Abstract
Cathepsin C (CTSC) is a lysosomal cysteine protease belonging to the papain superfamily. Our previous study showed that CTSC precursor (zymogen) is localized exclusively in cortical rods (CRs) of mature oocyte in the kuruma prawn Marsupenaeus japonicus, suggesting that CTSC might have roles on regulating release and/or formation of a jelly layer. In this study, enzymically active CTSC of the kuruma prawn was prepared by recombinant expression in the High Five insect cell line. The recombinant enzyme with a polyhistidine tag at its C-terminus was considered to be initially secreted into the culture medium as an inactive form of zymogen, because Western blot with anti-CTSC antibody detected a 51 kDa protein corresponding to CTSC precursor. After purification by affinity chromatography on nickel-iminodiacetic acid resin, the enzyme displayed three forms of 51, 31, and 30 kDa polypeptides. All of the forms can be recognized by antiserum raised against C-terminal polyhistidine tag, indicating that the 31 and 30 kDa forms were generated from 51 kDa polypeptide by removal of a portion of the N-terminus of propeptide. Following activation at pH 5.5 and 37°C for 40 hours under native conditions, the recombinant CTSC (rCTSC) exhibited increased activity against the synthetic substrate Gly-Phe-β-naphthylamide and optimal pH at around 5. The purified rCTSC will be useful for further characterization of its exact physiological role on CRs release and/or formation of a jelly layer in kuruma prawn.
1. Introduction
Cathepsin C (CTSC), also named cathepsin J, dipeptidyl transferase and dipeptidyl aminopeptidase I (DPPI) (EC 3.4.14.1), is a member of the papain superfamily of lysosmal cysteine protease [1, 2]. The amino acid sequences of CTSC share 30%–40% homology with those of other cysteine protease members including cathepsin B, H, L [3, 4], and moreover, CTSC contains the same three highly conserved regions of catalytic active sites (Cys25, His159, and Asn197 by papain numbering) as in all papain family members. Nevertheless, CTSC has unique characteristics in structure and proteolytic specificity as follows: (1) CTSC is oligomeric protein (about 200 kDa) consisting of four identical subunits (tetramer), differing from monomeric relatives of the papain family. Each subunit is proteolytically processed into three chains: a significantly long propeptide, and heavy and light chains. (2) CTSC is only papain-like cysteine protease that requires halide ions to achieve maximum enzyme activity. (3) Unlike the endoproteolytic activity of cathepsins B, S, and L, CTSC has broad exopeptidase activity and can progressively remove dipeptides from the free amino terminus of various protein and polypeptides substrates except when the amino terminal residue is lysine or arginine, or proline residues exist on the either side of the cleavage site [5, 6]. Based on its unique exoproteolytic specificity, CTSC has been applied to amino acid sequencing of protein and excision of fusion peptides from the amino termini of recombinant proteins with CTSC break-point [7].
In mammals, CTSC was extensively distributed in many tissues. High levels were found in lung, liver, kidney and spleen. CTSC is not only capable of degrading intracellular protein but also involved in cell growth [8] and the activation of several granule-associated serine proteases, such as cathepsin G [9], lymphocyte granzymes A and B [10–12], neutrophil elastase and chymases [9], and mast cell chymase and tryptase [13–17]. CTSC plays a requisite role in the posttranslational processing and activation of these granule serine proteases by the removal of one or more dipeptides. CTSC is thus thought to be one of the major processing enzymes known so far [18]. Mutations of the CTSC gene are responsible for Papillon-Lefevre syndrome in human [19].
In penaeid shrimps, we first cloned and characterized the kuruma prawn (Marsupenaeus japonicus) CTSC [20]. High expressions of the CTSC transcripts were found in the ovary at final stages of oocyte maturation. The CTSC precursor (proCTSC) was localized exclusively in cortical rods (CRs) in extracellular cortical crypts of oocytes. CRs appear after completion of yolk accumulation in oocytes, and are, upon fertilization, released from the crypts to form a jelly layer surrounding an egg. Accordingly, the proCTSC was suggested to be activated during fertilization and has a crucial role on regulating CR release and/or formation of a jelly layer. This study attempted to obtain an enzymically active CTSC of the kuruma prawn. Since the deduced amino acid of kuruma prawn CTSC possessed a potential glycosylation site [20], the production of recombinant protein using High Five insect cell line was applied to obtain an active enzyme with an arthropod-type sugar chain. The purified recombinant product of CTSC provides sufficient active enzyme to further characterize its potential physiological role of regulating CR release and/or formation of a jelly layer.
2. Materials and Methods
2.1. Insect Cells Culture
The High Five insect cell line (BTI-TN-5B1-4), originated from the ovarian cells of the cabbage looper, Trichoplusia ni, was commercially purchased from Invitrogen. The insect cells were routinely grown and subcultured at 27°C in serum-free medium Express Five SFM (Invitrogen), supplemented with L-glutamine to the final concentration of 18 mM.
2.2. Construction of Expression Plasmid Vector
Total RNA was isolated from the kuruma prawn ovary with Trizol Reagent (Invitrogen) and reverse transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen) following the manufacturer's instructions. The cDNA fragments encoding the prawn proCTSC with its signal peptide were PCR-amplified from the ovary cDNA pool using the following specific primers: sense primer (5′-ACACTCCTGTGGGTCGTAGCA-3′) and antisense primer (5′- ATGGTGATGGTGATGATGTGGGATGGGCACAGCTTCCA-3′), while the cDNA fragments encoding the proCTSC without its signal peptide were amplified using the sense primer 5′-GCAGACACGCCCGCGAACTGCAC-3′ and the same antisense primer as mentioned above. These primers were based on the sequence of prawn proCTSC cDNA, as published by Qiu et al. [20]. The sense and antisense primers were supplemented with a start codon (double underline) and a polyhistidine tag (underline) followed by a terminal codon (box), respectively. The PCR products were directly cloned to expression plasmid pIB/V5-His-TOPO (Invitrogen), which contains baculovirus promoters and need no other viral factors to activating protein expression in insect cell lines [21]. The recombinant plasmid vectors were correspondingly designated as pIB/CTSC1 (with signal peptide) and pIB/CTSC2 (without signal peptide). The constructed plasmids were transformed into TOP10 competent cells and the positive clones were sequenced to verify the correct orientation and open reading frame of the insert.
2.3. Expression of Recombinant CTSC in Insect Cell Line
By using the lipid-mediated transfection reagent Cellfectin (Invitrogen), approximately 1 μg of constructed plasmids DNA were transfected into log-phase insect High Five cells (1.8–2.3 × 106 cells/mL, >95% viability) in serum-free media Express Five SFM (Invitrogen) according to the standard protocol provided by the manufacturer. The expression of recombinant protein was assayed with the cultured medium 1–4-day posttransfection.
2.4. Purification of Recombinant CTSC
The recombinant protein was purified from the cultured medium by affinity chromatography on Ni-chelating resin using ProBond Purification System (Invitrogen). In order to remove any possible media components that may strip Ni+2 from metal-chelating resins, the harvested supernatant of medium was ultrafiltered and buffer exchanged through a Centriplus Centrifugal Filter Unite (Millipore) with Native Purification Buffer (NPB, 50 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole, pH 8.0) prior to affinity chromatography. The condensed sample was loaded onto a Ni-chelating resin column equilibrated with the buffer NPB. After binding for 60 minutes at room temperature, the column was washed and eluted with buffer NPB containing 20 mM and 250 mM imidazole, respectively. The eluent was collected in 1 mL fractions and analyzed with Western blot. Fractions with immuno-positive eluent were pooled and adjusted to pH 5.5 with 1 M citric acid followed by incubation at 37°C for 40 hours to complete the proteolytic maturation.
2.5. Western Blot
Ten microliters of the culture media and purified recombinant protein was resolved on a 5–20% gradient SDS-PAGE gel under reducing conditions and electroblotted onto a polyvinylidene difluoride membrane (PVDF, Amersham-Pharmacia Biotech). The membrane was treated with blocking solution (Roche Diagnostics Corporation) overnight at 4°C and incubated with primary antibody at 1 : 1000 for 1 hour at room temperature. The antibody to the C-terminus of the kuruma prawn CTSC was raised as described previously [20], and the anti-histidine-tag antibody was purchased from Invitrogen. After washing with Tris-buffered saline (20 mM Tris-HCl, 0.9% NaCl, pH 7.4) containing 0.1% Tween-20, the membrane was incubated with a second antibody (goat anti-rabbit serum) conjugated with alkaliphosphatase (Bio-Rad) at 1 : 10 000 for 1 hour at room temperature. NBT/BCIP (Roche Diagnostics Corporation) was used as a substrate for color development.
2.6. Activity Assay
Activity was assayed by the hydrolysis of Gly-Phe-β-naphthylamide (Sigma) at 37°C essentially as described by McGuire et al. [1]. The assay buffer mixture (600 μL) contained 100 μM substrate, 50 mM sodium acetate, pH 5.5, 30 mM NaCl, 1 mM DTT, and 0.5 mM EDTA. The reaction was stopped by the addition of 500 μL of 50 mM glycine-NaOH, pH 10.5. One unit of enzymatic activity was defined as the amount of enzyme required to release 1 μmoL of β-naphthylamine from the hydrolyzed substrate per minute as monitored the fluorescence at 335 nm excitation, 405 nm emission.
To determine the optimal pH value, substrate hydrolysis was performed at 37°C over a pH range of 3.0–8.0. The assay buffer used was 50 mM citrate (pH 3.0), 50 mM sodium acetate (pH 4.0–5.5), 50 mM sodium phosphate (pH 6.0–7.0), and 50 mM Tris-HCl (pH 7.5–8.0), respectively. Protein concentrations were determined by the Bradford assay [22], using bovine serum albumin as a standard.
3. Results and Discussion
3.1. PCR Amplification and Expression Vector Construction
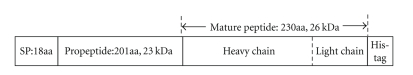
Based on the full-length cDNA sequence of the proCTSC in our previous work [20], the proCTSC-encoding regions were PCR-amplified from the kuruma prawn ovarian cDNA pool. Since CTSC was predicted as a secreted protein, to verify whether the CTSC secretion does rely on the native secretion signal, two forward primers were designed to separately generate a PCR product either with or without the shrimp CTSC signal peptide. After T-A cloning of PCR products, the insert sequence was confirmed to hold a correct open reading frame and no mutations. In the expression vectors pIB/CTSC1 (with signal peptide), as expected, the insert cDNA sequence encodes a proCTSC of 449 amino acids with a calculated molecular weight of 51 kDa, comprising a putative signal peptide of 18 amino acids, a notably long propeptide of 201 amino acids and a putative mature peptide region of 230 amino acids (Figure 1). Additionally, a 6-histidine tag was introduced to the C-terminal of the light chain to simplify purification of expression product. Likewise, pIB/CTSC2 (without signal peptide) was confirmed to encode a CTSC comprising the same propeptide and mature peptide with a 6-histidine tag.
Figure 1.
Schematic presentation of the rCTSC precursor encoded by pIB/CTSC1. The deduced amino acid sequence comprises of 449 amino acid residues with calculated molecular masses as below: signal peptide (SP): 18aa, 1.9 kDa; propeptide: 201aa, 23 kDa, and matured peptide: 230aa, 26 kDa with heavy chain: 163aa, 19 kDa, and light chain: 67aa, 7 kDa, respectively. The 6-histidine tag sequence is fused to the C-terminus of the light chain. The rCTSC precursor encoded by pIB/CTSC2 lacks 18aa of the signal peptide.
3.2. Expression and Purification of the Recombinant CTSC
Considering that the High Five cells were apt to form clumps in suspension culture, adhere-wall culture method was employed to produce the recombinant CTSC (rCTSC) in stable expression. A serum-free medium was chosen for easier purication and downstream processing of secreted recombinant products. By means of immunoblotting analysis using antibody to the C-terminus of CTSC, a target band of a 51 kDa protein was detected in the culture media for High Five cells infected by pIB/CTSC1 with signal peptide but not in those by pIB/CTSC2 without signal peptide and uninfected High Five cells as well (Figure 2). These results illustrate that the shrimp rCTSC signal peptide has functioned properly in insect cells and is required by the insect cells for an efficient secretion of the recombinant protein.
Figure 2.
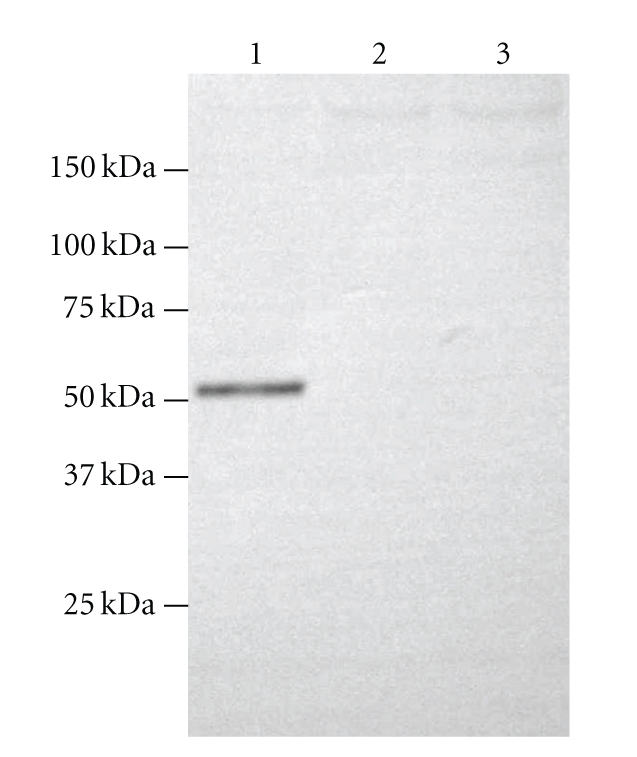
Immunoblot analysis on recombinant expression of CTSC in the culture media from 24 hours postinfected High Five cells by pIB/CTSC1 (with signal peptide, lane 1) and pIB/CTSC2 (without signal peptide, lane 2). The collected media from uninfected High Five cells was also loaded as a negative control (lane 3). Ten microliters of culture media in each lane was resolved on SDS-PAGE gel under reducing conditions and was electroblotted onto a PVDF membrane. Recombinant CTSC fusion proteins were probed with anti-His-tag antibody.
To find the optimal time of harvest, a time course was set up for expression of proCTSC in High Five insect cells. The culture medium was harvested at 24 hours intervals from 24-hour to 96-hour posttransfection. The amount of proCTSC reached the maximum at 72-hour posttransfection, since there is no significant increase of the amount of proCTSC after the expression progressed further than 72 hours of infection, as shown by Western blot analysis of the cultured medium (Figure 3). Therefore, 72 hours infection was chosen as the optimal harvest time for subsequent experiments in order to avoid any possible degradation of recombinant product at longer time points.
Figure 3.
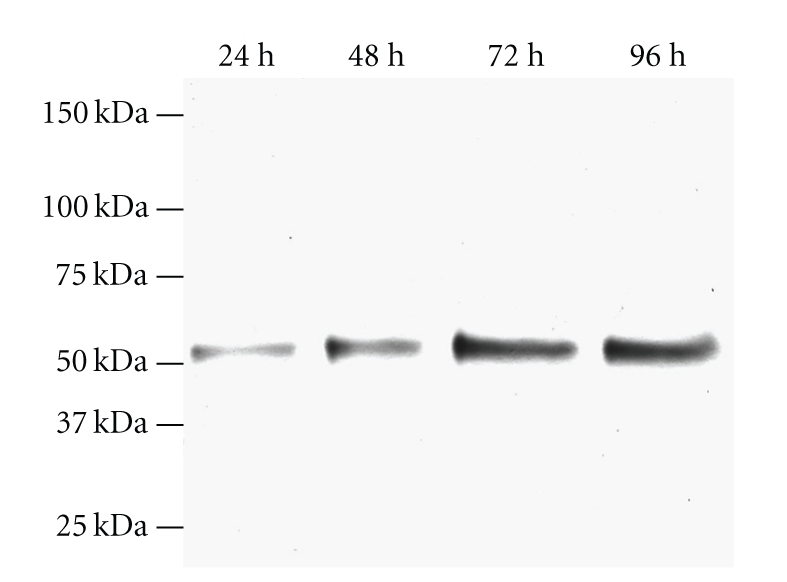
Time course expression of a recombinant protein in the culture medium from pIB/CTSC1 infected High five cells as revealed by Western blot. The culture media was collected at 24-, 48-, 72-, and 96-hour (h) postinfection. The expression yield of target protein was estimated using an anti-His-tag antibody.
Immunoblot analysis of the rCTSC purified by Ni-chelating affinity chromatography revealed three polypeptides with molecular masses of approximate 51 kDa, 31 kDa, and 30 kDa (Figure 4). The amount of 51 kDa polypeptides is much larger than that of the other two. Flatworm CTSC expressed in insect cells also exhibited similar three bands of 55 kDa, 39 kDa, and 38 kDa polypeptides after purification. N-terminal sequencing demonstrated that the 39 kDa and 38 kDa polypeptides shared identical N-terminal sequence and were produced by removal of 126 residues from N-terminus of 55 kDa proenzyme [23]. Thus, it is likely that 31 kDa and 30 kDa polypeptides are derived from 51 kDa polypeptide by differential processing of N-terminus of peptides as demonstrated in flatworm rCTSC. It is worth noting that the 30 or 31 kDa is apparently larger than the calculated molecular weight of the mature CTSC of 26 kDa, although the existence of a potential glycosylation (position 228) and an extra 6-histidine tag attached to the C-terminus of the mature peptide might increase the expected size of mature peptide by ~2 kDa. The best explanation for the increased size is that a small partial propeptide still remained on the mature peptide as revealed in flatworm rCTSC. The cleavage site in the proCTSC is near the C-terminus of the propeptide region instead of the joint between propeptide and mature peptide. Like flatworm rCTSC, more than one processed NH2 terminis were identified in the expected heavy chain peptide of insect-expressed shrimp rCTSC. Interestingly, similar result was also reported in the native CTSC purified from the dog mast cell, in which the heavy chain is heterogeneously processed to three different forms [17] and has multiple heavy chain NH2 termini. The functional significance of variable heavy chain N-terminal processing is unknown. Whether the unusual processing might result in specific activity remains to be further investigated.
Figure 4.
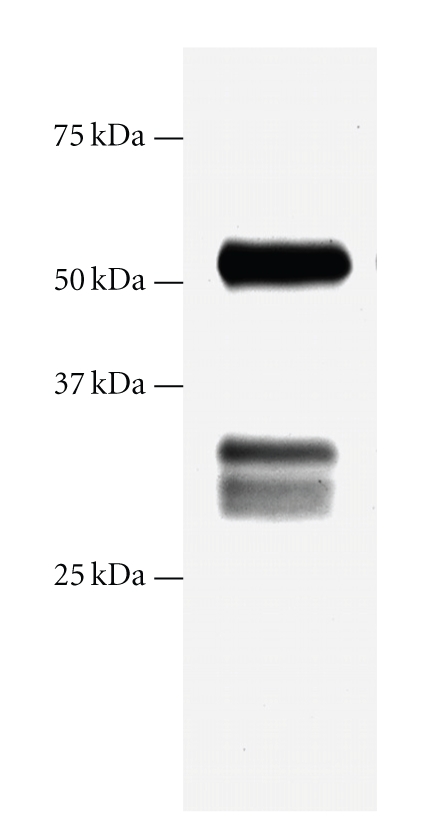
Immunoblot analysis on purified rCTSC eluted from the Ni-chelating resin column. The purified rCTSC contained peptides with molecular weights of 51, 30, and 31 kDa, which was detected by a polyclonal antibody against C-terminus of kuruma prawn CTSC.
3.3. Activity of rCTSC
The rCTSC activity was assayed in the medium before and after incubation. The rCTSC exhibited very low activity against the substrate Gly-Phe-β-naphthylamide before incubation. Interestingly, an increased activity occurred after incubation of the rCTSC for 40 hours at 37°C at pH 5.5 (Table 1), indicating that the insect cells initially secreted inactive zymogen proCSTC (51 kDa) in the medium and then the inactive proCTSC underwent autoactivation during the incubation. The enzymatic activity could be from the processed forms of 30 and 31 kDa that were produced during purification as noted above. The removal of the propeptide is prerequisite for the rCTSC to acquire enzymatic activity. Several lines of evidence supported that a partial processed propeptide attached to the oligomeric form was required for proper folding of human and rat native CTSC [24, 25]. But it remains unknown whether the processed propeptide is also necessary for proper folding of the prawn rCTSC or not when expressed in insect cells.
Table 1.
Purification and incubation of rCSTC secreted by insect cells.
| Total volume (mL) | Activity (U/mL) | Total activity (U) | Yield (%) | |
|---|---|---|---|---|
| Concentrated culture medium | 50 | 0.0036 | 0.18 | 100 |
| Purified with Ni-chelating resin | 5 | 0.0040 | 0.020 | 11.1 |
| Incubated for 40 hours at pH5.5 | 5 | 0.0753 | 0.3765 | 209 |
It has been shown that the mature peptide of rat rCSTC expressed in insect cells was cleaved into heavy chain (24 kDa) and light chain (5.5 kDa) as its native CTSC [26]. However, like the flatworm mature rCSTC [23], there was no cleavage between heavy and light chains in the mature peptide of the kruma prawn rCTSC and the Western blot profile of active rCTSC did not change in the incubated medium as compared with the nonincubated (data no shown). Similarly, only a single chain of the mature peptide (26 kDa) of bovine rCTSC was found with enzymatic activity in methylotrophic yeast expression system [27]. The active CTSC purified natively from human spleen is also composed of only a subunit of 24 kDa (intact mature peptide) rather than two subunits (heavy and light chains) [1]. Therefore, it can be deduced that the cleavage between heavy and light chains might be not required for the maturation and activation of rCTSC.
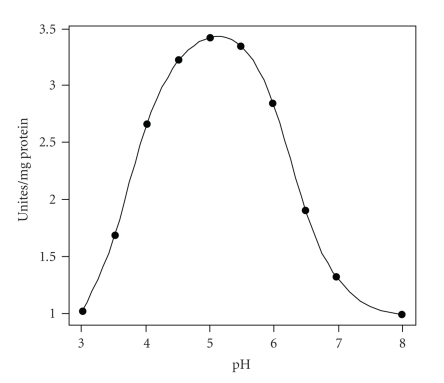
As shown in Figure 5, the optimal pH of the rCTSC was around 5, which was basically the same with that of the native prawn CTSC [28]. This value is also consistent with that obtained from cathepsin L in the shrimp (Penaeus vannamei) [29] but lower than that of human CTSC (pH 6.0) [30]. Only slight activity was measured at neutral pH and in extreme pH regions, which is a typical characteristic of the lysosomal cysteine proteinases.
Figure 5.
Effect of pH on the activity of the prawn rCTSC. Purified rCTSC was assayed for the substrate hydrolysis at 37°C over a pH range of 3.0–8.0. Results are the mean values of maximum activity from three experiments.
In summary, we have successfully produced the kuruma prawn rCTSC using the High Five insect cell expression system. The purified rCTSC exhibited high enzymical activity after incubation at pH 5.5. To our knowledge this is the first time that active rCTSC of crustacean species is produced. The active rCTSC will be useful for further characterization of its exact physiological role on CRs release and/or formation of a jelly layer in penaeid shrimp.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Project no. 30471348), the Marine Biology Program of Shanghai Leading Academic Discipline (Project no. J50701), and a postdoctoral fellowship from Japan Society for Science Promotion (JSPS).
References
- 1.McGuire MJ, Lipsky PE, Thiele DL. Purification and characterization of dipeptidyl peptidase I from human spleen. Archives of Biochemistry and Biophysics. 1992;295(2):280–288. doi: 10.1016/0003-9861(92)90519-3. [DOI] [PubMed] [Google Scholar]
- 2.Kominami E, Ishido K, Muno D, Sato N. The primary structure and tissue distribution of cathepsin C. Biological Chemistry Hoppe-Seyler. 1992;373(7):367–373. doi: 10.1515/bchm3.1992.373.2.367. [DOI] [PubMed] [Google Scholar]
- 3.Ishidoh K, Muno D, Sato N, Kominami E. Molecular cloning of cDNA for rat cathepsin C. Cathepsin C, a cysteine proteinase with an extremely long propeptide. Journal of Biological Chemistry. 1991;266(25):16312–16317. [PubMed] [Google Scholar]
- 4.Paris A, Strukelj B, Pungercar J, Renko M, Dolenc I, Turk V. Molecular cloning and sequence analysis of human preprocathepsin C. FEBS Letters. 1995;369(2-3):326–330. doi: 10.1016/0014-5793(95)00777-7. [DOI] [PubMed] [Google Scholar]
- 5.McDonald JK, Zeitman BB, Reilly TJ, Ellis S. New observations on the substrate specificity of cathepsin C (dipeptidyl aminopeptidase I). Including the degradation of beta-corticotropin and other peptide hormones. Journal of Biological Chemistry. 1969;244(10):2693–2709. [PubMed] [Google Scholar]
- 6.McDonald JK, Reilly TJ, Zeitman BB, Ellis S. Cathepsin C: a chloride-requiring enzyme. Biochemical and Biophysical Research Communications. 1966;24(5):771–775. doi: 10.1016/0006-291x(66)90392-5. [DOI] [PubMed] [Google Scholar]
- 7.Martensen I, Koolman J, Mentlein R. Proline-specific dipeptidyl peptidase from the blue blowfly Calliphora vicina hydrolyzes in vitro the ecdysiostatic peptide trypsin-modulating oostatic factor (Neb-TMOF) Archives of Insect Biochemistry and Physiology. 1998;37(2):146–157. doi: 10.1002/(SICI)1520-6327(1998)37:2<146::AID-ARCH3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Doughty MJ, Gruenstein EI. Cell growth and substrate effects on characteristics of a lysosomal enzyme (cathepsin C) in Duchenne muscular dystrophy fibroblasts. Biochemistry and Cell Biology. 1987;65(7):617–625. doi: 10.1139/o87-082. [DOI] [PubMed] [Google Scholar]
- 9.McGuire MJ, Lipsky PE, Thiele DL. Generation of active myeloid and lymphoid granule serine proteases requires processing by the granule thiol protease dipeptidyl peptidase I. Journal of Biological Chemistry. 1993;268(4):2458–2467. [PubMed] [Google Scholar]
- 10.Smyth MJ, McGuire MJ, Thia KYT. Expression of recombinant human granzyme B1: a processing and activation role for dipeptidyl peptidase I. Journal of Immunology. 1995;154(12):6299–6305. [PubMed] [Google Scholar]
- 11.Kummer JA, Kamp AM, Citarella F, Horrevoets AJG, Hack CE. Expression of human recombinant granzyme A zymogen and its activation by the cysteine proteinase cathepsin C. Journal of Biological Chemistry. 1996;271(16):9281–9286. doi: 10.1074/jbc.271.16.9281. [DOI] [PubMed] [Google Scholar]
- 12.Wilharm E, Parry MAA, Friebel R, et al. Generation of catalytically active granzyme K from Escherichia coli inclusion bodies and identification of efficient granzyme K inhibitors in human plasma. Journal of Biological Chemistry. 1999;274(38):27331–27337. doi: 10.1074/jbc.274.38.27331. [DOI] [PubMed] [Google Scholar]
- 13.Murakami M, Karnik SS, Husain A. Human prochymase activation. A novel role for heparin in zymogen processing. Journal of Biological Chemistry. 1995;270(5):2218–2223. [PubMed] [Google Scholar]
- 14.Sakai K, Ren S, Schwartz LB. A novel heparin-dependent processing pathway for human tryptase: autocatalysis followed by activation with dipeptidyl peptidase I. Journal of Clinical Investigation. 1996;97(4):988–995. doi: 10.1172/JCI118523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McEuen AR, Ashworth DM, Walls AF. The conversion of recombinant human mast cell prochymase to enzymatically active chymase by dipeptidyl peptidase I is inhibited by heparin and histamine. European Journal of Biochemistry. 1998;253(1):300–308. doi: 10.1046/j.1432-1327.1998.2530300.x. [DOI] [PubMed] [Google Scholar]
- 16.Pham CTN, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolters PJ, Raymond WW, Blount JL, Caughey GH. Regulated expression, processing, and secretion of dog mast cell dipeptidyl peptidase I. Journal of Biological Chemistry. 1998;273(25):15514–15520. doi: 10.1074/jbc.273.25.15514. [DOI] [PubMed] [Google Scholar]
- 18.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO Journal. 2001;20(17):4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart TC, Hart PS, Bowden DW, et al. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. Journal of Medical Genetics. 1999;36(12):881–887. [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu G-F, Yamano K, Unuma T. Cathepsin C transcripts are differentially expressed in the final stages of oocyte maturation in kuruma prawn Marsupenaeus japonicus . Comparative Biochemistry and Physiology, Part B. 2005;140(2):171–181. doi: 10.1016/j.cbpc.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Mirzaei M, Xu Y, Elias CB, Prakash S. Nonviral production of human interleukin-7 in Spodoptera frugiperda insect cells as a soluble recombinant protein. Journal of Biomedicine and Biotechnology. 2009;2009:8 pages. doi: 10.1155/2009/637942. Article ID 637942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Hola-Jamriska L, King LT, Dalton JP, Mann VH, Aaskov JG, Brindley PJ. Functional expression of dipeptidyl peptidase I (cathepsin C) of the oriental blood fluke Schistosoma japonicum in Trichoplusia ni insect cells. Protein Expression and Purification. 2000;19(3):384–392. doi: 10.1006/prep.2000.1261. [DOI] [PubMed] [Google Scholar]
- 24.Cigić B, Kriźaj I, Kralj B, Turk V, Pain RH. Stoichiometry and heterogeneity of the pro-region chain in tetrameric human cathepsin C. Biochimica et Biophysica Acta. 1998;1382(1):143–150. doi: 10.1016/s0167-4838(97)00173-8. [DOI] [PubMed] [Google Scholar]
- 25.Wiederanders B, Kaulmann G, Schilling K. Functions of propeptide parts in cysteine proteases. Current Protein and Peptide Science. 2003;4(5):309–326. doi: 10.2174/1389203033487081. [DOI] [PubMed] [Google Scholar]
- 26.Lauritzen C, Pedersen J, Madsen MT, Justesen J, Martensen PM, Dahl SW. Active recombinant rat dipeptidyl aminopeptidase (cathepsin C) produced using the baculovirus expression system. Protein Expression and Purification. 1998;14(3):434–442. doi: 10.1006/prep.1998.0976. [DOI] [PubMed] [Google Scholar]
- 27.Komeda T, Tazumi K, Shimada H, et al. Production of active bovine cathepsin C (dipeptidyl aminopeptidase I) in the methylotrophic yeast Candida boidinii . Applied Microbiology and Biotechnology. 2002;59(2-3):252–258. doi: 10.1007/s00253-002-1010-z. [DOI] [PubMed] [Google Scholar]
- 28.Qiu GF, Zhu Q, Cai SL, Yamano K. Enzymatic activity measurement of cathepsin C during the oogenesis of kuruma shrimp Marsupenaeus japonicus . Journal of Fisheries of China. 2009;33(3):417–423. [Google Scholar]
- 29.Le Boulay C, van Wormhoudt A, Sellos D. Cloning and expression of cathepsin L-like proteinases in the hepatopancreas of the shrimp Penaeus vannamei during the intermolt cycle. Journal of Comparative Physiology B. 1996;166(5):310–318. doi: 10.1007/BF02439917. [DOI] [PubMed] [Google Scholar]
- 30.Dolenc I, Turk B, Pungercic G, Ritonja A, Turk V. Oligomeric structure and substrate induced inhibition of human cathepsin C. Journal of Biological Chemistry. 1995;270(37):21626–21631. doi: 10.1074/jbc.270.37.21626. [DOI] [PubMed] [Google Scholar]