolated alleles of Su(z)2 and have identified a special class that display negative complementation with Su(z)21. Consistent with the implication that instances of negative complementation result from protein–protein interactions (Fincham 1966), we find that three of the alleles that display negative complementation with Su(z)21 contain missense mutations in the RF, a protein motif known to mediate such interactions. Two other alleles that display negative complementation identify two domains in the HR that lie outside the RF and HTH and are conserved in both Bmi-1 and Mel-18. Finally, we have looked more closely at the CTRs of Su(z)2 and Psc and find that both contain many regions of intrinsic protein disorder, which may speak further to the functional similarities between these two proteins.
MATERIALS AND METHODS
Culture conditions and stocks:
All crosses were conducted at 25° on standard Drosophila cornmeal, yeast, sugar, and agar medium with p-hydroxybenzoic acid methyl ester added as a mold inhibitor. In general, crosses were carried out with approximately three females and approximately three males in vials and brooded daily to prevent crowding. All chromosomes carrying Su(z)2 mutations were isogenic and kept in stock heterozygous with the CyO-19 GFP-bearing balancer chromosome (Bloomington Stock Center). Su(z)24 is an unstable allele, as we have identified two isolates with distinct molecular signatures: both isolates contain an 8-bp deletion in exon 5, but one contains an ∼9-kb insertion in exon 1 while the other does not. As we cannot state with certainty that either isolate corresponds to the original Su(z)24 mutation, we have renamed the insert-bearing allele Su(z)24-34, in recognition of its recovery from stock 34abl.1$, and the allele lacking the insert Su(z)24-31, in recognition of its recovery from stock 31ar.1$. Note that stocks 34ab1.1$ and 31ar.1$ are related by lineage to a single originating stock of Su(z)24. Su(z)2sM was discovered in a series of control crosses designed to confirm the full viability of Su(z)21 in trans to wild-type second chromosomes derived from a variety of standard laboratory strains. To our great surprise, we discovered that one of our Canton-S stocks displayed nearly complete lethality when crossed to Su(z)21. Single chromosomes extracted from this stock displayed similar lethality when heterozygous with Su(z)21, indicating that this Canton-S stock was homozygous for a mutation on the second chromosome that was lethal when heterozygous Su(z)21. This spontaneous mutation was subsequently called Su(z)2sM. The stock of dp cn bw; + that was used in the mutagenesis is isogenic for chromosomes II and III.
Mutagenesis:
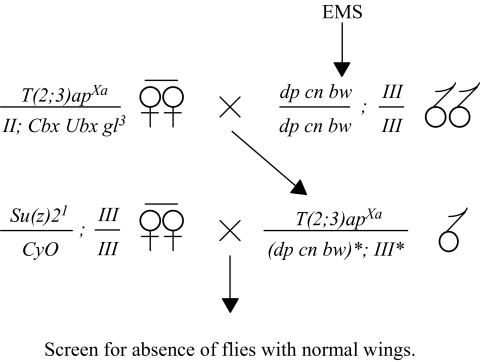
The Su(z)2 complex, including Psc and Su(z)2, is located on chromosome II at meiotic map position 67.3 and polytene position 49E (Wu and Howe 1995). In our mutageneses designed to recover Su(z)21-interacting mutations (SIMs), males of the genotype dp cn bw ; + were fed EMS as previously described (Wu and Howe 1995) and crossed to T(2;3)apXa/CyO virgin females. Approximately 17,500 F1 males heterozygous for T(2;3)apXa and mutagenized (*) chromosomes II* and III* were then individually mated in vials with three Su(z)21/CyO virgin females. F2 progeny were scored for the absence of flies with normal wings (i.e., Cy+ and ap+), indicating that chromosome II* and/or chromosome III* carried a SIM mutation that was lethal in a Su(z)21 background.
To verify that such lethality was due to Su(z)21, and not to an extraneous mutation on the Su(z)21 chromosome, we assessed the linkage of the capacity of Su(z)21 to suppress z1 to that of the lethal interaction between Su(z)21 and the SIMs. This analysis was applied to six (s14, s15, s20, s21, s36, and s84) of the seven SIMs; the s95 allele was not tested because it suppresses z1 on its own. We crossed z1 wis; Su(z)21/+ virgin females to putative SIM/CyO males and looked for z1 wis/Y; putative SIM/Su(z)21 recombinant F1 males, which would be predicted to be viable with red eyes and straight wings if the lesion on the Su(z)21 chromosome that was responsible for the lethal interaction with the SIMs were separable from Su(z)21. No z1 wis/Y; putative SIM/Su(z)21males were observed for s20 (0 recombinants/195 total flies scored), s36 (0/343), and s84 (0/361), while few were observed for s15 (1/257) and s21 (5/147). The frequency of red-eyed straight-winged males carrying s15 and s21 can be explained by the low but significant viability of s15/Su(z)21 and s21/Su(z)21 flies (Table 1). A few recombinants were also recovered in the analysis of s14, consistent with other data indicating it to be an allele of Psc.
TABLE 1.
Genetic analysis of SIMs
| Su(z)2s15 | Su(z)2s20 | Su(z)2s21 | Su(z)2s36 | Su(z)2s84 | Su(z)2s95 | Su(z)2sM | ||
|---|---|---|---|---|---|---|---|---|
| Su(z)21 | P | 14 (353) | 0 (339) | 2 (261) | 0 (202) | 0 (200) | 0 (259) | 8 (371) |
| M | 16 (223) | 0 (182) | 3 (184) | 0 (135) | 0 (138) | 0 (154) | 8 (195) | |
| Su(z)21.b8 | P | 10 (123) | 31 (196) | 26 (165) | 25 (134) | 0 (218) | 0 (113) | 34 (149) |
| M | 25 (186) | 30 (174) | 30 (252) | 35 (135) | 0 (108) | 0 (213) | 32 (249) | |
| Su(z)21.b7 | P | 26 (136) | 23 (251) | 24 (187) | 16 (216) | 0 (182) | 4 (253) | 25 (177) |
| M | 30 (128) | 32 (253) | 34 (207) | 35 (224) | 0 (157) | 3 (272) | 29 (214) | |
| Su(z)24-31 | P | 18 (257) | 29 (270) | 28 (183) | 33 (238) | 0 (132) | 0 (219) | 34 (270) |
| M | 21 (260) | 26 (253) | 33 (229) | 33 (262) | 0 (127) | 0 (186) | 30 (287) | |
| Su(z)24-34 | P | 13 (111) | 35 (210) | 36 (199) | 0 (238) | 0 (144) | 0 (174) | 32 (176) |
| M | 10 (201) | 27 (205) | 31 (285) | 0 (205) | 0 (248) | 0 (145) | 30 (114) | |
| Su(z)2h29 | P | 15 (185) | 27 (210) | 23 (337) | 23 (157) | 0 (107) | 0 (221) | 33 (166) |
| M | 34 (144) | 33 (196) | 39 (163) | 32 (117) | 0 (123) | 0 (144) | 29 (100) |
The first column lists the allele contributed by one parent, while the first row lists the allele contributed by the other. The paternal (P) or maternal (M) origin of the allele in the first column is indicated. For each cross, the viability of the mutant class heterozygous for the allele in column one and the allele in row one is indicated as a percentage, followed by the total number of flies scored in parentheses. Two alleles that are completely viable when heterozygous with one another are expected to have a viability equal to 33% under ideal conditions (see materials and methods).
All crosses to test viability were conducted in the following way: w−; mutant allele 1/CyO males or females were crossed to w−; mutant allele 2/CyO females or males, respectively. We defined viability as the number of Cy+ flies/total progeny. Under ideal conditions, viability should equal 33% when mutant allele 1/mutant allele 2 heterozygotes are 100% viable and transmission rates of all chromosomes are equal. We have avoided calculating viability in terms of expected viability (i.e., the relative percentage of 33%) because the mutant allele 1/CyO and mutant allele 2/CyO classes could not be distinguished in the majority of our crosses, precluding our ability to determine the relative transmission rates for the two mutant alleles.
Molecular analysis of mutant alleles:
Southern analysis was performed on DNA extracted from flies heterozygous for a mutant allele and the Cy0-19 balancer. Thirty flies of each genotype were frozen at −80° overnight, and their DNA was extracted using the Berkeley Drosophila Genome Project crude fly protocol (Spradling et al. 1999). Aliquots of DNA corresponding to 2.5 flies were then digested with EcoRI, NotI, and EcoRI/NotI, separated on an agarose gel, transferred to nylon filters via standard Southern blot protocols, and probed with 32P-labeled Su(z)2 cDNA.
Sequence analysis was conducted on embryos homozygous for a mutant allele as previously described (King et al. 2005) by using primer sets specific for all Su(z)2 exons. Double-strand sequence was obtained for all exons of all alleles, except the three structurally rearranged alleles: Su(z)21.b7, for which only exons 4 and 5 were sequenced; Su(z)2s95, for which only exons 1, 2, and 5 were sequenced; and Su(z)24-34, for which only exons 1–5 were sequenced. Note that our sequencing strategy for Su(z)21.b7 did not allow us to confirm the presence of the L120F missense mutation found in Su(z)21, from which Su(z)21.b7 was derived. The key molecular lesions associated with Su(z)21, Su(z)24-31, Su(z)24-34, Su(z)2h29, Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2s84, Su(z)2sM, Su(z)31, and Su(z)2Deos were verified by obtaining genomic DNA from flies heterozygous for a mutant allele and CyO-19, by amplifying the relevant region by PCR, and by sequencing the resulting fragment. We found the Su(z)2 locus to be highly polymorphic between mutant and wild-type laboratory strains from different backgrounds. In fact, the high frequency of strain-specific polymorphisms required the use of strain-specific primer sets. Polymorphic changes within the exonic regions are noted in the supporting information in Table S1.
Identification of the roo element insert in Su(z)2s95 resulted from our inability to amplify either exon 3 or exon 4 of this allele. Because the primer sets for these two exons overlap, we anticipated that Su(z)2s95 would contain foreign sequence that either had inserted between the sites homologous to the upstream primer for exon 4 and the downstream primer for exon 3 or had disrupted one of these two sites. This localized the putative insertion to a 196-bp region that spanned the third intron. We then used inverse PCR to identify the distal breakpoint of the insertion, followed by sequence analysis to identify sequences homologous to a roo element LTR. Primers internal to the roo element were then used with the upstream primer for exon 3 and the downstream primer for exon 4 to amplify the two ends of the insertion, producing amplicons of the expected size and sequence. Additional analysis suggested that the insertion may not be a full-length wild-type roo element.
The Su(z)21.b7 deficiency breakpoints were amplified by PCR from Su(z)21.b7/CyO genomic DNA using the upstream primer (95delus) 5′-TGTTCGGTCCCAAAGAAGC-3′ and the downstream primer (95delds4) 5′-TGATCAAGGAAAATGTGTATTTTAGC-3′. While these primers are predicted to generate a 5262-bp PCR product from wild-type DNA, they instead amplified a 1.5-kb fragment, consistent with the results of our Southern analyses of Su(z)21.b7. This amplicon was subcloned into the TOPO-TA vector (Invitrogen), and 10 independent clones were end-sequenced with the M13 forward and reverse primers to identify the sequence at the junction of the deficiency breakpoints. The sequence, 5′-CCAAGGTTCTTAGTTCT-3′, contains a 4-bp insertion at the junction (underlined).
Sequence data for Su(z)2 mutations have been deposited in GenBank and correspond to accession nos. FJ897446–FJ897460. The roo element/genomic DNA junction sequences for Su(z)2s95 and the breakpoint sequence for Su(z)21.b7 correspond to GenBank accession nos. FJ876147–FJ876149.
The s14 mutation is caused by a G-to-A mutation in Psc that abolishes the exon 5 splice site. This mutation is predicted to truncate the Psc protein and may result in a protein that is similar in size to that encoded by Psc1, which is also lethal in trans to Su(z)21. The Pscs14 sequence data have been given accession no. FJ917397.
RESULTS
Our studies began with five extant alleles: Su(z)21, Su(z)21.b7, Su(z)24-31, Su(z)24-34, and Su(z)2h29 (Gelbart 1971; Kalisch and Rasmuson 1974; Wu and Howe 1995). Su(z)21, described above, was induced by EMS, suppresses z1, and shows SSNC with Psc1. Su(z)21.b7 was recovered as an X-ray-induced l-o-f derivative of Su(z)21 and neither suppresses z1 nor shows SSNC with Psc1 (Wu 1984; Adler et al. 1991; Wu and Howe 1995). Su(z)24-31 and Su(z)24-34 represent distinct isolates derived from our stock of Su(z)24, which appears to be an unstable allele (materials and methods). Su(z)24 had been induced by X rays and behaved as a g-o-f allele that was lethal in trans to Su(z)21. It also suppressed z1 and showed SSNC with Psc1 although, in both cases, its phenotype was weaker than that of Su(z)21 (Gelbart 1971; Wu et al. 1989; Wu and Howe 1995). The Su(z)24-31 isolate remains a suppressor of z1 but shows a degree of SSNC with Psc1 that exceeds that observed with Su(z)24 (data not shown); whereas Psc1 +/+ Su(z)24 animals had a viability of ∼33% as compared to wild type, Psc1 +/+ Su(z)24-31 animals are not viable. The second isolate, Su(z)24-34, differs from Su(z)24-31 in that it is a weaker suppressor of z1 and shows only weak, if any, SSNC with Psc1 (data not shown). Finally, Su(z)2h29 is an EMS-induced l-o-f allele that is also lethal when heterozygous with Su(z)21 but neither suppresses z1 nor exhibits SSNC with Psc1 (Wu and Howe 1995).
A genetic screen for new alleles of Su(z)2:
We undertook a mutagenesis to generate additional Su(z)2 alleles, anticipating that the molecular genetic analysis of such alleles would identify important protein domains and elucidate how the structure of Su(z)2 contributes to its function. Previous attempts to generate Su(z)2 alleles by screening for mutations that failed to complement deficiencies deleting both Psc and Su(z)2 were largely unsuccessful (Wu and Howe 1995). These screens tested >17,000 mutagenized second chromosomes and recovered eight alleles of Psc but only one of Su(z)2, indicating a large bias against the recovery of Su(z)2 alleles. To shift this bias toward Su(z)2, we conducted an F2 screen for EMS-induced mutations that are lethal when heterozygous with Su(z)21 (Figure 2; materials and methods). Since Su(z)21 is lethal when heterozygous with deficiencies of the locus, the l-o-f Su(z)21.b7 allele, both isolates of the g-o-f Su(z)24 allele, and the l-o-f Su(z)2h29 allele, we reasoned that this strategy would allow recovery of both l-o-f and g-o-f alleles of Su(z)2. Furthermore, since Su(z)21 displays SSNC with Psc1, this strategy also had the potential of recovering extragenic mutations in genes such as Psc, whose products may interact with the Su(z)2 protein.
Figure 2.—
Screen for SIMs. dp cn bw/dp cn bw; III/III males, isogenic for chromosomes II, marked with dp cn and bw, and III, were fed EMS and mated to T(2;3)apXa/II; Cbx Ubx gl3 virgin females. Single T(2;3)apXa/(dp cn bw)*; III* F1 males bearing mutagenized (*) autosomes were then mated in vials to Su(z)21/CyO; III/III virgin females. The vials were subsequently scored for the absence Su(z)21/(dp cn bw)*; III/III* F2 progeny, indicating that at least one of the mutagenized autosomes may carry a SIM. Because T(2;3)apXa causes a dominant notched wing phenotype, and the CyO balancer causes a dominant curly wing phenotype, vials lacking Su(z)21/(dp cn bw)*; III/III* F2 progeny were identified by the absence of flies with normal (non-notched, straight) wings. Note that use of T(2;3)apXa allowed for the simultaneous testing of both autosomes because it is a translocation between chromosomes II and III.
We screened >14,000 mutagenized dp cn bw second chromosomes and identified seven Su(z)21 interacting mutations (SIMs): s14, s15, s20, s21, s36, s84, and s95. In addition, we independently identified a spontaneous mutation (sM) in our Canton-S wild-type stock that behaved like a SIM (materials and methods). Taken together, the eight mutations showed a range of reduced viability when heterozygous with Su(z)21 (Table 1; data for s14 are not shown). Note that we calculate the viability of flies heterozygous for the two alleles of Su(z)2 as the percentage of such flies emerging from a cross in which females heterozygous for one allele are crossed to males heterozygous for the other allele. Under ideal conditions, wild-type viability is expected to give a score of 33% with this mating scheme (materials and methods; legend to Table 1).
To characterize these mutations further, we crossed each to the Su(z)21.b8 deletion that removes both Psc and Su(z)2, as well as to Su(z)21.b7, Su(z)24-31, Su(z)24-34, and Su(z)2h29 (Table 1). These crosses revealed that we had identified putative mutations in both Psc and Su(z)2, as predicted. These are exemplified by s14, s84, and s95, which are all lethal when heterozygous with Su(z)21.b8. s14 proved to be a new allele of Psc, as it fails to complement l-o-f alleles of Psc but is viable in trans to Su(z)21.b7, Su(z)24-31, Su(z)24-34, and Su(z)2h29 (data not shown; materials and methods). In contrast, s84 and s95 fail to, or only minimally, complement Su(z)21.b7 (Table 1), suggesting that they are new alleles of Su(z)2. Consistent with this, both mutations fail to complement one another (Table 2) as well as Su(z)24-31, Su(z)24-34, and Su(z)2h29 (Table 1). The recovery of these three alleles, one identifying Psc and two identifying Su(z)2, validated the efficacy of our mutagenesis and suggested that the remaining mutations would be informative.
TABLE 2.
Inter se crosses of SIMs
| Su(z)2s15 | Su(z)2s20 | Su(z)2s21 | Su(z)2s36 | Su(z)2s84 | Su(z)2s95 | ||
|---|---|---|---|---|---|---|---|
| Su(z)2s20 | P | 5 (341) | |||||
| M | 8 (350) | ||||||
| Su(z)2s21 | P | 11 (152) | 18 (179) | ||||
| M | 10 (230) | 17 (212) | |||||
| Su(z)2s36 | P | 5 (383) | 17 (327) | 15 (259) | |||
| M | 3 (466) | 10 (325) | 21 (195) | ||||
| Su(z)2s84 | P | 11 (425) | 16 (524) | 19 (258) | 17 (256) | ||
| M | 11 (303) | 20 (480) | 22 (224) | 20 (435) | |||
| Su(z)2s95 | P | 11 (401) | 17 (364) | 22 (221) | 19 (621) | 0 (329) | |
| M | 6 (494) | 19 (407) | 26 (235) | 20 (615) | 0 (404) | ||
| Su(z)2sM | P | 18 (231) | 27 (206) | 23 (222) | 28 (251) | 24 (156) | 30 (280) |
| M | 18 (152) | 32 (106) | 29 (350) | 30 (162) | 33 (214) | 37 (135) |
See Table 1 legend for explanation of format.
The behavior of the remaining five mutations (s15, s20, s21, s36, and sM) was notable. First, although they all show reduced viability when heterozygous with Su(z)21, they differ in the strength of their lethal interaction: s20 and s36 are completely lethal, s21 and sM are weakly viable, and s15 shows significant viability (Table 1). Second, their behavior in trans to Su(z)24-31 demonstrates that these alleles do not represent a simple allelic series; whereas s15 proved to be the most viable of the five when heterozygous with Su(z)21, it is not among the four (s20, s21, s36, and sM) that show significantly increased viability in trans to Su(z)24-31. Third, all heterozygous combinations of these five alleles show some degree of viability, s15 again distinguishing itself as the least able among the five to promote viability (Table 2). Note that these alleles are also viable in trans to s84 and s95 (Table 2) and that s36 is homozygous viable (data not shown). Fourth, and perhaps most surprisingly, each is markedly viable in trans to the l-o-f alleles Su(z)21.b8, Su(z)21.b7, and Su(z)21.h29 (Table 1). Taken together, these data made it difficult to assign these mutations unambiguously to Su(z)2. However, as described below, molecular analysis revealed that all except s15 either grossly disrupted the structure of Su(z)2 or contained lesions within the exons of the gene.
Molecular analysis of Su(z)2 alleles:
We carried out a molecular analysis of the Su(z)2 locus for Su(z)21, Su(z)21.b7, Su(z)24-31, Su(z)24-34, Su(z)2h29, and the seven SIMs that we believed would prove to be alleles of Su(z)2 (s15, s20, s21, s36, s84, s95, and sM). Southern analyses revealed that Su(z)21.b7, Su(z)24-34, and s95 contain gross structural changes. Except for s15, all of the remaining eight alleles were found to be structurally normal by Southern analysis but to contain discrete lesions within Su(z)2 as identified by sequencing of the exons (below; see materials and methods and Table S1 for additional details). We have therefore formalized the nomenclature for s20, s21, s36, s84, s95, and sM by giving them a base name of Su(z)2: Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2s84, Su(z)2s95, and Su(z)2sM. Although we were unable to find any change associated with s15, we have tentatively named this SIM Su(z)2s15 on the basis of its behavior in complementation analyses. Below we describe the lesion associated with Su(z)21, the founding allele of the locus, after which we detail the structure of the three grossly rearranged alleles and then the six alleles resulting from point mutations.
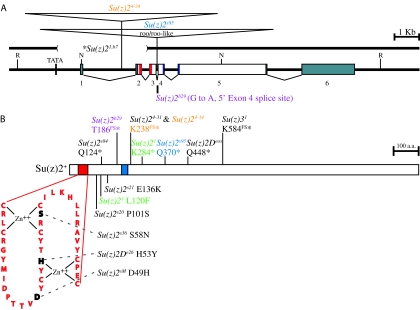
Su(z)21 contains an L120F missense mutation and a K284* nonsense mutation, which occurs shortly after the HTH domain (Figure 3) and is expected to produce a truncated protein. The L120F missense mutation is located in a region between the RF and HTH domains that will be discussed further below. The Su(z)21.b7 derivative of Su(z)21 carries the K284* mutation of Su(z)21 as well as an ∼3.5-kb deletion of the first two exons and a 4-bp insertion (TTCT) at the site of the deletion (Figure 3; materials and methods; see Table S1 for sequence data regarding regions of Su(z)21 and Su(z)21.b7 lying C-terminal to K284*). This finding differs from that of a previous study, which reported a deletion of only ∼2 kb (Brunk et al. 1991b). Further analysis of Su(z)21.b7 (materials and methods) identified a putative TATA box promoter sequence 8 bp upstream of the Su(z)21.b7 breakpoint. The presence of this putative promoter sequence is consistent with observations that Su(z)21.b7 is competent for transcription (Ali and Bender 2004).
Figure 3.—
Analysis of Su(z)2 alleles. (A) Structure of the wild-type Su(z)2 locus and insertions and deletions associated with Su(z)2 mutations. Psc (not shown) is located to the left. Exons 1–6 are shown as numbered rectangles. The RF is in red, and the HTH is in blue (exon 4–5 junction). Noncoding sequence is in gray. The TATA notation shown upstream of exon 1 is the putative TATA box identified 8 bp upstream of the Su(z)21.b7 breakpoint. R, EcoRI; N, NotI. (B) Frameshift, nonsense, and missense mutations associated with Su(z)2 mutations. Nonsense mutations are shown above the protein and are divided into two classes: those associated with a frameshift followed by a stop codon (FS*, top) and those associated with only a stop codon (*, bottom). Missense mutations are shown below the protein. The RF is shown in an exploded view, with bases that are altered by Su(z)2 mutations shown in black. Zn++-coordinating residues are indicated. Su(z)21 (green), Su(z)24-34 (orange), Su(z)2h29 (purple), and Su(z)2s95 (blue) are complex and have been color coded to highlight the multiple mutations that they contain. Su(z)21 contains an L120F missense mutation and a K284* nonsense mutation. Both Su(z)24-31 and Su(z)24-34 contain an 8-bp deletion in exon 5 (not shown) that generates a three-amino-acid frameshift ending in a K238* nonsense codon (B). Su(z)24-34 differs from Su(z)24-31 in that it also contains an insertion (A) that has been localized to a 1.6-kb ClaI/BamHI fragment in the distal half of intron 1. Su(z)2h29 contains a G-to-A transition that disrupts a 5′ acceptor site for exon 4 (A) and is predicted to result in a frameshift ending in a T186* nonsense codon (B). Su(z)2s95 contains a roo or roo-like (A) and a Q370* nonsense codon (B). *, Su(z)21.b7, a derivative of Su(z)21, carries a deletion (A), a 4-bp insertion (TTCT, not shown) at the site of the deletion, and the K284* nonsense codon that is also present in Su(z)21 (B). We have not determined whether Su(z)21.b7 also contains the L120F missense mutation that is found in Su(z)21. Both A and B are drawn to scale.
Southern and sequence analyses revealed that Su(z)24-34 and Su(z)2s95 are complex mutations. Su(z)2s95 contains a roo or roo-like element inserted in the third intron and a Q370* nonsense mutation in exon 5 (Figure 3, materials and methods). Similarly, Su(z)24-34 contains an ∼9-kb insertion in the first intron and an 8-bp deletion in exon 5 that results in a three-amino-acid frameshift followed by a nonsense codon (E235K, Q236K, T237R, and K238*) (Figure 3). The other Su(z)24 isolate, Su(z)24-31, retains the 8-bp deletion but does not carry the insertion (materials and methods; see Table S1 for sequence data regarding regions of Su(z)24-31 and Su(z)24-34 lying C-terminal to K238*). Consistent with this structural difference between the two alleles, Su(z)24-31 displays a genetic behavior that differs from that of Su(z)24-34. In particular, Su(z)24-31 complements Su(z)2s36, while Su(z)24-34 does not (Table 1) and, as mentioned earlier, is a stronger suppressor of z1 and shows a stronger interaction with Psc1.
The genetic behavior of Su(z)2s84 and Su(z)2h29 indicated that they would have alterations in Su(z)2, and this proved to be true. The Su(z)2s84 allele is caused by a Q124* nonsense mutation just after the RF (Figure 3). The small size of the predicted Su(z)2s84 protein suggests that its phenotype should be severe, consistent with observations that its capacity to complement other alleles is poor relative to that of several other alleles (Table 1). Su(z)2h29 results from a G-to-A transition that abolishes the 5′ splice acceptor site for exon 4. If exon 3 is able to splice over exon 4 to exon 5, this allele is predicted to cause a frameshift that extends from amino acid 139 to 186 after which a nonsense codon is encountered (Figure 3).
The four remaining alleles [Su(z)2s20, Su(z)2s21, Su(z)2s36, and Su(z)2sM] did not at first appear to be alleles of Su(z)2 because they complement Su(z)21.b8, Su(z)21.b7, and Su(z)2h29. However, as recombination analyses placed all four in the vicinity of the Su(z)2 complex (data not shown), we proceeded with sequence analyses and discovered that all four contain missense mutations in the HR of Su(z)2 (Figure 3). The lesions associated with Su(z)2s36 and Su(z)2sM alter the structure of the RF. The Su(z)2s36 allele contains a S58N missense mutation located within the first Zn++-coordinating domain of the RF, while Su(z)2sM contains a D49H missense mutation in the loop between the two Zn++-coordinating domains. Although each of these mutations is predicted to destabilize the RF, both are hemizygous viable (Table 1). The mutations associated with Su(z)2s20 and Su(z)2s21 are located between the RF and HTH domains. The Su(z)2s20 allele contains a P101S change, while the Su(z)2s21 allele contains a E136K change.
We did not find any change associated with the Su(z)2s15 allele. This allele may contain an alteration outside the coding sequence of Su(z)2 that affects either the regulation of the gene or the stability of its mRNA. Both of these possibilities would be consistent with the genetic behavior of this allele. Alternatively, Su(z)2s15 may represent a mutation in a gene that interacts with Su(z)2. Unfortunately, the semilethality of this mutation complicates an accurate mapping of its location.
Su(z)2D mutations are alleles of Su(z)2:
The discovery that Su(z)2s20, Su(z)2s21, Su(z)2s36, and Su(z)2sM are alleles of Su(z)2 prompted us to reconsider our prior genetic analyses of three alleles that had been previously proposed to represent a third complementation group of the Su(z)2 complex (Wu and Howe 1995). The existence of this third complementation group, called Su(z)2D, had been suggested primarily by the behavior of Su(z)25, which suppresses z1 in a dominant fashion despite the fact that it deletes both Psc and Su(z)2. Additional support for the existence of Su(z)2D came from the complementation patterns of Su(z)2De26, Su(z)2Deos, and Su(z)31, all three of which were believed to represent Su(z)2D (Wu and Howe 1995). Interestingly, the behavior of these three alleles is reminiscent of the SIMs. Our findings show that Su(z)2De26, Su(z)2Deos, and Su(z)31 are all viable when heterozygous with SIM mutations (Table 3), and yet all carry mutations within Su(z)2.
TABLE 3.
Genetic analysis of Su(z)2D alleles
| Su(z)2s15 | Su(z)2s20 | Su(z)2s21 | Su(z)2s36 | Su(z)2s84 | Su(z)2s95 | Su(z)2sM | ||
|---|---|---|---|---|---|---|---|---|
| Su(z)2De26 | P | 9 (102) | 33 (250) | 36 (214) | 38 (193) | 21 (165) | 27 (240) | 36 (118) |
| M | 15 (156) | 33 (115) | 31 (318) | 35 (226) | 20 (183) | 25 (199) | 33 (222) | |
| Su(z)2Deos | P | 14 (160) | 33 (334) | 27 (142) | 27 (327) | 20 (282) | 16 (190) | 37 (159) |
| M | 10 (175) | 29 (241) | 34 (218) | 39 (157) | 23 (167) | 24 (173) | 33 (130) | |
| Su(z)31 | P | 15 (193) | 27 (171) | 30 (332) | 22 (149) | 19 (156) | 25 (207) | 33 (190) |
| M | 23 (100) | 25 (194) | 29 (117) | 37 (111) | 26 (188) | 23 (332) | 39 (152) |
See Table 1 legend for explanation of format.
First, we noted that the genetic behavior of Su(z)2De26 strongly resembles that of Su(z)2s36 (Wu and Howe 1995; R. B. Emmons and C.-t. Wu, unpublished results): Su(z)2De26 is lethal when heterozygous with either Su(z)21 or Su(z)24-34, but shows significant viability when homozygous or heterozygous with Su(z)21.b8 or Su(z)21.b7. Remarkably, we found that Su(z)2De26 is similar to Su(z)2s36 at the molecular level as well, containing a missense mutation (H53Y) in the RF (Figure 3). As His53 is required to form the second Zn++-coordinating domain in the RF, this mutation would be expected to severely disrupt the RF and compromise Su(z)2 function. Indeed, mutations disrupting the Zn++-coordinating domains within the RF of Bmi-1 disrupt the ability of Bmi-1 to interact with other proteins and to localize to subnuclear regions (Alkema et al. 1997; Hemenway et al. 1998). Interestingly, the Su(z)2De26 mutation predicts a protein that would be structurally similar to that produced by Psce23, which contains a C268Y change expected to disrupt the RF of Psc. However, unlike Psce23, which is homozygous and hemizygous lethal, Su(z)2De26 shows significant homozygous and hemizygous viability. This finding suggests that the RF is not required for Su(z)2 function, which is in stark contrast to the requirement of the RF for wild-type Psc function, or that the mutated RF of the Su(z)2De26 protein retains some wild-type function.
Su(z)2Deos and Su(z)31 also display some similarities with the SIMs; like Su(z)2s20, Su(z)2s21, and Su(z)2sM, they show reduced viability in trans to Su(z)21 and higher viability in trans to Su(z)21.b7 and Su(z)24-34. However, they differ from these three SIM alleles in that they display complete or nearly complete lethality in trans to Su(z)21.b8 (Wu and Howe 1995; R. B. Emmons and C.-t, Wu, unpublished results), with separate studies suggesting that Su(z)2Deos is the more severe of the two (Wu and Howe 1995). We found that both Su(z)2Deos and Su(z)31 contain mutations predicted to truncate Su(z)2 after the HR and more C-terminal to the K284* nonsense mutation of Su(z)21 (Figure 3). Su(z)2Deos contains a Q448* nonsense mutation while Su(z)31 has a G inserted after nucleotide position 1873, resulting in a seven-amino-acid frameshift followed by a nonsense codon (E577R, E578G, A579G, R580A, S581E, I582Y, N583Q, S584*) (Figure 3; materials and methods; see Table S1 for sequence data regarding regions of Su(z)31 lying C-terminal to S584*). That Su(z)31, Su(z)2Deos, and Su(z)21 are predicted to produce increasingly shorter proteins and increasingly more severe phenotypes (Tables 1 and 2; Wu and Howe 1995; R. B. Emmons and C.-t. Wu, unpublished results) suggests that the lethality associated with Su(z)2Deos and Su(z)31 stems at least in part from the loss of critical functions encoded by the CTR sequences, perhaps specifically by amino acids 285–576. Furthermore, the viability of Su(z)2Deos and Su(z)31 in trans to Su(z)21.b7 may indicate a capacity of Su(z)21.b7 to provide some function and/or reflect the contribution Psc, which remains intact upstream of Su(z)21.b7 but is lacking from the Su(z)21.b8 deletion.
Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2sM, and Su(z)2De26 display negative complementation with Su(z)21, Su(z)24-31, and/or Su(z)24-34:
Our molecular confirmation that Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2sM, and Su(z)2De26 contain missense mutations within the HR of Su(z)2 was interesting, given their genetic behavior. While all five are quite viable when hemizygous in trans to Su(z)21.b8, three of the alleles, Su(z)2s20, Su(z)2s21, and Su(z)2sM, are lethal or semilethal when in trans to Su(z)21, and the remaining two, Su(z)2s36 and Su(z)2De26, are lethal in trans to both Su(z)21 and Su(z)24-34, with Su(z)2De26 also being lethal in trans to Su(z)24-31 (R. B. Emmons and C.-t. Wu, unpublished results). Taken together, the genetic behavior of these alleles is consistent with negative complementation, a type of interallelic interaction in which the activity of one allele is specifically poisoned by another (Fincham 1966). Typically, negative complementation is seen when two alleles, m1 and m2, display a phenotype that is stronger when they are heterozygous with each other than when either is homozygous (i.e., m1/m2 is worse than m1/m1 and m2/m2) (Fincham 1966) or when m1 and/or m2 is heterozygous with a deficiency (i.e., m1/m2 is worse than m1/Df and/or m2/Df) (Bickel et al. 1996). This latter situation mirrors the behavior of Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2sM, and Su(z)2De26 with respect to Su(z)21. For example, we find complete lethality when Su(z)2s36 is heterozygous with Su(z)21 even though Su(z)2s36 is viable when hemizygous in trans to Su(z)21.b8. This negative complementation cannot be attributed to an interaction with another mutation on the Su(z)21 chromosome because Su(z)21.b8, which does not display negative complementation with Su(z)2s36, is a derivative of Su(z)21. Note the additional levels of negative complementation associated with Su(z)2s36 and Su(z)2De26 (Table 1; R. B. Emmons and C.-t. Wu, unpublished results); whether these additional levels of negative complementation stem from the lesions of Su(z)2s36 and Su(z)2De26 falling directly in the first and second, respectively, Zn++-coordinating domains of the RF is as yet unclear.
Su(z)2s20 and Su(z)2s21 identify conserved subregions within the HR:
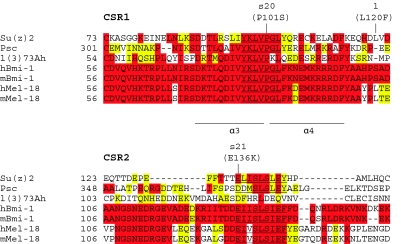
The mutations associated with Su(z)2s20 and Su(z)2s21 are interesting because they are located in the HR, but do not affect either the RF or HTH domains. To better understand this region, we generated ClustalX alignments (Thompson et al. 1997; Chenna et al. 2003) among the predicted protein products of Su(z)2, Psc, a third Drosophila homolog called lethal(3)73Ah [l(3)73Ah; Irminger-Finger and Nothiger 1995], as well as human and mouse bmi-1 and mel-18. Although it is not clear whether l(3)73Ah is a PcG gene, it contains the HR, but not the CTR (Figure 1). We found that the lesions associated with Su(z)2s20 and Su(z)2s21 fall within two highly conserved subregions located between the RF and HTH domains (Figure 4). We will refer to these conserved subregions as CSR1 and CSR2.
Figure 4.—
ClustalX alignment of Su(z)2, Psc, and L(3)73Ah from Drosophila with Bmi-1 and Mel-18 from both Homo sapiens and Mus musculus. The alignment generated by ClustalX is focused on the region of the proteins located between the RF and HTH domains and begins with the last conserved cysteine residue in the RF. Amino acid positions are indicated on the left. Amino acids highlighted in red are identical (equivalent to “*” in ClustalX) among the proteins indicated, while strongly conserved amino acids (equivalent to “:” in ClustalX) are highlighted in yellow. More weakly conserved amino acids (equivalent to “.” in ClustalX) are not highlighted. The CSR1 core sequence is underlined twice, with the Su(z)2s20 mutation indicated above. The Su(z)21 missense mutation is shown to the right of CSR1. Residues that correspond to α-helical regions 3 and 4 in the Bmi-1/Ring1B structure are shown. The CSR2 core sequence is underlined once, with the Su(z)2s21 mutation indicated above.
CSR1 contains a core sequence of Y K L V P G L that is conserved in all seven proteins examined and mutated in the Su(z)2s20 protein, where the proline is replaced by a serine (Figure 4). Database searches (ELM, ProSite, Pfam, SMART) did not identify any known protein motifs within this region and, while a number of potential sites for post-translational modification are present, there is currently no evidence for post-translational modification of this region. CSR2 is less well defined than is CSR1 and contains a core sequence of [E/D] Ψ Ψ S L S [I/L] [E/Q] [F/Y]. Database searches (ELM, ProSite, Pfam, SMART) did not identify any protein motif, but did reveal a putative CK2 phosphorylation site that is conserved in Bmi-1, whose localization to chromatin fluctuates throughout the cell cycle in a phosphorylation-dependent manner (Voncken et al. 1999). The lesion associated with Su(z)2s21 substitutes a lysine for the glutamic acid in this putative phosphorylation site, which is also the first glutamic acid in the core sequence.
It is interesting to note that the L120F missense mutation associated with Su(z)21 falls in the region between CSR1 and CSR2 (Figure 4). Although this mutation does not appear to identify a region of strong conservation, it does alter a leucine that is conserved in both human and mouse mel-18 (Figure 4). Therefore, although our consideration of the structural basis for the Su(z)21 phenotype has centered on the K284* nonsense mutation and the CTR truncation that it predicts, it remains possible that the L120F missense mutation also contributes to the severity of Su(z)21.
The CTR of Su(z)2 is intrinsically disordered:
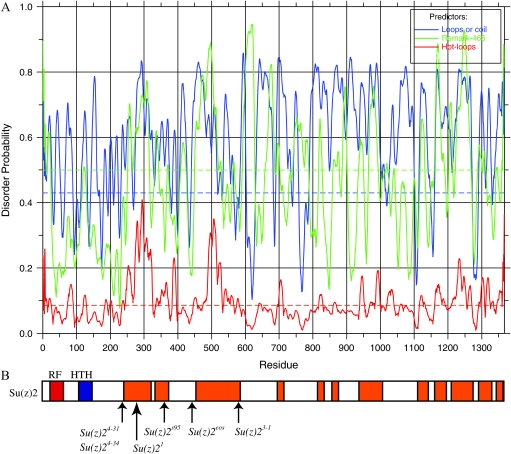
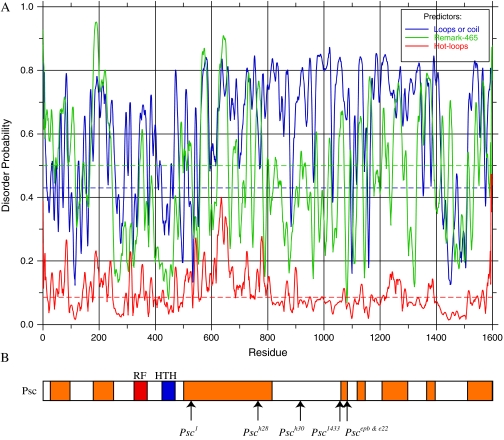
The CTRs of Su(z)2 and Psc are important domains as they are essential for the function of these proteins in vivo (this report and King et al. 2005) as well as in vitro (King et al. 2005; Lo et al. 2009). However, consistent with other studies (Brunk et al. 1991a; van Lohuizen et al. 1991; Lo et al. 2009), we were unable to identify significant regions of homology or conserved functional domains, although both CTRs contain a myriad of sites for potential post-translational modification. Using SMART analysis, however, we discovered that each CTR is predicted to contain high levels of intrinsic protein disorder. Figure 5 shows a disorder probability plot using the default parameters of DisEMBL (Figure 5A) and the regions of the Su(z)2 CTR that are predicted to be disordered by all three parameters (Loops/coil, Remark-465, and Hot-loops) (Figure 5B) (Linding et al. 2003). Disordered regions were merged in instances where peak distances were <20 amino acids apart for one of the predictors. Note that these predictions are conservative because they require a statistically significant score from all three parameters, and the level of predicted disorder in the CTR increases dramatically if only two of the three parameters are considered. We obtained similar results for Psc (Figure 6).
Figure 5.—
Disorder analysis of Su(z)2. (A) DisEMBL analysis of Su(z)2 showing the predictions for loops or coil (blue), remark-465 (green), and hot loops (red). (B) Su(z)2 protein is shown at a scale matching that of the plot above. The RF is in red, and the HTH is in blue. Regions of protein disorder predicted by all three methods are shown in orange. The predicted points of truncation of the truncation alleles are shown below.
Figure 6.—
Disorder analysis of Psc. See Figure 5 for explanation of format.
DISCUSSION
Of the many PcG genes known, several belong to gene pairs: Psc and Su(z)2 (Adler et al. 1989; Wu et al. 1989; Wu and Howe 1995), ph-p and ph-d (Dura et al. 1987), pho and phol (Brown et al. 2003), and esc and escl (Wang et al. 2006). These gene pairs show some degree of redundancy or similarity between the two members of a pair and are generally typified by double-mutant combinations in which the phenotype of flies carrying a mutation in each gene is worse than that of flies carrying a mutation in only one of the genes. Here we have focused on the Su(z)2 gene of the Psc-Su(z)2 gene pair. In particular, we have carried out a molecular genetic analysis of 14 Su(z)2 alleles, 7 of which [Su(z)2s15, Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2s84, Su(z)2s95, and Su(z)2sM] were newly generated for this study and 3 of which [Su(z)2De26, Su(z)2Deos, and Su(z)31] were previously thought to represent a third region of the Su(z)2 complex called Su(z)2D. Here, we discuss negative complementation at the locus and then compare the structure of Su(z)2 to Psc in the context of the CTR and its disordered domains.
Negative complementation at the Su(z)2 locus:
The allele-specific noncomplementation of Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2sM, and Su(z)2De26 with Su(z)21, Su(z)24-31, and/or Su(z)24-34 represents a rare type of genetic interaction called negative complementation (Fincham 1966; Bickel et al. 1996). In Drosophila, negative complementation has been described at Notch (Foster 1975; Portin 1975), dEGFR (Raz et al. 1991; Clifford and Schupbach 1994), ord (Bickel et al. 1996, 1997), α-tubulin84B (Matthews and Kaufman 1987), and Mos1 (Lohe et al. 1996). All of the proteins encoded by these genes require protein–protein interactions for wild-type function.
Negative complementation at Su(z)2 highlights the multidomain structure of the Su(z)2 protein because it occurs between alleles that contain missense mutations in the HR and Su(z)21, Su(z)24-31, or Su(z)24-34, all three of which are predicted to generate proteins lacking nearly all of the CTR. As this interaction is not associated with other Su(z)2 alleles predicted to delete CTR sequences, negative complementation at the locus may be specific for Su(z)21, Su(z)24-31, and Su(z)24-34 and not a general consequence of partial or complete CTR loss. Alternatively, CTR sequences including and lying C-terminal to lysine 284, which is the point of truncation in the longest of these three truncation alleles, may antagonize the capacity of longer proteins, such as those predicted by Su(z)2Deos and Su(z)31, to effect negative complementation, reminiscent of proposals of intramolecular regulation for Psc and Su(z)2 (Sharp et al. 1994; Wu and Howe 1995; Platero et al. 1996; King et al. 2005). Regardless, as the Su(z)21 and Su(z)24-31 proteins are predicted to contain little more than the HR, these observations suggests that much, if not all, of the HR (including the RF, CSR1, CSR2, and the HTH) can function independently of the rest of the protein. This interpretation likely applies also to Su(z)24-34 because the negative complementation observed between this allele and Su(z)2e26 argues that it produces a product even though it carries a large insertion. These observations are consistent with the in vitro assays of truncated Su(z)2 proteins (Lo et al. 2009) and reminiscent of the structural organization of Psc, which also consists of more than one functional domain (Wu and Howe 1995; King et al. 2005).
Although the potential participation of Su(z)2 in a larger complex can complicate models explaining negative complementation, one interpretation is that an antimorphic nature (Wu and Howe 1995) of the Su(z)21, Su(z)24-31, and Su(z)24-34 proteins compromises or poisons the function encoded by the missense alleles which, however, are able to support wild-type or nearly wild-type viability on their own (Table 1). In light of models for negative complementation that invoke protein–protein interactions, it may be that the amino acid substitutions within the HR that are encoded by the missense alleles may compromise the ability of the resulting mutant Su(z)2 protein to interact properly with itself or other factors, protein or otherwise, either transiently or as part of a more stable complex. For example, the RF in the Su(z)2De26 protein may be compromised such that it cannot compete effectively against the Su(z)21 protein in a Su(z)21/Su(z)2De26 heterozygote, resulting in complexes that are nonfunctional or abnormal, simultaneously reducing the amount of functional Su(z)2De26-containing complexes. The scenario in which Su(z)2 interacts with another protein is supported by the behavior of Psc, which interacts with Ph, Pc, and Sce (Kyba and Brock 1998; Francis et al. 2001), and by in vitro observations that the HR of Psc and Su(z)2 is important for complex formation (King et al. 2005; Lo et al. 2009). Furthermore, colocalization of Su(z)2 with Psc and Pc in polytene chromosomes suggests that Su(z)2 can associate with PRC1 or another PcG complex in vivo (Rastelli et al. 1993; Platero et al. 1996; Sharp et al. 1997).
If Su(z)2 functions as a homodimer, our observations would be consistent with the missense mutations causing the affinity of the resulting proteins for themselves to be less than their affinity for Su(z)21, thereby creating inactive Su(z)21-containing dimers. Su(z)21 could compromise this dimer in many ways, including acting in a prion-like fashion to inactivate Su(z)2De26, mislocalizing Su(z)2De26 to a subcellular region that does not support Su(z)2 activity, or creating a dimer with abnormal activity. On the other hand, if Su(z)2 functions as a heterodimer, our findings would be consistent with the missense mutations causing the affinity of the resulting Su(z)2 protein for its partner to be less than that of Su(z)21. In this scenario, dimerization would favor the inclusion of Su(z)21, which would again compromise the dimer. Both scenarios assume that the missense mutations decrease the capacity of the resulting proteins for intermolecular interactions, consistent with their location in the RF.
Su(z)2s20 and Su(z)2s21 are especially noteworthy in that the causative lesions of these two alleles fall within the HR but outside of the RF and HTH motifs, reminiscent of studies suggesting that sequences just C-terminal to the RF of Bmi-1 are important for Bmi-1 function (Hemenway et al. 1998; Satijn and Otte 1999). Su(z)2s20 is predicted to alter a proline residue in the CSR1 core sequence YKLVPGL, which is completely conserved in Bmi-1 and Mel-18. This change, in conjunction with the negative complementation observed with Su(z)2s20, suggests that CSR1 mediates protein–protein interactions. This interpretation is supported by the crystal structure of the Bmi-1/Ring1B heterodimer, which reveals that the region that we designate as CSR1 lies at the interface between these two proteins. Specifically, the proline residue appears to establish the three-dimensional geometry of two α-helical regions in Bmi-1, α3 and α4, which contain residues that form salt bridges with Ring1B as well as residues that stabilize these interactions (Figure 4; Buchwald et al. 2006; Li et al. 2006).On the basis of this, we believe that a substitution of a serine for this proline would alter the capacity of Su(z)2 to interact with other factors.
Su(z)2s21 substitutes a lysine for the first glutamic acid of the [E/D] Ψ Ψ S L S [I/L] [E/Q] [F/Y] motif in a region that we refer to as CSR2. The conservation of CSR2 is not as prominent as that of CSR1 and, perhaps consistent with this, the negative complementation of Su(z)2s21 with Su(z)21 is not as strong as that of Su(z)2s20 (Table 1). Because the region of Bmi-1 that is orthologous to CSR2 was not included in the crystal structures mentioned above (Buchwald et al. 2006; Li et al. 2006), we cannot postulate how the amino acid change directed by Su(z)2s21 would affect the specificity and/or avidity of any potential interaction between the Su(z)2 protein and other factors. However, since this change resides within a putative CK2 phosphorylation site and the activity of Bmi-1 is modulated by phosphorylation (Voncken et al. 1999), our findings suggest that such modulation could function by mediating the regulation of interactions between Bmi-1 and other factors.
At first glance, the negative complementation of Su(z)2 alleles would appear to be in stark contrast to the genetic behavior of structurally similar alleles of Psc. Psce23 predicts a C268Y missense mutation in the RF that is analogous to that of Su(z)2De26, yet it displays intragenic complementation with Psch28, Psch30, and Psce22, all three of which delete significant portions of the CTR (Wu and Howe 1995; King et al. 2005). In fact, it is this complementation and subsequent biochemical and molecular analyses that indicated that Psc contains multiple domains that are functionally separable. Psce23 does not, however, complement Psc1 (Wu and Howe 1995), which encodes the truncation of Psc that is most similar in structure to the truncation of Su(z)2 encoded by Su(z)21 (King et al. 2005). Our analysis suggests this failure could be due to negative complementation.
The CTRs of Su(z)2 and Psc:
Our prediction that several recessive lethal Su(z)2 alleles truncate Su(z)2 within the CTR recalls our earlier report that truncations removing ∼40% or more of the CTR of Psc reduce viability (King et al. 2005). These findings argue for the in vivo importance of the CTR of both proteins and provide further support that the function of the CTR of Psc and Su(z)2 may be conserved despite differences in their primary amino acid sequences. We have also found that ≥45% of the CTRs of both Su(z)2 and Psc are contained within the domains of predicted intrinsic disorder scattered throughout the CTR (Figures 5 and 6). As such, the CTRs are reminiscent of intrinsically disordered proteins (IDPs), which are proteins containing regions that do not possess a defined conformation under native conditions but adopt specific conformations when they interact with ligands, DNA, protein, or other factors or when they self-associate, as is seen with prions (reviewed by Dyson and Wright 2005; Hansen et al. 2006). IDPs are generally enriched for particular amino acids, such as arginine, glutamine, glutamic acid, lysine, proline, serine, and occasionally alanine and glycine, and their tendency for disorder can be computationally predicted with a high degree of accuracy (Vucetic et al. 2003; reviewed by Dunker et al. 2001, 2002). Indeed, on the basis of the amino acid composition of the CTRs of Psc and Su(z)2, Lo et al. (2009) also recently hypothesized the potential of these two proteins to contain regions of disorder. Importantly, analyses of IDPs show that intrinsic disorder in and of itself can be sufficient for function. For example, the long C-terminal regions of linker histones are essential for their functions even though they are intrinsically disordered and functionally interchangeable among evolutionarily diverged species of linker histones (reviewed by Hansen et al. 2006). In light of these features of IDPs, it may be that the role of the CTRs of Su(z)2 and Psc in vivo and their capacity to inhibit transcription and/or chromatin remodeling in vitro rests on regions of disorder and the capacity of such regions to transition to an ordered state (also see Lo et al. 2009).
The structural nature of the CTRs may also pertain to the capacity of mutations in Su(z)2 and Psc to suppress the effect of z1 on white gene expression. Of the alleles that truncate the protein within the CTR, all suppress z1 and, of these, the strongest, Su(z)21, is predicted to delete nearly all of the CTR and, hence, nearly all of the blocks of intrinsic protein disorder. In this way, Su(z)21 resembles Psc1, which is the strongest suppressor of z1 at Psc and also leads to a severe truncation of the CTR. Although we cannot assess the involvement of the L120F missense mutation of Su(z)21 in suppression, the two simple truncation alleles, Su(z)2Deos and Su(z)31, rule out any requirement of L120F for z1 suppression even as they emphasize the importance of the CTR. Further support for a role of the CTR in the z1 phenotype comes from three truncation alleles of Psc that, curiously, enhance z1 (Wu and Howe 1995; King et al. 2005). These observations may be particularly relevant, as the zeste protein also contains regions of disorder (R. B. Emmons, unpublished results), has runs of glutamine and alanine in its CTR, and displays a strong tendency to self-associate or aggregate (reviewed by Pirrotta 1991; also see Chen and Pirrotta 1993; Rosen et al. 1998). The positions of the lesions of z1 and two z1-like alleles are clustered within this CTR (Pirrotta et al. 1987; Rosen et al. 1998), further implicating the CTR in Zeste function. These findings raise the possibility that cooperative and/or competitive interactions between the disordered regions of Su(z)2, Psc, and/or Zeste may underlie the ability of z1 to repress white and the capacity of Su(z)2 and Psc mutations to modify the z1 phenotype. Finally, we have found that the short CTRs of both Bmi-1 and Mel-18 also contain regions that are likely to be intrinsically disordered (data not shown), suggesting that the long CTRs of Psc and Su(z)2 may be closely related in structure and function to the minimal CTRs of their mammalian homologs despite their very different lengths.
Acknowledgments
We thank N. Francis and members of her laboratory, in particular S. Lo, for sharing unpublished data and many thought-provoking discussions. We also thank M. Ashburner, W. Bender, S. Hawley, J. Kennison, and E. Weischaus for insightful discussions, with special thanks to S. Hawley regarding negative complementation. Finally, we acknowledge the anonymous reviewers of our manuscript for key comments, members of the Wu laboratory for many helpful conversations and technical assistance, and D. Schwartz for input on protein sequence alignment. This work was supported by a Kirschstein National Research Service Award to R.B.E. and National Institutes of Health grant RO1GM61936 to C.-t.W. and Harvard Medical School.
References
- Adler, P. N., J. Charlton and B. Brunk, 1989. Genetic interactions of the suppressor 2 of zeste region genes. Dev. Genet. 10 249–260. [DOI] [PubMed] [Google Scholar]
- Adler, P. N., E. C. Martin, J. Charlton and K. Jones, 1991. Phenotypic consequences and genetic interactions of a null mutation in the Drosophila Posterior Sex Combs gene. Dev. Genet. 12 349–361. [DOI] [PubMed] [Google Scholar]
- Ali, J. Y., and W. Bender, 2004. Cross-regulation among the polycomb group genes in Drosophila melanogaster. Mol. Cell. Biol. 24 7737–7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkema, M. J., J. Wiegant, A. K. Raap, A. Berns and M. van Lohuizen, 1993. Characterization and chromosomal localization of the human proto-oncogene BMI-1. Hum. Mol. Genet. 2 1597–1603. [DOI] [PubMed] [Google Scholar]
- Alkema, M. J., M. Bronk, E. Verhoeven, A. Otte, L. J. van't Veer et al., 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 11 226–240. [DOI] [PubMed] [Google Scholar]
- Beuchle, D., G. Struhl and J. Muller, 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128 993–1004. [DOI] [PubMed] [Google Scholar]
- Bickel, S. E., D. W. Wyman, W. Y. Miyazaki, D. P. Moore and T. L. Orr-Weaver, 1996. Identification of ORD, a Drosophila protein essential for sister chromatid cohesion. EMBO J. 15 1451–1459. [PMC free article] [PubMed] [Google Scholar]
- Bickel, S. E., D. W. Wyman and T. L. Orr-Weaver, 1997. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics 146 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin, M. D., S. Bickel, M. Benson, V. Pirrotta and R. Tjian, 1988. Zeste encodes a sequence-specific transcription factor that activates the Ultrabithorax promoter in vitro. Cell 53 713–722. [DOI] [PubMed] [Google Scholar]
- Breiling, A., L. Sessa and V. Orlando, 2007. Biology of polycomb and trithorax group proteins. Int. Rev. Cytol. 258 83–136. [DOI] [PubMed] [Google Scholar]
- Brock, H. W., and C. L. Fisher, 2005. Maintenance of gene expression patterns. Dev. Dyn. 232 633–655. [DOI] [PubMed] [Google Scholar]
- Brown, J. L., C. Fritsch, J. Mueller and J. A. Kassis, 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130 285–294. [DOI] [PubMed] [Google Scholar]
- Brunk, B. P., E. C. Martin and P. N. Adler, 1991. a Drosophila genes Posterior Sex Combs and Suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature 353 351–353. [DOI] [PubMed] [Google Scholar]
- Brunk, B. P., E. C. Martin and P. N. Adler, 1991. b Molecular genetics of the Posterior sex combs/Suppressor 2 of zeste region of Drosophila: aberrant expression of the Suppressor 2 of zeste gene results in abnormal bristle development. Genetics 128 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald, G., P. van der Stoop, O. Weichenrieder, A. Perrakis, M. van Lohuizen et al., 2006. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25 2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker, C. A., and R. E. Kingston, 1994. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 14 1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, R., and Y. Zhang, 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14 155–164. [DOI] [PubMed] [Google Scholar]
- Chen, J. D., and V. Pirrotta, 1993. Stepwise assembly of hyperaggregated forms of Drosophila zeste mutant protein suppresses white gene expression in vivo. EMBO J. 12 2061–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson et al., 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, R., and T. Schupbach, 1994. Molecular analysis of the Drosophila EGF receptor homolog reveals that several genetically defined classes of alleles cluster in subdomains of the receptor protein. Genetics 137 531–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin, J., and G. Cavalli, 2004. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 23 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker, A. K., J. D. Lawson, C. J. Brown, R. M. Williams, P. Romero et al., 2001. Intrinsically disordered protein. J. Mol. Graph. Model. 19 26–59. [DOI] [PubMed] [Google Scholar]
- Dunker, A. K., C. J. Brown, J. D. Lawson, L. M. Iakoucheva and Z. Obradovic, 2002. Intrinsic disorder and protein function. Biochemistry 41 6573–6582. [DOI] [PubMed] [Google Scholar]
- Dura, J. M., N. B. Randsholt, J. Deatrick, I. Erk, P. Santamaria et al., 1987. A complex genetic locus, polyhomeotic, is required for segmental specification and epidermal development in D. melanogaster. Cell 51 829–839. [DOI] [PubMed] [Google Scholar]
- Dyson, H. J., and P. E. Wright, 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6 197–208. [DOI] [PubMed] [Google Scholar]
- Fincham, J. R., 1966. From Mendel's factors to genes as functional units. Adv. Sci. 23 124–128. [PubMed] [Google Scholar]
- Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis et al., 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17 1870–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, G. G., 1975. Negative complementation at the notch locus of Drosophila melanogaster. Genetics 81 99–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, N. J., A. J. Saurin, Z. Shao and R. E. Kingston, 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8 545–556. [DOI] [PubMed] [Google Scholar]
- Fritsch, C., D. Beuchle and J. Müller, 2003. Molecular and genetic analysis of the Polycomb group gene Sex combs extra/Ring in Drosophila. Mech. Dev. 120 949–954. [DOI] [PubMed] [Google Scholar]
- Gelbart, W. M., 1971. Cytogenetics of zeste expression in Drosophila melanogaster. Ph.D. Thesis, University of Wisconsin, Madison.
- Gorfinkiel, N., L. Fanti, T. Melgar, E. García, S. Pimpinelli et al., 2004. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 121 449–462. [DOI] [PubMed] [Google Scholar]
- Guo, W. J., S. Datta, V. Band and G. P. Dimri, 2007. a Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol. Biol. Cell 18 536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. J., M. S. Zeng, A. Yadav, L. B. Song, B. H. Guo et al., 2007. b Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breast cancer cells. Cancer Res. 67 5083–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J. C., X. Lu, E. D. Ross and R. W. Woody, 2006. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281 1853–1856. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., and W. D. Gilliland, 2006. Sometimes the result is not the answer: the truths and the lies that come from using the complementation test. Genetics 174 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway, C. S., B. W. Halligan and L. S. Levy, 1998. The Bmi-1 oncoprotein interacts with dinG and MPh2: the role of RING finger domains. Oncogene 16 2541–2547. [DOI] [PubMed] [Google Scholar]
- Hur, M. W., J. D. Laney, S. H. Jeon, J. Ali and M. D. Biggin, 2002. Zeste maintains repression of Ubx transgenes: support for a new model of Polycomb repression. Development 129 1339–1343. [DOI] [PubMed] [Google Scholar]
- Irminger-Finger, I., and R. Nothiger, 1995. The Drosophila melanogaster gene lethal(3)73Ah encodes a ring finger protein homologous to the oncoproteins MEL-18 and BMI-1. Gene 163 203–208. [DOI] [PubMed] [Google Scholar]
- Ishida, A., H. Asano, M. Hasegawa, H. Koseki, T. Ono et al., 1993. Cloning and chromosome mapping of the human Mel-18 gene which encodes a DNA-binding protein with a new ‘RING-finger’ motif. Gene 129 249–255. [DOI] [PubMed] [Google Scholar]
- Jack, J. W., and B. H. Judd, 1979. Allelic pairing and gene regulation: a model for the zeste—white interaction in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 76 1368–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G., 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316 153–155. [Google Scholar]
- Kahn, T. G., Y. B. Schwartz, G. I. Dellino and V. Pirrotta, 2006. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J. Biol. Chem. 281 29064–29075. [DOI] [PubMed] [Google Scholar]
- Kal, A. J., T. Mahmoudi, N. B. Zak and C. P. Verrijzer, 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14 1058–1071. [PMC free article] [PubMed] [Google Scholar]
- Kalisch, W. E., and B. Rasmuson, 1974. Changes of zeste phenotype induced by autosomal mutations in Drosophila melanogaster. Hereditas 78 97–104. [DOI] [PubMed] [Google Scholar]
- Kassis, J. A., 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46 421–438. [DOI] [PubMed] [Google Scholar]
- King, I. F., R. B. Emmons, N. J. Francis, B. Wild, J. Muller et al., 2005. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 25 6578–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba, M., and H. W. Brock, 1998. The Drosophila polycomb group protein Psc contacts ph and Pc through specific conserved domains. Mol. Cell. Biol. 18 2712–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney, J. D., and M. D. Biggin, 1992. zeste, a nonessential gene, potently activates Ultrabithorax transcription in the Drosophila embryo. Genes Dev. 6 1531–1541. [DOI] [PubMed] [Google Scholar]
- Lee, J. Y., K. S. Jang, D. H. Shin, M. Y. Oh, H. J. Kim et al., 2008. Mel-18 negatively regulates INK4a/ARF-independent cell cycle progression via Akt inactivation in breast cancer. Cancer Res. 68 4201–4209. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 89 225–239. [Google Scholar]
- Li, Z., R. Cao, M. Wang, M. P. Myers, Y. Zhang et al., 2006. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 281 20643–20649. [DOI] [PubMed] [Google Scholar]
- Linding, R., L. J. Jensen, F. Diella, P. Bork, T. J. Gibson et al., 2003. Protein disorder prediction: implications for structural proteomics. Structure 11 1453–1459. [DOI] [PubMed] [Google Scholar]
- Lo, S. M., N. K. Ahuja and N. J. Francis, 2009. Polycomb group protein Suppressor 2 of Zeste is a functional homolog of posterior sex combs. Mol. Cell. Biol. 29 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A. R., D. T. Sullivan and D. L. Hartl, 1996. Subunit interactions in the mariner transposase. Genetics 144 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansukhani, A., A. Crickmore, P. W. Sherwood and M. L. Goldberg, 1988. DNA-binding properties of the Drosophila melanogaster zeste gene product. Mol. Cell. Biol. 8 615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Langerak, J., and G. Cavalli, 2008. Polycomb group proteins and long-range gene regulation. Adv. Genet. 61 45–66. [DOI] [PubMed] [Google Scholar]
- Matthews, K. A., and T. C. Kaufman, 1987. Developmental consequences of mutations in the 84B alpha-tubulin gene of Drosophila melanogaster. Dev. Biol. 119 100–114. [DOI] [PubMed] [Google Scholar]
- McKee, B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677 165–180. [DOI] [PubMed] [Google Scholar]
- Metz, C. W., 1916. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera and its significance. J. Exp. Zool. 21 213–279. [Google Scholar]
- Min, J., Y. Zhang and R. M. Xu, 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17 1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann, L., A. J. Kal and C. P. Verrijzer, 2002. Characterization of the extended Myb-like DNA-binding domain of trithorax group protein Zeste. J. Biol. Chem. 277 47385–47392. [DOI] [PubMed] [Google Scholar]
- Mulholland, N. M., I. F. King and R. E. Kingston, 2003. Regulation of Polycomb group complexes by the sequence-specific DNA binding proteins Zeste and GAGA. Genes Dev. 17 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen, A. M., and M. van Lohuizen, 2008. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 20 201–207. [DOI] [PubMed] [Google Scholar]
- Pietersen, A. M., B. Evers, A. A. Prasad, E. Tanger, P. Cornelissen-Steijger et al., 2008. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr. Biol. 18 1094–1099. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V., 1991. The genetics and molecular biology of zeste in Drosophila melanogaster. Adv. Genet. 29 301–348. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V., E. Manet, E. Hardon, S. E. Bickel and M. Benson, 1987. Structure and sequence of the Drosophila zeste gene. EMBO J. 6 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero, J. S., E. J. Sharp, P. N. Adler and J. C. Eissenberg, 1996. In vivo assay for protein-protein interactions using Drosophila chromosomes. Chromosoma 104 393–404. [DOI] [PubMed] [Google Scholar]
- Portin, P., 1975. Allelic negative complementation at the Abruptex locus of Drosophila melanogaster. Genetics 81 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli, L., C. S. Chan and V. Pirrotta, 1993. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 12 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz, E., E. D. Schejter and B. Z. Shilo, 1991. Interallelic complementation among DER/flb alleles: implications for the mechanism of signal transduction by receptor-tyrosine kinases. Genetics 129 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, C., D. Dorsett and J. Jack, 1998. A proline-rich region in the Zeste protein essential for transvection and white repression by Zeste. Genetics 148 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi, E., and M. R. Capecchi, 2008. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn, D. P., and A. P. Otte, 1999. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol. Cell. Biol. 19 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst and R. E. Kingston, 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412 655–660. [DOI] [PubMed] [Google Scholar]
- Schuettengruber, B., D. Chourrout, M. Vervoort, B. Leblanc and G. Cavalli, 2007. Genome regulation by polycomb and trithorax proteins. Cell 128 735–745. [DOI] [PubMed] [Google Scholar]
- Schwartz, Y. B., and V. Pirrotta, 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Schwartz, Y. B., and V. Pirrotta, 2008. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20 266–273. [DOI] [PubMed] [Google Scholar]
- Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu et al., 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98 37–46. [DOI] [PubMed] [Google Scholar]
- Sharp, E. J., E. C. Martin and P. N. Adler, 1994. Directed overexpression of suppressor 2 of zeste and Posterior Sex Combs results in bristle abnormalities in Drosophila melanogaster. Dev. Biol. 161 379–392. [DOI] [PubMed] [Google Scholar]
- Sharp, E. J., N. A. Abramova, W. J. Park and P. N. Adler, 1997. The conserved HR domain of the Drosophila suppressor 2 of zeste [Su(z)2] and murine bmi-1 proteins constitutes a locus-specific chromosome binding domain. Chromosoma 106 70–80. [DOI] [PubMed] [Google Scholar]
- Soto, M. C., T. B. Chou and W. Bender, 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann, A., and M. van Lohuizen, 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6 846–856. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, N., 1907. The chromosomes in Drosophila ampelophila. Proceedings of the 7th International Zoological Congress, Cambridge University Press, Cambridge, UK.
- Stevens, N., 1908. A study of the germ cells of certain Diptera, with reference to the heterochromosomes and phenomena of synapsis. J. Exp. Zool. 5 359–374. [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen, M., M. Frasch, E. Wientjens and A. Berns, 1991. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 353 353–355. [DOI] [PubMed] [Google Scholar]
- Voncken, J. W., D. Schweizer, L. Aagaard, L. Sattler, M. F. Jantsch et al., 1999. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J. Cell Sci. 112(Pt. 24): 4627–4639. [DOI] [PubMed] [Google Scholar]
- Vucetic, S., C. J. Brown, A. K. Dunker and Z. Obradovic, 2003. Flavors of protein disorder. Proteins 52 573–584. [DOI] [PubMed] [Google Scholar]
- Wang, L., N. Jahren, M. L. Vargas, E. F. Andersen, J. Benes et al., 2006. Alternative ESC and ESC-like subunits of a polycomb group histone methyltransferase complex are differentially deployed during Drosophila development. Mol. Cell. Biol. 26 2637–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederschain, D., L. Chen, B. Johnson, K. Bettano, D. Jackson et al., 2007. Contribution of polycomb homologues Bmi-1 and Mel-18 to medulloblastomapathogenesis. Mol. Cell. Biol. 27 4968–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. T., 1984. A genetic analysis of transvection. Ph.D. Thesis, Harvard University, Cambridge, MA.
- Wu, C. T., and M. L. Goldberg, 1989. The Drosophila zeste gene and transvection. Trends Genet. 5 189–194. [DOI] [PubMed] [Google Scholar]
- Wu, C. T., and M. Howe, 1995. A genetic analysis of the Suppressor 2 of zeste complex of Drosophila melanogaster. Genetics 140 139–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. T., R. S. Jones, P. F. Lasko and W. M. Gelbart, 1989. Homeosis and the interaction of zeste and white in Drosophila. Mol. Gen. Genet. 218 559–564. [DOI] [PubMed] [Google Scholar]