Abstract
Mechanistic investigations of the water-splitting reaction of the oxygen-evolving complex (OEC) of photosystem II (PSII) are fundamentally informed by structural studies. Many physical techniques have provided important insights into the OEC structure and function, including X-ray diffraction (XRD) and extended X-ray absorption fine structure (EXAFS) spectroscopy as well as mass spectrometry (MS), electron paramagnetic resonance (EPR) spectroscopy and Fourier transform infrared spectroscopy applied in conjunction with mutagenesis studies. However, experimental studies have yet to yield consensus as to the exact configuration of the catalytic metal cluster and its ligation scheme. Computational modeling studies, including density functional (DFT) theory combined with quantum mechanics/molecular mechanics (QM/MM) hybrid methods for explicitly including the influence of the surrounding protein, have proposed chemically satisfactory models of the fully ligated OEC within PSII that are maximally consistent with experimental results. The inorganic core of these models is similar to the crystallographic model upon which they were based but comprises important modifications due to structural refinement, hydration and proteinaceous ligation which improve agreement with a wide range of experimental data. The computational models are useful for rationalizing spectroscopic and crystallographic results and for building a complete structure-based mechanism of water-splitting in PSII as described by the intermediate oxidation states of the OEC. This review summarizes these recent advances in QM/MM modeling of PSII within the context of recent experimental studies.
Keywords: oxomanganese complexes, photosystem II, water oxidation, oxygen evolution, oxygen evolving center, photosynthesis, quantum mechanics/molecular mechanics (QM/MM), density functional theory (DFT)
Introduction
The oxygen-evolving complex (OEC) of photosystem II (PSII) catalyzes the four-electron oxidation of water in the thylakoid membrane of green plant chloroplasts, producing dioxygen and releasing protons to the lumen as described by the following electrochemical half-reaction:
| (1) |
Our current understanding of the water-oxidation reaction introduced by Eq. (1) is based on the catalytic cycle proposed by Joliot and Kok (see Fig. 1) (Joliot et al. 1969; Kok et al. 1970). The cycle has been extensively investigated for several decades by a variety of biochemical, spectroscopic and theoretical studies (Barber 2003; Diner & Babcock 1996; Renger 2001; Vrettos et al. 2001a; Yachandra et al. 1996). The solar energy required for water oxidation is harvested by a system of chlorophyll and carotenoid cofactors in PSII and subsequently transmitted to the special chlorophyll moiety P680, responsible for the primary electron transfer event. P680 is oxidized, translocating electrons across the membrane through a number of redox cofactors (McEvoy & Brudvig 2006; Yachandra 2005). On the electron-acceptor side of PSII, plastoquinone (Q) is doubly reduced by the translocated electrons and doubly protoned by protons from the stromal side of the membrane. It is subsequently displaced by another Q cofactor, leaving the membrane as plastoquinol (QH2). The overall reaction is:
| (2) |
On the electron-donor side of PSII, each highly oxidizing P680+ radical (E ≈ +1.25 V) (Grabolle & Dau 2005; Rappaport et al. 2002) abstracts an electron from substrate-water bound to the OEC. This is mediated by the tyrosine residue (YZ) that serves as a redox intermediary (Debus et al. 1988b; Diner & Britt 2005; Metz et al. 1989), probably alternating between the YZOH (reduced) and deprotonated YZO• (oxidized) forms (Hoganson & Babcock 1997; Hoganson et al. 1995).
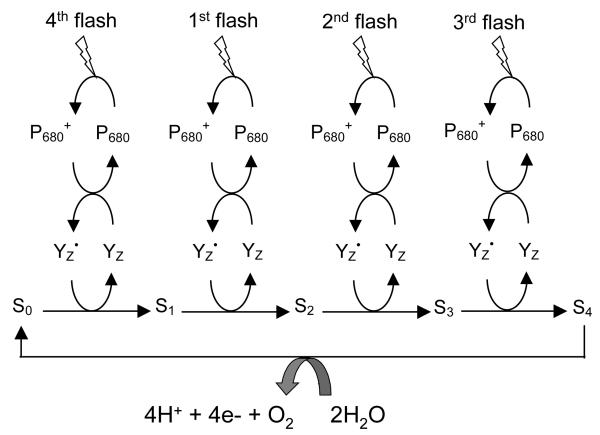
Figure 1.
Schematic description of the catalytic cycle of the OEC in PSII. P680 is repeatedly photo-oxidized, in turn oxidizing tyrosine Z (YZ), which is the immediate oxidant of the OEC. Four one-electron oxidations of the OEC generate the S4 state, which rapidly catalyzes the four-electron oxidation of water to produce O2 and reform the S0 state of the OEC.
As shown in Fig. 1, the OEC undergoes four consecutive light-driven one-electron oxidations and thereby stores four oxidizing equivalents that are rapidly used in the S4→S0 transition to oxidatively split two water molecules per cycle. Each redox state accumulates an oxidizing equivalent and is conventionally described as a ‘Storage-state’, or ‘S-state’, with S0 being the most reduced state and S4 the most oxidized. While the S-states are well established, the structure of the catalytic metal cluster along the cycle and its ligation scheme remain poorly understood. Computational modeling studies (Blomberg et al. 1997; Lundberg et al. 2003; McEvoy & Brudvig 2004; Siegbahn 2002; Siegbahn & Crabtree 1999; Siegbahn & Lundberg 2005; Siegbahn & Lundberg 2006; Sproviero et al. 2006a; Sproviero et al. 2006b; Sproviero et al. 2007c), based on density functional (DFT) theory and quantum mechanics/molecular mechanics (QM/MM) hybrid methods, have recently proposed energetically minimized and chemically satisfactory models of the fully ligated OEC within PSII, maximally consistent with experimental results. The computational models are useful for rationalizing a structure-based mechanism of water splitting in PSII as described by the OEC oxidation intermediate states. Here, we summarize recent advances in QM/MM modeling of the OEC of PSII.
Structure and Mechanism of the OEC
The mechanism of the OEC-catalyzed water-splitting reaction has long been a subject of investigation (Barber 2003; Diner & Babcock 1996; Renger 2001; Vrettos et al. 2001a; Yachandra et al. 1996). Besides its intrinsic biological interest, efficient electrochemical water-oxidizing catalysts are urgently required for the development of artificial photosynthetic systems (Lewis & Nocera 2006; Whitesides & Crabtree 2007). Understanding the operation of the OEC is therefore important for the development of novel solar energy transduction and storage systems with the potential to revolutionize renewable energy technology.
Much biochemical work has been pursued to elucidate the mechanism of water splitting at the OEC of PSII (Allahverdiyeva et al. 2004; Babcock et al. 1989; Bergmann et al. 1998; Berthomieu et al. 1998; Britt et al. 1994; Chu et al. 2001; Clausen et al. 2004; Debus 1992; Debus 2001; Debus et al. 2003; Debus et al. 1988a; Debus et al. 1988b; Debus et al. 2001; Debus et al. 2000; Debus et al. 2005; Diner 2001; Diner & Babcock 1996; Diner et al. 2004; Diner et al. 1998; Diner & Nixon 1998; Diner & Rappaport 2002; Faller et al. 2002; Hays et al. 1998; Hays et al. 1999; Hillier & Messinger 2005; Hoganson et al. 1995; McEvoy & Brudvig 2006; Messinger 2004; Metz et al. 1989; Nixon & Diner 1992; Nixon et al. 1992; Rappaport et al. 2002; Strickler et al. 2005; Tang et al. 1994; Tang et al. 1996; Tommos et al. 1995). A variety of techniques have been applied, including electron paramagnetic resonance (EPR) spectroscopy (Boussac et al. 1998; Boussac et al. 1989; Britt et al. 2004; Britt et al. 2000; Kulik et al. 2005a; Kulik et al. 2005b; Kulik et al. 2005c; Matsukawa et al. 1999; Messinger et al. 1997a; Messinger et al. 1997b; Miller & Brudvig 1991; Mino & Kawamori 2001; Nugent et al. 1997; Peloquin & Britt 2001; Peloquin et al. 1998; Peloquin et al. 2000; Poluektov et al. 2005; Razeghifard & Pace 1999), X-ray absorption spectroscopy (XAS) (Dau et al. 2001; Dau et al. 2003; Dau et al. 2004; Grabolle et al. 2006; Hasegawa et al. 1999; Haumann et al. 2005a; Haumann et al. 2005b; Iuzzolino et al. 1998; Liang et al. 2000; Mishra et al. 2007; Riggs-Gelasco et al. 1996; Robblee et al. 2002; Sauer & Yachandra 2004; Sauer et al. 2005; Stemmler et al. 1997; Yachandra 2002; Yachandra et al. 1986; Yachandra et al. 1987; Yano et al. 2006; Yano et al. 2005b), Fourier transform infrared (FTIR) spectroscopy (Chu et al. 2004; Debus et al. 2005; Kimura et al. 2005b; Strickler et al. 2005) and mass spectrometry (MS) (Hillier et al. 1998; Hillier & Wydrzynski 2000; Hillier & Wydrzynski 2001; Hillier & Wydrzynski 2004; Messinger et al. 1995) that have all played particularly important roles. In recent years, mechanistic investigations have been allied with a progressively clearer idea of the structure of the OEC, supported by X-ray diffraction studies (Biesiadka et al. 2004; Ferreira et al. 2004; Kamiya & Shen 2003; Loll et al. 2005; Zouni et al. 2001) and extended X-ray absorption fine structure (EXAFS) spectroscopy (Haumann et al. 2005a; Haumann et al. 2005b; Sauer et al. 2005; Yachandra 2005; Yano et al. 2005b). It appears that the OEC comprises a cluster of four manganese ions and one calcium ion, connected by bridging oxide ions. Surrounding amino-acids provide further carboxylic acid (oxygen) and histidine (nitrogen) ligation of the metals. However, there is still little agreement on the detailed configuration of the OEC Mn4Ca cluster. Current X-ray crystallographic results are available only at moderate resolution (3.0 Å – 3.5 Å) (Biesiadka et al. 2004; Ferreira et al. 2004; Loll et al. 2005) and have been called into question because of the possible reductive damage of the cluster due to the high doses of X-ray required for data collection (Grabolle et al. 2006; Yano et al. 2005a). EXAFS spectroscopy, although able to provide accurate bond distances within the intact cluster, do not provided a complete picture of the OEC architecture, or the description of the arrangement of ligands (Yano et al. 2006). Therefore, fundamental aspects of structure/function relations in the OEC remain poorly understood, including the structure of the catalytic metal cluster, the coordination of substrate water molecules, the nature and functional role of structural rearrangements in the metal cluster along the catalytic cycle, and the role played by surrounding protein environment and cofactors on the catalytic mechanism.
QM/MM studies have developed chemically sensible models of the OEC compatible with XRD and EXAFS measurements of PSII (Sproviero et al. 2006b; Sproviero et al. 2007c; Sproviero et al. 2007e). Starting from the empirical XRD structure at 3.5 Å resolution (Ferreira et al. 2004), computational models were developed by DFT QM/MM structural refinement of PSII in oxidation states consistent with EPR (Peloquin & Britt 2001; Peloquin et al. 2000) and X-ray spectroscopy (Dau et al. 2001; Yachandra et al. 1993) as well as recent XANES (Haumann et al. 2005b) and 55Mn ENDOR (Kulik et al. 2005a; Kulik et al. 2005b; Kulik et al. 2005c) experiments. Therefore, the resulting models circumvent the effect of radiation damage caused by reduction of the Mn centers in the description of the OEC configuration. The implemented computational methodology has been rigorously tested with well-characterized manganese model compounds, verifying its ability to predict highly accurate XRD geometries (Sproviero et al. 2006a). Therefore, the resulting DFT-QM/MM structures are expected to benefit the construction of a complete model of the water splitting catalytic cycle in terms of detailed molecular structures of the OEC catalytic intermediates (Sproviero et al. 2007e).
DFT Studies
DFT benchmark studies have analyzed the ability of the Becke-3-Lee-Yang-Parr (B3LYP) hybrid density functional to predict the geometric, electronic and magnetic properties of well characterized polynuclear high-valent oxomanganese complexes (Sproviero et al. 2006a). This work has been essential for validating the DFT level of theory, as applied to Mn complexes, since hybrid density functionals were known to exhibit shortcomings in the description of low-lying spin states of open-shell transition-metal complexes (Ghosh et al. 2003; Ghosh & Steene 2001; Ghosh & Taylor 2003; Ghosh & Taylor 2005; Koch & Holthausen 2001; Lundberg & Siegbahn 2005b; Reiher et al. 2001a; Reiher et al. 2001b). Spin-polarized calculations on benchmark model compounds included the synthetic di-μ-oxo MnIIIMnIV complexes [MnIIIMnIV(μ-O)2 (H2O)2(terpy)2]3+ (terpy = 2,2′:6,2″-terpyridine) and [MnIIIMnIV(μ-O)2(phen)4]3+ (phen = 1,10-phenanthroline), the [Mn3O4(bpy)4(H2O)2]4+ (bpy=2,2′-bipyridine) trimer, and the [Mn4O4L6]+ tetramer, with L=Ph2PO-2 (Sproviero et al. 2006a). These complexes involve unpaired spins with frustrated antiferromagnetic couplings that required to be addressed in terms of the broken symmetry (BS) DFT method (Noodleman 1981; Noodleman & Case 1992; Noodleman & Davidson 1986; Noodleman et al. 1995). The BS-DFT approach also allowed for calculations of exchange magnetic coupling constants and the ligand field analysis for metal d→d, charge transfer (ligand→metal, metal→ligand), and intervalence charge transfer (metal→metal or ligand→ligand) transitions (Sproviero et al. 2006a; Sproviero et al. 2006b; Sproviero et al. 2007e).
Earlier computational studies of oxomanganese complexes had found that the DFT/B3LYP methodology typically overestimated Mn-Mn distances, with errors in the 0.10–0.15 Å range (Lundberg & Siegbahn 2004; Petrie & Stranger 2004), when comparing fully relaxed configurations of model complexes with μ-oxo bridges with XRD data. In addition, it was reported that B3LYP overestimated Mn-ligand distances along the Jahn-Teller axis of Mn3+ ions by as much as 0.23 Å (Lundberg & Siegbahn 2004; Petrie & Stranger 2004). These errors were comparable to typical structural rearrangements in the OEC metal cluster, induced by oxidation of the constituent ions (i.e., changes of Mn ligand bond-lengths in the 0.1-0.2 Å), and therefore required further analysis. In addition, hybrid functionals were known to overestimate the relative stability of high-spin over low-spin states of transition metal complexes (Koch & Holthausen 2001; Reiher et al. 2001a), a difficulty that could be critical in the process of identifying the nature of ground electronic states, or in studies of spin-crossover phenomena in transition metal complexes (Harvey 2001; Harvey et al. 2003; Holthausen 2005; Poli & Harvey 2003; Reiher et al. 2001a; Schroder et al. 2000; Shaik et al. 2002).
Findings of the most recent benchmark studies can be summarized as follows (Sproviero et al. 2006a): (1) The DFT/B3LYP level of theory can accurately predict the geometry of polynuclear oxomanganese complexes, even when applied in conjunction with rather modest basis sets so long as oxo-bridges include sufficiently expanded basis functions, rendering bond-lengths within 0.05 Å and angles within a few degrees from XRD models; (2) The magnetic properties of oxomanganese complexes in predefined spin-electronic states are properly described by the DFT B3LYP level of theory, including exchange coupling constants and spin populations indicating antiferromagnetic couplings in agreement with magnetic experimental data; (2) The relative energies of states of different spin multiplicities are not accurately described by the DFT/B3LYP level (often overestimating the stability of high-spin states) because of the limitations of the B3LYP hybrid density functional to properly balance exchange and correlation contributions to the energies of the states.
Other recently reported benchmark studies have examined the accuracy of the DFT/B3LYP method in calculations of O—H bond dissociation enthalpies in a variety of redox-active mononuclear and polynuclear manganese complexes (Lundberg & Siegbahn 2005a). In general, the functional was found to perform well, with a mean absolute deviation of about 3.0 kcal mol-1 among most of the complexes, although the error in an outlier complex was considerably greater. These studies also pointed out that DFT/B3LYP has proved to be rather unreliable in modeling the bonding of O2 (and NO) to transition metals, giving an error of at least 5 kcal mol-1 in several instances (Siegbahn 2006). This weakness might be critical in investigations of the mechanisms of water splitting strongly dependent on metal—O2 binding.
QM/MM Modeling
QM/MM modeling of the OEC of PSII has been based on the ONIOM (our own N-layered integrated molecular orbital plus molecular mechanics) method (Dapprich et al. 1999; Vreven & Morokuma 2000) with electronic embedding (EE) at the (UHF B3LYP/lacvp,6-31G(2df),6-31G:AMBER) level of theory. These computations required a combined approach exploiting the capabilities of ONIOM as implemented in Gaussian03 (Frisch et al.), including both the link-hydrogen atom scheme for efficient and flexible definitions of QM layers and the possibility of modeling open-shell systems by performing Unrestricted-DFT (e.g., UB3LYP) calculations, and the efficient generation of high-quality initial-guess spin-electronic states generated with Jaguar 5.5 (Jaguar 5.5. Schroedinger 1991-2003).
The QM layer of the OEC of PSII has been defined as a reduced system X, including the metal cluster, the proteinaceous ligands (E333, CP43-E354, D342, D170, E189 and H332), water and hydroxo ligated to metals and chloride.
The molecular structure beyond the QM layer is the extended region Y that has been described by the Amber MM force-field (Cornell et al. 1995; Cornell et al. 1996). Region Y includes all amino-acid residues with α-carbons within 15 Å from any atom in the OEC metal ion cluster and an additional buffer shell of amino-acid residues with α-carbons within 15 to 20 Å from any atom in the OEC ion cluster, with harmonic constraints to preserve the natural shape of the system (McEvoy et al. 2005a; McEvoy et al. 2005b; Sproviero et al. 2006b). The total energy E has been obtained from three independent calculations:
where EMM,X+Y is the energy of the complete system computed at the molecular mechanics level of theory, while EQM,X and EMM,X correspond to the energy of the reduced-system X computed at the QM and MM levels of theory, respectively. Electrostatic interactions between regions X and Y are included in the calculation of both EQM,X and EMM,X at the quantum mechanical and molecular mechanics levels, respectively. Thus, the electrostatic interactions computed at the MM level in EMM,X and EMM,full cancel and the resulting DFT QM/MM evaluation of the total energy involves a quantum mechanical description of the polarization of the reduced system, due to the electrostatic influence of the surrounding protein environment. Polarization of the protein active site induced by the distribution of charge in the QM layer has been introduced by correcting the atomic charges of amino-acid residues in close contact with the QM layer, according to the self-consistent polarization protocol MoD-QM/MM (Gascon et al. 2006; Sproviero et al. 2006b; Sproviero et al. 2007c; Sproviero et al. 2007e).
Structural Models of the OEC
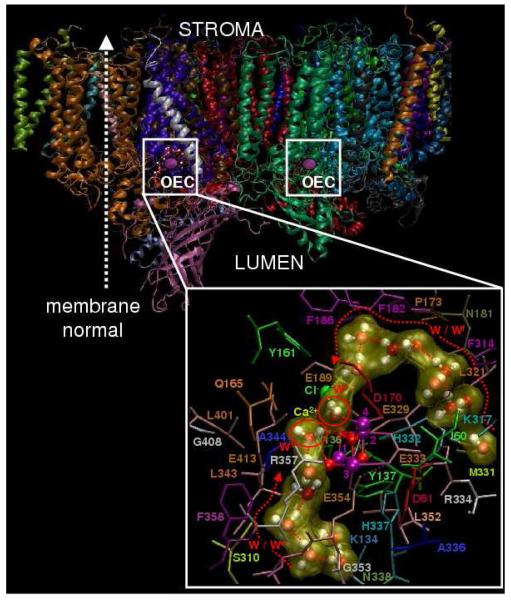
Recent DFT-QM/MM studies have introduced structural models of the OEC in PSII in the dark-stable S1 state, including the complete ligation of the metal cluster Mn4Ca by amino-acid residues, water, hydroxide, and chloride (see Fig. 2, top) (Sproviero et al. 2006b). These QM/MM models were developed by structural refinement of the XRD model of PSII from the cyanobacterium Thermosynechococcus elongatus (Ferreira et al. 2004), assuming a minimum displacement of the ligating residues from their crystallographic positions after completing the coordination spheres of the metals by hydration. These QM/MM structural models were constructed under the assumption that the crystallographic positions of the amino-acid residues in the XRD models were essentially correct, within the experimental resolution, with electronic density maps only broadened by structural disorder and radiation damage induced by photoreduction of Mn ions. Fully relaxed QM/MM configurations were obtained by energy minimization and were found to preserve the architecture of the Mn3CaO4 cuboidal core with a “dangler” Mn ligated to a corner μ4-oxide ion, suggested by the XRD model as previously proposed by EPR studies (Britt et al. 2004; Peloquin et al. 2000). The nature of the amino-acid residues directly ligated to the Mn4Ca cluster were also consistent with the ligands proposed by the XRD structure of Ferreira et al. (Ferreira et al. 2004), although with significant differences with regards to the actual ligation scheme. The resulting molecular structures are expected to provide biologically relevant insight into the OEC structure since room-temperature thermal fluctuations have negligible effects on the structure, protonation state, or charge localization effects of PSII, as indicated by XAS studies carried out at 20 K and room-temperature (Haumann et al. 2005b).
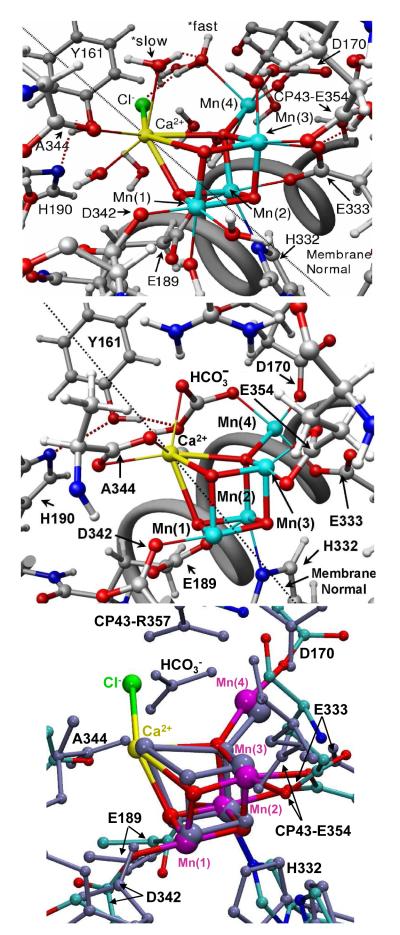
Figure 2.
The OEC and its surrounding molecular environment as described by the QM/MM model (Sproviero et al. 2006b, top), the XRD structure 1S5L (Ferreira et al. 2004, middle), and the superposition of the pentanuclear Mn4Ca clusters of the two models (bottom).
Water ligation
In contrast to the XRD structures, the QM/MM models have included manganese and calcium ions completely ligated consistently with standard coordination chemistry assumptions, supported by biochemical and spectroscopic data. This required hydration of the XRD models and addition of several small molecules as metal ligands to the OEC in order to satisfy the usual coordination numbers of high-valent Mn (5 or 6 ligands) and Ca ions (typically 6–8 coordinate) (Sproviero et al. 2006b). It was assumed that most (if not all) of these small ligands were either water bound or ‘water-derived’ species such as hydroxide or oxo ligands. Hydration involved an iterative procedure of “soaking” the XRD model in water and allowing the hydrated structure to relax in a process of energy minimization. This procedure led to the ligation of about six water and water-derived ligands to the metals of the OEC. Two of these waters, attached respectively to Ca and to Mn(4), were identified as possible substrate water molecules (see below), in agreement with earlier proposals (Haumann & Junge 1999; Hoganson & Babcock 1997; McEvoy & Brudvig 2004; Messinger 2004; Schlodder & Witt 1999), but in contrast to other models suggesting substrate water coordination as oxo-bridges between Mn ions (Brudvig & Crabtree 1986; Nugent et al. 2001; Pecoraro et al. 1994; Robblee et al. 2001; Yachandra et al. 1996). The two molecules accounted for the electronic density in the 1S5L XRD structure that was initially assigned to bicarbonate (Ferreira et al. 2004).
There is some important experimental information concerning water binding at and around the OEC. Pulsed EPR experiments using D2O-solvated PSII reveal the presence of several exchangeable deuterons near the OEC in the S0, S1 and S2 states, which is broadly consonant with the QM/MM model, although the best fit to the EPR data involves rather fewer waters than are seen in the QM/MM structure (Britt et al. 2004). Reducing the number of waters in contact with the cluster in the QM/MM model remains to be explored and would involve: 1) some or all of the ligating carboxylic amino-acid residues binding in a bridging, η2 mode (as in the crystallographic model of (Loll et al. 2005); and/or 2) the incorporation of non-water-derived small molecule ligands such as bicarbonate (Dasgupta et al. 2004; McEvoy & Brudvig 2004; Sproviero et al. 2006b), of which there is little evidence.
Time-resolved mass spectrometry experiments, analyzing the production of 18O-containing-dioxygen by the H218O-solvated OEC, have made important contributions to the question of substrate water-binding at the OEC (Hendry & Wydrzynski 2002; Hendry & Wydrzynski 2003; Hillier et al. 1998; Hillier & Wydrzynski 2000; Hillier & Wydrzynski 2001; Hillier & Wydrzynski 2004; Messinger et al. 1995). These observation have been recently addressed by calculations of transition state energy barriers for water exchange in structural models of the OEC in the S1 and S2 states while progressively detaching substrate water molecules from Ca2+ and the dangling Mn(4) (Sproviero et al. 2007f). The resulting structural rearrangements provided insight on the water exchange mechanisms and the relative binding strengths, considering that elongation of the metal—oxygen bond is likely the primary step in water exchange and rate-determining (Lundberg et al. 2003; Rotzinger 1997; Rotzinger 2005). These calculations complemented earlier studies of water exchange in transition metal complexes (Cady et al. 2006; Helm & Merbach 1999; Houston et al. 2006; Rotzinger 1997; Rotzinger 2005; Tagore et al. 2006; Tagore et al. 2007), including theoretical studies of manganese complexes, based on Hartree—Fock and complete active-space self-consistent field theories (Lundberg et al. 2003; Rotzinger 1997; Rotzinger 2005; Tsutsui et al. 1999) as well as DFT studies of water exchange in other transition metal complexes (Deeth & Elding 1996; Hartmann et al. 1999; Hartmann et al. 1997; Lundberg et al. 2003; Vallet et al. 2001).
It was found that the QM/MM models are consistent with two substrate waters in different environments within the OEC and so exchanging at different rates, with one of the substrate waters bound to calcium (Hendry & Wydrzynski 2003). Stretching the Ca2+—Wslow bond was energetically more demanding than stretching the Mn(4)—Wfast bond due to charge delocalization in the oxomanganese complex, where charge transfer between manganese ions and ligand/oxo-bridges can affect the net ionic charges of metal centers. Charge delocalization partially neutralizes the net ionic charges of the Mn centers, leaving a smaller positive charge on Mn(4) (q=+1.35) than on Ca2+ (q=+1.77). These results are thus consistent with Wslow attached to Ca2+, even when such a metal center has a smaller oxidation number than Mn(4) (Hendry & Wydrzynski 2003). The underlying charge delocalization is also common to synthetic oxomanganese complexes (Sproviero et al. 2006a) and complicates the correlation between partial ionic charges and formal oxidation numbers (Sproviero et al. 2006a; Sproviero et al. 2006b; Sproviero et al. 2007c).
In agreement with experiments (Hillier & Wydrzynski 2004), the QM/MM models also predict that the exchange rate of the slowly exchangeable water molecule increases upon S1→S2 oxidation (Sproviero et al. 2007f). Important mechanistic evidence for this observation also comes from metal-substitution studies at the calcium binding site (Vrettos et al. 2001b). QM/MM calculations indicated that charge transfer interactions between Ca2+ and D1-A344 decreased the partial ionic charge of calcium (Δq=-0.21), weakening the Ca2+—Wslow bond. Therefore, the exchange rate of Wslow was predicted to increase upon S1→S2 oxidation of the QM/MM model, due to the corresponding changes in partial ionic charges modulated by charge transfer interactions.
Chloride
QM/MM structures included Cl- in the model since it is known to be required for transitions beyond the S2 state (Wincencjusz et al. 1997). The binding position was found by replacing each water molecule by Cl- and selecting the lowest energy configuration (McEvoy et al. 2005b; Sproviero et al. 2006b). Chloride was found to ligate loosely to the metal cluster, between calcium and YZ, in accordance with pulsed EPR data obtained from the acetate-inhibited OEC. EPR experiments indicate that acetate (known to bind competitively with chloride) (Kühne et al. 1999; Sinclair 1984) binds at 3.1 Å from YZ (Force et al. 1997), in agreement with in silico substitution of calcium-bound chloride by acetate (McEvoy et al. 2005b). Therefore, the QM/MM structures predict that chloride is not directly bound to a Mn center but rather loosely bound to the metal cluster by electrostatic interactions, at approximately 5 Å from the nearest Mn.
Protein ligands
The QM/MM models predict that the proteinaceous ligation of the OEC involves several amino-acid residues already thought to be ligands to the metal cluster on the basis of XRD (Biesiadka et al. 2004; Ferreira et al. 2004; Loll et al. 2005), site-directed mutagenesis and spectroscopic studies (Boerner et al. 1992; Chu et al. 2004; Chu et al. 1994; Chu et al. 1995; Clausen et al. 2004; Debus 2001; Debus et al. 2005; Diner 2001; Kramer et al. 1994; Nixon & Diner 1992; Nixon et al. 1992; Roffey et al. 1994; Strickler et al. 2006; Strickler et al. 2005). The proposed QM/MM coordination is similar to the proposal by Ferreira et al. (Ferreira et al. 2004), except that D1-Glu333 in the QM/MM models bridges Mn(2) and Mn(3) in a η2 mode, instead of monodentally binding Mn(4). In addition, CP43-Glu354 is protonated in the QM/MM models and binds in a monodentate fashion to Mn(3) instead of binding as a bidentate, chelating fashion as suggested by the crystal structure. These changes were found to be consistent not only with the overall electronic density maps associated with XRD models but also with EXAFS spectra and mechanistic data, as discussed in the following sections. A detailed description of metal-metal distances and oxidation states is presented in Tables 1 and 2.
Table 1.
Interionic distances and bond angles relative to the membrane normal in the DFT QM/MM structural models of the OEC of PSII in the S0, S1(a), S1 (a) with P-EXAFS optimization, S2, S3 and S4 states (Sproviero et al. 2007d), including comparisons to the 1S5L XRD model (Ferreira et al. 2004).
| S0 | S1 (a) | S1 (a) Opt. P-EXAFS |
S2 | S3 | S4 | X-Ray | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bond Vector |
Length [Å] |
Angle | Length [Å] |
Angle | Length [Å] |
Angle | Length [Å] |
Angle | Length [Å] |
Angle | Length [Å] |
Angle | Length [Å] |
Angle |
| Mn(1)-Mn(2) | 2.65 | 59° | 2.76 | 57° | 2.72 | 51° | 2.78 | 58° | 2.69 | 57° | 2.69 | 54° | 2.65 | 59° |
| Mn(1)-Mn(3) | 2.92 | 76° | 2.76 | 85° | 2.71 | 75° | 2.77 | 81° | 2.81 | 73° | 2.82 | 74° | 2.67 | 79° |
| Mn(2)-Mn(3) | 2.96 | 78° | 2.82 | 63° | 2.78 | 68° | 2.86 | 65° | 2.82 | 77° | 2.58 | 72° | 2.72 | 71° |
| Mn(2)-Mn(4) | 3.79 | 54° | 3.34 | 54° | 3.23 | 58° | 3.29 | 59° | 3.84 | 58° | 3.55 | 61° | 3.25 | 58° |
| Mn(3)-Mn(4) | 3.04 | 21° | 3.72 | 29° | 3.33 | 34° | 3.55 | 35° | 2.81 | 21° | 2.81 | 27° | 3.26 | 38° |
| Ca-Mn(2) | 3.59 | 63° | 3.31 | 53° | 3.39 | 62° | 3.78 | 57° | 3.63 | 63° | 3.61 | 71° | 3.40 | 59° |
| Ca-Mn(3) | 3.51 | 50° | 3.95 | 35° | 3.43 | 40° | 4.00 | 36° | 3.74 | 53° | 3.58 | 57° | 3.38 | 39° |
Table 2.
Mulliken spin population analysis and ESP atomic charges in the DFT QM/MM models of the OEC of PSII in the S0, S1(a), S2, S3and S4 states (Sproviero et al. 2007d)
| Ion Center |
S0 | S1 | S2 | S3 | S4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spin Pop. |
Ox. # |
ESP Charge |
Spin Pop. |
Ox. # |
ESP Charge |
Spin Pop. |
Ox. # |
ESP Charge |
Spin Pop. |
Ox. # |
ESP Charge |
Spin Pop. |
Ox. # |
ESP Charge |
|
| Mn(1) | -2.88 | +4 | +1.30 | -2.80 | +4 | +1.11 | -2.79 | +4 | +1.14 | -2.87 | +4 | +1.38 | -2.85 | +4 | +1.32 |
| Mn(2) | +3.83 | +3 | +1.20 | +2.75 | +4 | +1.08 | +2.92 | +4 | +1.02 | +3.15 | +4 | +1.16 | +3.19 | +4 | +1.72 |
| Mn(3) | -3.87 | +3 | +1.27 | -3.82 | +3 | +1.26 | -2.74 | +4 | +1.59 | -2.97 | +4 | +1.62 | -2.84 | +4 | +1.72 |
| Mn(4) | +3.80 | +3 | +1.15 | +3.80 | +3 | +1.35 | +3.79 | +3 | +1.49 | +2.98 | +4 | +1.13 | +3.10 | +4 | +0.97 |
| O(5) | +0.00 | -2 | -0.75 | +0.05 | -2 | -0.60 | +0.09 | -2 | -0.53 | +0.03 | -2 | -0.68 | +0.03 | -2 | -0.76 |
| O(6) | +0.05 | -2 | -0.92 | +0.02 | -2 | -0.80 | +0.02 | -2 | -0.81 | +0.03 | -2 | -0.84 | +0.01 | -2 | -0.99 |
| O(7) | +0.00 | -2 | -0.74 | +0.02 | -2 | -0.67 | -0.03 | -2 | -0.78 | +0.10 | -2 | -0.72 | +0.07 | -2 | -0.72 |
| O(8) | -0.03 | -2 | -0.95 | -0.07 | -2 | -0.98 | -0.09 | -2 | -0.86 | -0.04 | -2 | -1.11 | -0.05 | -2 | -1.49 |
| Ca | -0.00 | +2 | +1.60 | -0.01 | +2 | +1.77 | -0.00 | +2 | +1.56 | -0.00 | +2 | +1.65 | -0.00 | +2 | +1.66 |
| Cl | -0.04 | -1 | -0.54 | -0.00 | -1 | -0.71 | +0.00 | -1 | -0.67 | -0.00 | -1 | -0.68 | -0.27 | -1 | -0.48 |
Oxidation States
Two redox isomers were found to have comparable stability in the S1 resting state, including model (a) shown in Fig. 2 (top), where the dangling manganese Mn(4) is pentacoordinated and the Mn oxidation states are Mn(1) = IV, Mn(2) = IV, Mn(3) = III, Mn(4) = III, also referred as Mn4(IV,IV,III,III); and model (b) (not shown in Fig. 2), where the dangling manganese is hexacoordinated with an additional water and the oxidation states are Mn4(IV,III,III,IV). These results are consistent with EPR (Peloquin & Britt 2001; Peloquin et al. 2000) and X-ray spectroscopy (Dau et al. 2001; Yachandra et al. 1993) as well as recent XANES (Haumann et al. 2005b) and 55Mn ENDOR (Kulik et al. 2005a; Kulik et al. 2005b; Kulik et al. 2005c) experiments, but disagree with low-valent Mn4(III,III,III,III) proposals (Kuzek & Pace 2001; Zheng & Dismukes 1996). The two models are neutral and predict anti-ferromagnetic coupling between Mn(1) and Mn(2), between Mn(2) and Mn(3), and between Mn(3) and Mn(4), but frustrated spin-coupling between Mn(1) and Mn(3) in the cuboidal structure. Both models include complete coordination of the high-valent Mn centers, with the preferential spin state III or IV of Mn(4) determined by the number of ligands (five or six, respectively). The coordination of H332 to the Mn cluster stabilizes the oxidation state IV for Mn(2) when Mn(4) is pentacoordinated, while the oxidation state III (with a Jahn-Teller elongation along the Mn-H332 axis) when the coordination sphere of Mn(4) is complete.
EXAFS Spectra
The XRD data has yielded molecular structural models of PSII resolving nearly all cofactors and most of the amino-acid residues in the protein complex structure at 3.0–3.5 Å resolution (Ferreira et al. 2004; Loll et al. 2005). The precise positions of the Mn ions, substrate water molecules, and proteinaceous ligands, however, remain uncertain since the coordinate error in the density maps is usually as high as 1 Å (Kamiya & Shen 2003) and the resolution of bridging ligands is typically out of reach (Dau et al. 2003; Dau et al. 2004).
The XRD models of the OEC metal centers have, therefore, been tentatively proposed consistently with the overall electronic density maps and the Mn-Mn distances determined by XAS studies (Dau et al. 2001; Robblee et al. 2001). In fact, the best structural data on the OEC comes from high-resolution EXAFS spectroscopy (Dau et al. 2001; Dau et al. 2003; Dau et al. 2004; Haumann et al. 2005b; Yachandra et al. 1987; Yano et al. 2006; Yano et al. 2005b). Therefore, the QM/MM models have been carefully analyzed as compared to both XRD models and the most recent high-resolution EXAFS data.
The comparisons of calculated and experimental EXAFS data required first-principle simulations of the EXAFS spectra based on the QM/MM structural models by solving the multiscattering problem associated with the photoelectrons emitted by the Mn ions upon X-ray absorption. Calculations have been carried out according to the Real Space Green function approach as implemented in the program FEFF8 (version 8.2) (Ankudinov et al. 2002; Bouldin et al. 2001). The method is based on the theory of the oscillatory structure due to multiple-scattering originally proposed by Kronig (Kronig 1931; Kronig & Penney 1931) and worked out in detail by Sayers (Sayers et al. 1971), Stern (Stern 1974), Lee and Pendry (Lee & Pendry 1975), and by Ashley and Doniach (Ashley & Doniach 1975). The oscillatory part of the dipole transition matrix element, or EXAFS data, has been obtained with the module FEFF83, explicitly considering atoms within 10 Å from any metal in the OEC. The quantum mechanical interference of outgoing photoelectrons generated by X-ray absorption, with the scattered waves from atoms surrounding the Mn ions, generates oscillations of EXAFS intensities in momentum ‘k-space’. The Fourier transform of these oscillations determine the metal-metal distances, the coordination of Mn ions, and the coordination bond lengths with ligands, including oxo-bridges, water/hydroxo species and amino-acid residues directly ligated to Mn ions (see Figs. 3, 5 and 8).
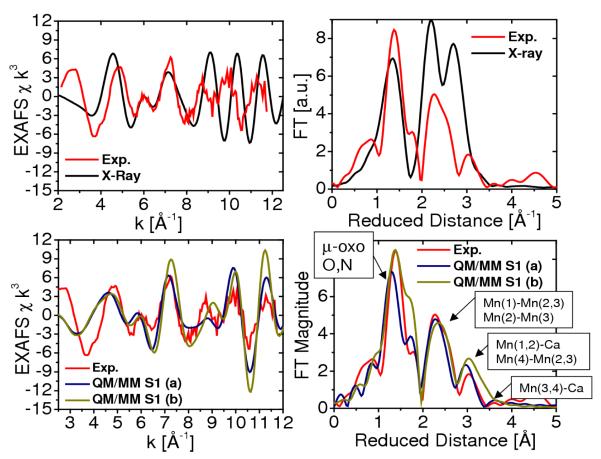
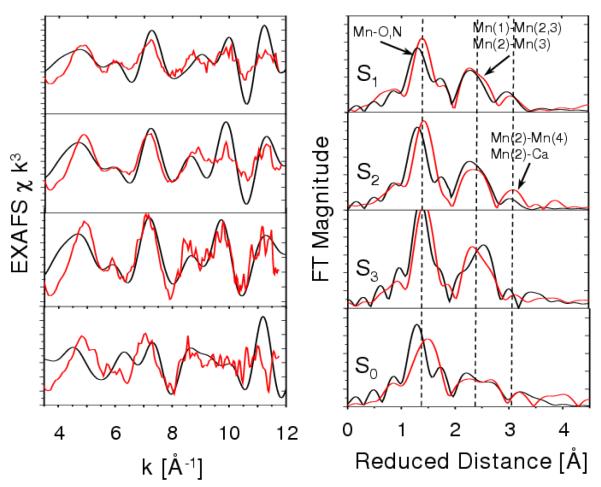
Figure 3.
Comparison between experimental (Haumann et al. 2005b, red) and calculated (Sproviero et al. 2006b) (blue,green and black) EXAFS spectra in k-space (left) and Fourier transform of the EXAFS spectra in R-space (right) for the OEC of PSII, as described by the 1S5L X-ray diffraction model (Ferreira et al. 2004, top) and the DFT QM/MM models of the S1 state, obtained at the ONIOM-EE (UHF B3LYP/lacvp,6-31G(2df),6-31G:AMBER) level (Sproviero et al. 2006b), including model (a) where the dangling manganese is pentacoordinated and the oxidation states are Mn(1) = IV, Mn(2) = IV, Mn(3) = III, Mn(4) = III; and (b) where the dangling manganese is hexacoordinated with an additional water and the oxidation states are Mn(1)=IV, Mn(2)=III, Mn(3)=III, Mn(4)=IV. Copyright 2006 American Chemical Society.
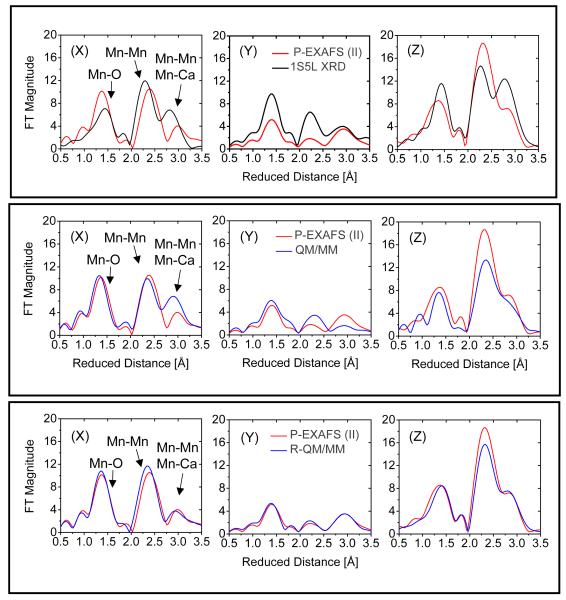
Figure 5.
Fourier transforms of polarized Mn EXAFS spectra from single crystals of PSII in the S1 state as described by model IIa from (Yano et al. 2006) (red), 1S5L XRD model (Ferreira et al. 2004, black upper panel) and the QM/MM models (blue), including the unrefined QM/MM model (blue, middle panel) (Sproviero et al. 2006b) and the refined-QM/MM (blue, bottom panel) along the axis normal to the membrane Z and the two orthogonal axis in the plane of the membrane (X and Y), as defined in Fig. 3 of (Yano et al. 2006).
Figure 8.
Comparison between experimental (Sproviero et al. 2006d, red) and calculated (Haumann et al. 2005b, black) EXAFS spectra of OEC S-state intermediates of water splitting. Left: k-weighted EXAFS spectra. Right: Fourier-transformed spectra in r-space, showing three prominent peaks corresponding to scattering centers in the first (O,N), second (Mn in the core), and third (dangling Mn, Ca) coordination shells of Mn, respectively. Vertical dashed lines are included to facilitate the comparison of peaks for different S-states. Copyright 2006 American Chemical Society.
Isotropic EXAFS Spectra
Figure 3 shows the comparison of calculated and experimental EXAFS spectra in k-space (left) as well as the corresponding spectra in reduced distances (right). The experimental spectra in k-space were kindly provided by Prof. Holger Dau. The calculated spectra correspond to the two redox isomers obtained at the DFT-QM/MM level of theory and the X-ray model structure 1S5L. The comparison shows that there is very good agreement in the description of the peaks associated with multiscattering from the N and O Mn-ligand centers at 1.8 Å (reduced distances 1.41 Å), the short Mn-Mn distances at 2.7 Å (reduced distances 2.32 Å) characteristic of PSII and backscattering due to the dangling Mn and Ca2+ at >3 Å. The main difference between the two simulated EXAFS spectra is the slightly different structure of the peak at reduced distance 1.6 Å, where model (b) has a more pronounced shoulder due to the slightly shorter 2.1 Å coordination bond length between Mn(2) and the carboxylate oxygens of E333, while the corresponding bond length in model (a) is 2.2 Å.
The quantitative analysis of interatomic bond lengths and bond orientation angles relative to the membrane normal in QM/MM models has allowed for rigorous comparisons with high-resolution EXAFS spectra and X-ray diffraction models (see Table 1) (Sproviero et al. 2006b; Sproviero et al. 2007c; Sproviero et al. 2007e). The interatomic distances compatible with EXAFS measurements are: 2 × 2.7 Å Mn-Mn distances, 1 × 2.8 Å Mn-Mn distance, 1 × 3.3 Å Mn-Mn distance, and 2 × 3.4 Å Mn-Ca distances. Interatomic distances greater than 3.4 Å are difficult to measure using EXAFS spectroscopy, and in fact the most reliable measured distances are those below 3 Å. Model (a) yields the following distances: 2 × 2.76 Å Mn Mn distances, 1 × 2.82 Å Mn Mn distance, 1 × 3.34 Å Mn Mn distance, and 1 × 3.31 Å Mn Ca distance. It is, therefore, concluded that the agreement between QM/MM models and EXAFS data is very good, with the most significant discrepancy being that the two shortest Mn-Mn distances are slightly too long. In contrast, the calculated EXAFS spectrum, based on the X-ray model 1S5L structure shows much worse agreement with EXAFS data given by the 3.5 Å resolution X-ray structure, which incorporates 3 × 2.7 Å Mn Mn distances, 2 × 3.3 Å Mn Mn distances and 2 × 3.4 Å Mn Ca distances. This is mainly due to the incomplete coordination of the metal centers. Also, the second and third peaks are shifted and with higher amplitude due to the slightly different geometry of the metal cluster.
In addition to the observed agreement between calculated and experimental isotropic EXAFS spectra, the configuration of the cuboidal Mn4Ca complex with a dangler Mn in the QM/MM hybrid model shares common structural features with the X-ray diffraction model. In fact, due to the limited resolution of the electronic density maps associated with X-ray diffraction data, it is difficult to judge whether the oxomanganese cores in the QM/MM model and in the 3 X-ray diffraction structures are truly identical or whether there are any significant differences. The QM/MM model is also consistent with the original analysis of 55Mn-ENDOR measurements on the S2 state (Britt et al. 2000). The significance of these results with respect to the S1 state is determined by the fact that the S1 and S2 states have almost identical geometry, as discussed later in this paper. The 55Mn-ENDOR measurements disfavored the ‘dimer of dimers’ model over models with a trinuclear-Mn core and a fourth Mn set off from the core by a longer Mn-Mn internuclear distance, or ‘dangler’ models. Recent work on polarized EXAFS models, however, favored the ‘dimer of dimers’ models over the cuboidal Mn4Ca complex with a dangler Mn (Yano et al. 2006).
Polarized EXAFS Spectra
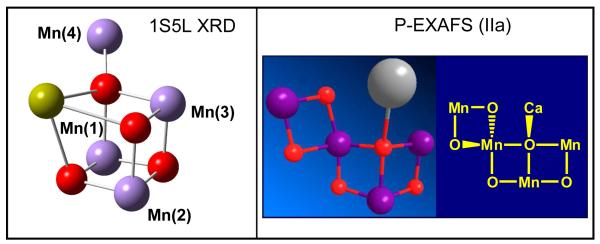
The preparation of 3-dimensionally ordered single crystals of PSII with dimensions of ∼0.3 × 0.3 × 0.9 mm has allowed the analysis of the orientational dependence of EXAFS amplitudes that provide information on the orientations of the Mn-Mn and Mn-Ca vectors. These studies reduced an original set of 11 possible empirical EXAFS models (Derose et al. 1994) of the Mn4Ca cluster, including the ‘dimer of dimers’ model extensively discussed in the past (Yachandra et al. 1996), to one model although with four possible positions for Ca2+ (i.e., models I, II, IIa and III described in (Yano et al. 2006)). None of these so-called ‘polarized-EXAFS models’ agree with the cuboidal Mn4Ca complex with a dangler Mn suggested by XRD, EPR and QM/MM studies. Figure 4 shows a comparison of the structure of the metal cluster Mn4Ca proposed by the polarized-EXAFS model IIa and the cuboidal Mn4Ca cluster with a dangler Mn model suggested by 55Mn-ENDOR measurements as well as XRD and QM/MM studies.
Figure 4.
Structure of the Mn4Ca inorganic core of the OEC of PSII suggested by the X-ray diffraction structure 1S5L (Ferreira et al. 2004) and model IIa of (Yano et. al, 2006).
The polarized-EXAFS models I, II, IIa and III of (Yano et al. 2006) are currently considered to be the most reliable empirical models of the inorganic core of PSII developed to date. However, placing any of these models into the X-ray diffraction structures results in unsatisfactory metal-ligand distances, coordination numbers and geometries (Yano et al. 2006). In addition, it is important to mention that the EXAFS spectroscopy does not provide a description of the arrangement of ligands. Furthermore, model I is less likely to be correct since the Mn-Ca vectors are not oriented along the membrane normal.
One possible explanation (favored by the Berkeley group) for the lack of consistency between the polarized-EXAFS models and the XRD model is that XRD data might be fundamentally flawed because of radiation-induced damage, although the cuboidal model of the OEC with a dangler Mn is at least partially consistent with 55Mn-ENDOR measurements as well as QM/MM studies. Another possibility is that more complete structural models might be able to reconcile the polarized-EXAFS and XRD data, as recently investigated with DFT-QM/MM models (Sproviero et al. 2007d).
Theoretical studies (Sproviero et al. 2007d) have analyzed the polarized-EXAFS spectra based on the DFT-QM/MM model of the OEC in the S1 state, as compared to the corresponding spectra based on models I, II, IIa and III of (Yano et al. 2006). These studies are particularly important for the reconciliation of XRD and EXAFS models since the DFT-QM/MM model provides isotropic EXAFS spectra in much better agreement with experimental data than the XRD models (see Figs. 3 and 5), and yet it is fully consistent with the protein ligands proposed by the 2 independently developed X-ray diffraction structures (Ferreira et al. 2004; Loll et al. 2005).
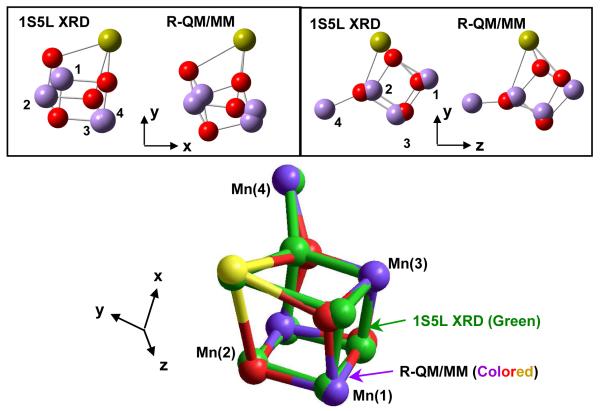
Figure 5 shows the comparison of polarized EXAFS spectra for model IIa, proposed by Yano et al. in (Yano et al. 2006) and the corresponding polarized EXAFS spectra based on the 1S5L XRD model (top) and the DFT-QM/MM model (middle) in the S1 state. These results show that the polarized-EXAFS spectra of model IIa agree much better with the polarized-EXAFS spectra of the DFT-QM/MM model than with the corresponding spectra of the 1S5L XRD model. In addition, Fig. 5 (bottom) shows that quantitative agreement with the polarized-EXAFS spectra of model IIa can be obtained by slightly refining the orientation of some metal-O vectors in the QM/MM model, without significantly affecting the metal-metal or metal-ligand distances. Figure 6 shows a superposition of the inorganic Mn4Ca core in the refined QM/MM (R-QM/MM)(Sproviero et al. 2007d) and 1S5L XRD(Ferreira et al. 2004) models. Furthermore, Table 1 shows the metal-metal and metal-ligand bond lengths and orientations, relative to the membrane normal, in the QM/MM and R-QM/MM models. These results show that the QM/MM cuboidal Mn4Ca cluster with a dangler Mn model and ligated consistently with XRD models can provide polarized EXAFS spectra that are fully consistent with experimental data.
Figure 6.
Comparison of the Mn4Ca metal cluster of PSII, as described by the 1S5L XRD (Ferreira et al. 2004) and R-QM/MM (Sproviero et al. 2007c) models of the OEC.
FTIR Spectra
Some of the strongest evidence against the proteinaceous ligation scheme suggested by the QM/MM model comes from recent papers published by Debus and co-workers, examining the vibrational properties of amino-acid residues in close contact with the OEC by using FTIR spectroscopy (Strickler et al. 2006; Strickler et al. 2005). These FTIR studies indicated that the vibrational frequencies of carboxylate groups associated with amino-acid residues D1-D170, D1-D342 and D1-E189 are not shifted as the OEC is oxidized from the S0 to the S3 states (Strickler et al. 2006; Strickler et al. 2005). The experimental evidence is given by the S-state difference FTIR spectra (i.e. Sn+1 - Sn spectra) obtained both from the wild-type PSII and from a site-directed mutant in which the amino-acid residue under scrutiny has been replaced by a different residue. If significant differences were observed between the wild-type and mutant spectra then the mutated residue would exhibit vibrational frequencies which are S-state-dependent as expected for a ligand of a redox-active ion. However, no significant differences were observed in S-state difference spectra obtained from wild-type PSII and those from PSII with the mutated residues. The simplest possible interpretation has been that none of these amino-acid residues are ligated to the Mn ions oxidized during the S0 —S3 transitions, including D1-Asp342 (Strickler et al. 2007), D1-Glu189 (Strickler et al. 2006) and D1-Asp170 (Debus et al. 2005), in marked disagreement with QM/MM and X-ray diffraction models. Furthermore, the only residue which is identified by FTIR spectroscopy as ligating a redox-active Mn ion (and definitely not Ca) is the D1 carboxyl terminus, D1-Ala344 (Chu et al. 2004; Strickler et al. 2005), which in the QM/MM model provides the only protein ligand of calcium. The sensitivity of D1-Ala344 to the S1→S2 and S3→S0 transitions has also been documented with FTIR spectroscopy by Ono and co-workers (Kimura et al. 2005b). The interpretation of these results has been that D1-A344 is ligated to the Mn ion oxidized during the S1-S2 transition (Chu et al. 2004), in disagreement with QM/MM and X-ray diffraction models. Unfortunately, more rigorous interpretations of these experiments have been hindered by the lack of systematic studies on the influence that the oxidation of Mn centers has on the vibrational frequencies of carboxylate ligands in high-valent oxomanganese complexes. These contradictions between the FTIR spectroscopy and QM/MM descriptions the proteinaceous ligation scheme are also latent in the XRD data of (Ferreira et al. 2004) and (Loll et al. 2005), identifying D1-Asp342, D1-Glu189 and D1-Asp170 as manganese ligands, and D1-Ala344 as a possible ligand to calcium.
Several possible scenarios have been considered in order to account for the widespread discrepancies between the FTIR data and the XRD and QM/MM models. One possible explanation (favored by Debus and coworkers) is that the XRD data are flawed because of radiation damage resulting in the gross derangement of nearby amino-acids. Another possibility is that it is hard to decisively assign small differences between FTIR S-state difference spectra, produced by the mutation of only one residue, to either direct ligation effects or to indirect structural perturbations. This difficulty has been recently highlighted by the markedly different interpretations of similar FTIR data by two groups investigating the structural role of D1-Glu189 (Kimura et al. 2005a; Strickler et al. 2006). Another possibility (favored by QM/MM studies) is that the apparent disagreement between FTIR and X-ray diffraction models is due to the intrinsic difficulties associated with the interpretation of the FTIR frequency shifts as resulting from the electronic and structural rearrangements in the complex biomolecular environment, including changes in hydrogen bonding, protonation states of the ligands, and formation of oxo-bridges (Sproviero et al. 2007b).
QM/MM studies have suggested that the vibrational modes of carboxylate ligands to manganese ions might be insensitive to changes in the formal oxidation states of the ions, because of electron delocalization within the cluster (Sproviero et al. 2007b). At the same time, the smearing out of electrostatic charge might induce vibrational frequency shifts of carboxylate groups not directly ligated to redox-active manganese ions due to charge rearrangments associated with S-state transitions. In support of this argument, recent calculations have shown that the observed shift in carboxylate vibrational frequency of the terminal D1-Ala344 residue upon S1→S2 transition is compatible with ligation of D1-Ala344 to calcium, rather than to the manganese ion which undergoes a change in its formal oxidation state (Sproviero et al. 2007b). It has been been recently suggested, however, that charge delocalization is unlikely to account for all of the numerous changes in FTIR spectra which are observed to accompany S-state transitions (Strickler et al. 2007).
Preliminary computational studies of oxomangenese complexes have also addressed the effect of oxidation state transitions on the vibrational frequencies of carboxylate ligands directly attached to redox active Mn ions (Sproviero et al. 2007a). These studies predict that the change of the frequency of the carboxylate asymmetric stretch is significant upon MnIII →MnIV oxidation only when the carboxylate group is coordinated along the Jahn-Teller axis of a MnIII. Otherwise, vibrations of carboxylate ligands can often be quite insensitive to Mn oxidation. It is important to note, however, that carboxylate groups are usually strong ligands and, therefore, seldom ligate along the elongated Jahn-Teller axis of MnIII. In fact, the QM/MM models suggest that neither D1-D170, D1-D342, nor D1-E189 ligate along the Jahn-Teller axis of a Mn center when the OEC is in the S0—S3 states. Therefore, these results are consistent with negligible changes in the vibrational frequencies of carboxylate ligands even when they are directly ligated to Mn centers that undergo oxidation state transitions as suggested by the DFT-QM/MM models. The only amino-acid residue ligated along the Jahn-Teller axis of a MnIII ion is CP43-E354 for which there should be an observable vibrational frequency shift as the OEC evolves from the S0 to the S3 state. This prediction, however, remains to be addressed by experiments.
QM/MM Mechanistic Model of Water Splitting
QM/MM computational modeling work has been focused mainly on the development of structures of the OEC of PSII that are consistent with a wide range of experiments. The resulting structural models have distinct mechanistic implications (Sproviero et al. 2006b; Sproviero et al. 2007c; Sproviero et al. 2007e). Most notably, the QM/MM model of the OEC in the S1 state includes two terminal ligand water molecules that plausibly represent the two substrate water molecules responsible for oxygen evolution by catalytic water splitting, one bound to Ca and the other one bound to Mn(4) with their respective oxygen atoms 2.7 Å apart. Furthermore, the QM/MM structural models allowed for the investigation of structural changes induced by oxidation/reduction of the OEC and the effect of such electronic changes on the underlying coordination/protonation state of the ligands along the catalytic cycle of water oxidation.
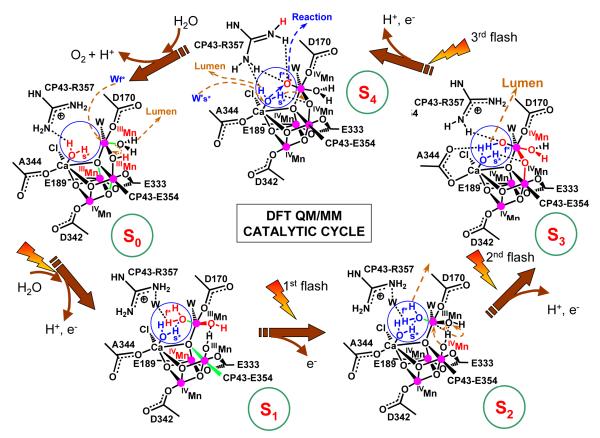
The resulting QM/MM mechanistic model is shown in Fig. 7. The quantitative analysis of structural and spin-electronic state rearrangements is presented in Tables 1 and 2, showing that Mn(2), Mn(3) and Mn(4) accumulate oxidizing equivalents while Mn(1) remains redox inactive throughout the cycle. The fourth oxidizing equivalent is accumulated as a terminal oxyl radical MnIV-O· of the dangling manganese (Siegbahn & Crabtree 1999) (an oxidized form of a substrate water molecule, deprotonated and ligated to Mn) and is essential for O—O bond formation. Dioxygen evolution, during the S4→S0 transition, involves a nucleophilic attack of the calcium-bound water molecule on the electrophilic oxyl radical MnIV —O·. This reaction is similar to earlier proposals by Pecoraro (Pecoraro et al. 1998) and Brudvig (Vrettos et al. 2001a), with the difference that in the QM/MM model the nucleophilic water attacks an oxyl radical rather than an oxo-MnV species. Another distinct aspect is that the reaction is promoted by water exchange in the coordination sphere of Ca2+ and that the nucleophilic water is activated not only by Ca2+ but also by two other basic species, including CP43-R357 and the basic μ-oxo bridge linking Mn(4) and Mn(3).
Figure 7.
Catalytic cycle of water splitting suggested by DFT QM/MM models of the OEC of PSII (Sproviero et al. 2007d). Dashed arrows in brown indicate transformations leading to the following S-state in the cycle. Changes caused by an S-state transition are highlighted in red. The blue circles highlight substrate water molecules (also shown in blue). Coordination bonds elongated by Jahn-Teller distorsions are marked in green. The oxidation states of Mn ions are indicated as Roman superscripts, while the orientation of the metal cluster corresponds to Fig. 2, where Mn(1), Mn(2), Mn(3) and Mn(4) are indicated.
The QM/MM reaction mechanism is also significantly different from other proposals where the oxidation reaction involves species near the cluster (Roelofs et al. 1996; Siegbahn 2002), or a manganese-bridging oxo group (Robblee et al. 2001). The overall reaction also disagrees with other proposals where manganese-bridging oxo-ligands react with one another during the O—O bond forming step (Brudvig & Crabtree 1986; Yachandra et al. 1996), where oxyl radicals react with μ-oxo bridges instead of reacting with terminal Ca2+-bound water molecules (Hillier & Messinger 2005; Messinger 2004; Siegbahn & Lundberg 2005), or where basic μ-oxo ligands deprotonate manganese-bound terminal water molecules (Dau et al. 2001). Importantly, a study of μ-oxide ligand exchange rates in high-valent manganese complexes disfavors binding of either substrate water as a μ-oxide bridge in the OEC, at least in the S2 and S3 states (Tagore et al. 2006).
The QM/MM mechanism is consistent with significant structural rearrangements in the metal cluster during the S0→S1 and S2→S3 transitions, as evidenced by the EXAFS spectroscopy (Robblee et al. 2002). During the S0→S1 transition, the protonated μ-oxo bridge between Mn(4) and Mn(3) is opened due to changes in oxidation and protonation states that stretch the Mn(4)-O distance, leaving the HO- group ligated to Mn(3). Strengthening the coordination between Mn(3) and its axial ligand changes the Jahn-Teller distortion (elongated coordination bond) from axial to equatorial, shortening the Mn(1)-Mn(3) distance. As shown in Table 1, these results are consistent with the EXAFS observation of a shortening of a Mn-Mn distance by approximately 0.15 Å (Robblee et al. 2002), with a single short (2.7 Å) Mn-Mn distance per Mn tetramer in the S0 state and two short (2.72 Å) Mn-Mn distances in the S1 state, with a third distance close to 2.8 Å. The interpretation of EXAFS data, however, has been based on deprotonation of one OH-bridge (Haumann et al. 2005b; Robblee et al. 2002) rather than on opening of a protonated μ-oxo bridge between Mn(4) and Mn(3).
During the S2→S3 transition, the substrate water molecule bound to Mn(4) is deprotonated, consistently with electrochromism data (Haumann & Junge 1996; Junge et al. 2002; Lavergne & Junge 1993; Schlodder & Witt 1999; Witt 1996), and the oxidation state of Mn(4) is advanced from III to IV. These changes induce inter-ligand proton transfer from the HO- ligand of Mn(3) to the hydrogen bonded HO- ligand of Mn(4), strengthening the attractive interactions between Mn(4) and the HO- ligand of Mn(3), forming a μ-oxo bridge between Mn(3) and Mn(4), and transforming the HO- ligand of Mn(4) into a water ligand (the HO- proton acceptor is regenerated by deprotonation of the water ligand in the S0→S1 transition). These rearrangements are consistent with a pH-dependent rate constant (Haumann et al. 1997), suggesting a transition ’kinetically steered’ by proton movements. The rearrangements are also consistent with the observation of significantly conformational changes in the structure of the cluster upon formation of the S3 state (Haumann et al. 2005b; Liang et al. 2000), as manifested in the EXAFS spectra (see Fig. 8) (Sproviero et al. 2007e). The proposed formation of the μ-oxo bridge, between Mn(3) and Mn(4) by proton transfer between HO- ligands is similar to the transition proposed by Siegbahn (Siegbahn & Lundberg 2005) where, in contrast to the QM/MM model, the bridge is formed by double deprotonation of a water ligand attached to Mn(3) that releases a proton to the lumen and transfers the other proton to an HO- ligand attached to the same Mn(3). In the QM/MM model, however, the substrate water molecule attached to the dangling Mn(4) is deprotonated, analogously to other proposals (Messinger 2004).
The proposed QM/MM molecular structures of catalytic intermediates have been partially validated through the comparison of experimental (Haumann et al. 2005b) and calculated EXAFS spectra as shown in Fig. 8 (Sproviero et al. 2007e). In addition to the comparison of the spectra in k-space, Fig. 8 (right panel) shows the Fourier transform EXAFS amplitudes characterizing the detailed structural rearrangements of the metal cluster along the catalytic cycle. These include changes in Mn-Mn and Mn-Ca distances as well as changes in the Mn-ligand coordination bond lengths.
The origin of some of the deviations between calculated and experimental spectra might be due to contributions from other redox isomers of comparable energy, or a slightly different arrangements of ligands (Sproviero et al. 2006b). However, the overall comparison indicates that the DFT QM/MM models are consistent with experimental data throughout the entire catalytic cycle. The first peak at reduced distance 1.41 Å (actual distance 1.8 Å) is determined by photoelectron scattering from N and O centers directly ligated to Mn ions. Scattering contributions from μ-oxo bridges and the ligated carboxylate group of E333 coordinated to Mn are responsible for a shoulder at reduced distance 1.6 Å (actual distance 2.1 Å). The second prominent peak at reduced distance 2.32 Å (actual distance 2.7 Å) corresponds to the characteristic short Mn-Mn distances in PSII. Finally, the third peak at reduced distance 3.0 Å and beyond corresponds to backscattering from the dangling-Mn and Ca2+ at distances >3.3 Å. In agreement with experimental data (Haumann et al. 2005b), the simulated spectra of the OEC in the S1 and S2 states indicate that such a distribution of scattering amplitudes is consistent with a dangler cuboidal cluster where two Mn-Mn distances (Mn(1)—Mn(2) and Mn(1)—Mn(3)) are shorter than 2.75 Å while the third distance Mn(2)-Mn(3) is close to 2.8 Å. The width of the second prominent peak at reduced distance 2.32 Å (actual distance 2.7 Å) indicates the distribution of short Mn-Mn distances in the manganese cuboidal core. In the S0 state, however, the Mn(1)-Mn(3) distance is longer and the Mn(1)-Mn(2) shorter, splitting and broadening the second prominent peak of the FT-EXAFS spectrum into a bimodal distribution. While this spectroscopic feature is properly described by the calculated spectrum of the S0 state, the first coordination sphere of Mn is found to be more structured in the simulated spectrum than in the experimental data. These deviations suggest the presence of other redox isomers, or structural disorder.
In the S3 state, the scattering peak at reduced distance 2.32 Å becomes narrower consistently with three Mn-Mn distances that become similar to each other. While this aspect is in agreement with the EXAFS data by Dau and co-workers (Dau et al. 2003; Dau et al. 2004), it is still in disagreement with data from the Berkeley group indicating that one of the Mn-Mn distances is longer in the S3 state (Sauer & Yachandra 2004; Yano et al. 2005b).
Proton Exit Channel
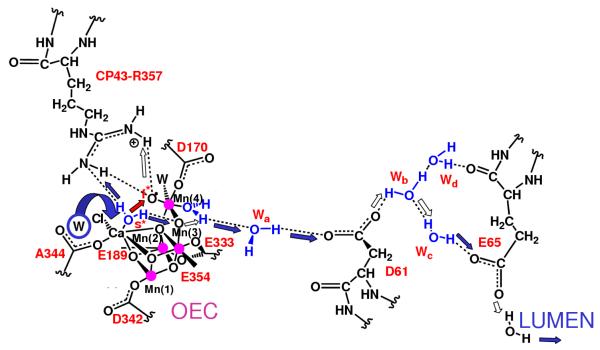
The QM/MM mechanistic model shown in Fig. 7 involves multiple changes in oxidation states. These transitions are coupled to proton transfer events balancing the overall charge of the cluster and rearranging the protonation states of water ligands. The QM/MM structural models also reveal a network of hydrogen bonds linking the proposed substrate water molecules with D1-Asp61 (see Fig. 9), the first residue of the proton exit pathway leading to the lumenal surface of PSII, suggested by XRD data (Ferreira et al. 2004). The sidechain of CP43-R357, along the hydrogen-bonding network including both substrate water molecules and the calcium-bound chloride ion, might be responsible for proton abstraction from the manganese-bound substrate water in the second half of the S-state cycle (McEvoy & Brudvig 2004; Sproviero et al. 2006b). In order to play this role satisfactorily it must not only be in close contact with this water (as it is in the QM/MM model), but its pKa must also be affected by the electrostatic charge of both the OEC and the YZ / D1-H190 pair. This would make CP43-R357 the redox-coupled base (McEvoy & Brudvig 2004), predicted to be thermodynamically necessary by Krishtalik (Krishtalik 1986), consistently with its indispensability for O2 evolution as demonstrated by mutagenesis studies of PSII from Synechocystis sp. PCC 6803 where mutation of the homologous CP43 arginine (CP43-R342) to serine suppressed O2 evolution (Knoepfle et al. 1999). In addition, arginine is a well-known component of hydrogen bonding networks in proteins (Puustinen & Wikstrom 1999; Qian et al. 2004).
Figure 9.
Proton exit channel suggested by the hydrated DFT QM/MM structural models, including a network of hydrogen bonds extended from substrate water molecules ‘s’ (slow) and ‘f’ (fast), via CP43-R357, to the first residue (D1-D61) leading to the lumenal side of the membrane (Sproviero et al. 2006d). Proton translocation events are indicated by blue and white arrows; the O=O bond formation event is indicated by a red arrow, as promoted by water exchange from Ca2+. Amino acid residues labeled with one-letter symbols correspond to the D1 protein subunit. Copyright 2006 American Chemical Society.
A recent theoretical study has shown that the pKa of CP43-R357 might indeed be particularly sensitive to an increase in the charge of the OEC (Ishikita et al. 2006). However, this remains to be examined in the QM/MM models. It is also possible that the pKa of CP43-R357 is indirectly affected by the rearrangements of protonation states and hydrogen bonds of the ligands along the catalytic cycle. In particular, the OH- ligand of Mn(3) experiences significant changes in protonation states along the cycle. It deprotonates by proton transfer to an OH- ligand of Mn(4), during the S2→S3 transition, transforming such a ligand into water and forming a μ-oxo bridge between Mn(3) and Mn(4). This event induces a rearrangement of hydrogen bonds: the hydrogen-bond between the two substrate water molecules is broken and two hydrogen bonds are formed, including a bond between Wslow and the μ-oxo bridge and another one between Wfast and CP43-R357. Upon further deprotonation of Wfast in the S3→→S4 state, Wslow forms a hydrogen bond with CP43-R357 in addition to its hydrogen bond with the μ-oxo bridge, establishing two deprotonation pathways. During the S4→S0 transition, one proton transfers to the lumen via CP43-R357 while the other one translocates to the μ-oxo bridge and subsequently forms the OH- ligand of Mn(3), upon opening of the bridge in the S0→S1 transition.
The QM/MM model is also consistent with the hypothesis that the YZ / D1-H190 pair might be responsible for electron transport, electrostatically affecting the pKa of CP43-R357 (McEvoy & Brudvig 2004). Especially, judging by the proximity of YZ to the Mn4Ca cluster. Simple inspection of the QM/MM structural models also indicates that the phenoxy oxygen of YZ is typically close to the chloride ligand (3.4 Å apart) and Cl- is positioned at 3.14 Å from Ca2+. Furthermore, the YZ phenol group is hydrogen bonded to the imidazole ⧠-N of the H190 side-chain. This hydrogen-bonding partnership is in line with mutational and spectroscopic studies (Hays et al. 1998; Hays et al. 1999; Roffey et al. 1994), supporting D1-Y161 as an electron transport cofactor.
Considering the potential functional roles of CP43-R357 and D1-Y161, it is natural to expect that proton coupled electron transfer (PCET) might take place, as the catalytic cycle progresses, passing protons to the lumen via CP43-R357 and electrons to the oxidized state P680+ via the redox-active tyrosine D1-Y161 (Debus et al. 1988b; Metz et al. 1989). These two charge-transfer processes might be coupled by long-range electrostatic interactions, where the oxidized form of D1-Y161 induces deprotonation of the cluster by modulating the pKa of CP43-R357, and the deprotonated form of the cluster induces oxidation by electron transfer to the oxidized form of D1-Y161. Such a PCET mechanism would be consistent with a recent study of the energetics of the proposed proton exit pathway (Ishikita et al. 2006), but would disagrees with earlier proposals where YZ was thought to abstract hydrogen atoms from the OEC cluster in every one of the S-state transitions (Hoganson et al. 1995), or simultaneously oxidize and deprotonate the hydrated OEC (Hoganson & Babcock 1997). Finally, we note that the proposed charge transfer mechanism in the presence of Cl- and Ca2+ ions is consistent with experimental evidence indicating that transitions beyond S2 are blocked by the absence of calcium, or chloride, or by the presence of acetate (Kühne et al. 1999; Szalai & Brudvig 1996; Wincencjusz et al. 1997).
Water Channels
Figure 10 shows that the DFT-QM/MM structural models include extended hydrogen bonds of water molecules approaching the Mn4Ca cluster from the lumen along two distinct pathways, none of which correspond to the postulated proton exit channel discussed in the previous section (Sproviero et al. 2006b; Sproviero et al. 2007e). One of these pathways leads to the substrate water molecule ligated to Ca2+ while the other channel leads to the substrate water molecule ligated to the dangling Mn. Water molecules are interconnected by hydrogen bonds in these channels but they make very few hydrogen bonds with the surrounding amino-acid residues, establishing on average only 2–3 hydrogen bonds per molecule. This limited structure of hydrogen bonds establishes optimum conditions for water mobility, minimizing the number of competitive hydrogen bonding interactions with the surrounding amino-acid residues, and enhancing the attachment of water molecules to the metal cluster. Such an efficient mechanism for water supply is essential to achieve typical turnovers of up to 100 water molecules attaching to the Mn4Ca per second. Upon dioxygen formation, the substrate water molecules attached to the cluster react and the next water molecules along the two channels are attached to the corresponding metal centers. The QM/MM mechanistic model indicates that one substrate water molecule ligates to Ca2+ in the S4→S0 transition, promoting dioxygen formation, while the other water molecule ligates to the dangling Mn, during the S0→S1 transition, opening a μ-oxo bridge between Mn(4) and Mn(3). Both of these events produce dramatic conformational changes in the cluster, as indicated by EXAFS spectroscopic measurements (Haumann et al. 2005b).
Figure 10.
Molecular structure of the photosystem II dimer proposed by the 1S5L X-ray diffraction model from Thermosynechococcus elongatus (Ferreira et al. 2004) and inset QM/MM description of the oxygen-evolving complex (OEC) (Sproviero et al. 2006d), including substrate water pathways from the lumen. Purple: Mn, Red: O; Yellow: Ca2+; and Green: Cl-. Copyright 2006 American Chemical Society.
Conclusions
After decades of biochemical and spectroscopic work, both the structure and mechanism of the OEC in PSII has eluded precise description. However, in recent years the structure of the Mn4Ca cluster has begun to be revealed by a variety of methods, most importantly biochemical and spectroscopic, including X-ray crystallography, EXAFS, EPR, MS and FTIR spectroscopic methods.
Computational studies, including DFT in conjunction with QM/MM modeling techniques, have used the models obtained by experimental studies as the starting point for development of detailed structural models and for the elucidation of mechanistic implications. It has been critical for the success of these approaches to assess the limitations of current electronic structure methodologies, as applied to the description of complex transition metal systems, and to evaluate the resulting computational models by direct comparisons with a wide range of readily available experimental data.
DFT-QM/MM models of the OEC of PSII in the various S-state intermediates, along the PSII catalytic cycle of water splitting, have been developed varying in oxidation and protonation states and the ligation of the Mn4Ca cluster by amino-acid residues, water, hydroxide and chloride. These complete and chemically sensible structural models are consistent with available mechanistic data, with XRD studies and with high-resolution EXAFS measurements and, therefore, provide fundamental insights into the mechanistic model of catalytic water splitting. As a result, there has been a productive integration of experimental and theoretical studies towards the elucidation of the catalytic mechanism of oxygen evolution in PSII.
Acknowledgments
V.S.B. acknowledges supercomputer time from the National Energy Research Scientific Computing (NERSC) center and financial support from Research Corporation, Research Innovation Award # RI0702, a Petroleum Research Fund Award from the American Chemical Society PRF # 37789-G6, a junior faculty award from the F. Warren Hellman Family, the National Science Foundation (NSF) Career Program Award CHE # 0345984, the NSF Nanoscale Exploratory Research (NER) Award ECS # 0404191, the Alfred P. Sloan Fellowship (2005-2006), a Camille Dreyfus Teacher-Scholar Award for 2005, and a Yale Junior Faculty Fellowship in the Natural Sciences (2005-2006). G.W.B acknowledges support from the National Institutes of Health grant GM32715. J.A.G acknowledges support from the Pittsburgh Supercomputer Center, teragrid project TG-CHEM060028T, and the Camille & Henry Dreyfus New Faculty Award for 2006.
References
- Allahverdiyeva Y, Deak Z, Szilard A, Diner BA, Nixon PJ, Vass I. The function of D1-H332 in Photosystem II electron transport studied by thermoluminescence and chlorophyll fluorescence in site-directed mutants of Synechocystis 6803. Eur. J. Biochem. 2004;271:3523–3532. doi: 10.1111/j.0014-2956.2004.04287.x. [DOI] [PubMed] [Google Scholar]
- Ankudinov AL, Bouldin CE, Rehr JJ, Sims J, Hung H. Parallel calculation of electron multiple scattering using Lanczos algorithms. Phys. Rev. B. 2002;65 [Google Scholar]
- Ashley CA, Doniach S. Theory of Extended X-Ray Absorption-Edge Fine-Structure (Exafs) in Crystalline Solids. Phys. Rev. B. 1975;11:1279–1288. [Google Scholar]
- Babcock GT, Barry BA, Debus RJ, Hoganson CW, Atamian M, McIntosh L, Sithole I, Yocum CF. Water Oxidation in Photosystem .2. From Radical Chemistry to Multielectron Chemistry. Biochemistry. 1989;28:9557–9565. doi: 10.1021/bi00451a001. [DOI] [PubMed] [Google Scholar]
- Barber J. Photosystem II: the engine of life. Quart. Rev. Biophys. 2003;36:71–89. doi: 10.1017/s0033583502003839. [DOI] [PubMed] [Google Scholar]
- Bergmann U, Grush MM, Horne CR, DeMarois P, Penner-Hahn JE, Yocum CF, Wright DW, Dube CE, Armstrong WH, Christou G, Eppley HJ, Cramer SP. Characterization of the Mn oxidation states in photosystem II by K beta X-ray fluorescence spectroscopy. J. Phys. Chem. B. 1998;102:8350–8352. [Google Scholar]
- Berthomieu C, Hienerwadel R, Boussac A, Breton J, Diner BA. Hydrogen bonding of redox-active tyrosine Z of photosystem II probed by FTIR difference spectroscopy. Biochemistry. 1998;37:10547–10554. doi: 10.1021/bi980788m. [DOI] [PubMed] [Google Scholar]
- Biesiadka J, Loll B, Kern J, Irrgang KD, Zouni A. Crystal structure of cyanobacterial photosystem II at 3.2 angstrom resolution: a closer look at the Mn-cluster. Phys. Chem. Chem. Phys. 2004;6:4733–4736. [Google Scholar]
- Blomberg MRA, Siegbahn PEM, Styring S, Babcock GT, Akermark B, Korall P. A quantum chemical study of hydrogen abstraction from manganese-coordinated water by a tyrosyl radical: A model for water oxidation in photosystem II. J. Am. Chem. Soc. 1997;119:8285–8292. [Google Scholar]
- Boerner RJ, Nguyen AP, Barry BA, Debus RJ. Evidence from Directed Mutagenesis That Aspartate-170 of the D1 Polypeptide Influences the Assembly and or Stability of the Manganese Cluster in the Photosynthetic Water-Splitting Complex. Biochemistry. 1992;31:6660–6672. doi: 10.1021/bi00144a005. [DOI] [PubMed] [Google Scholar]
- Bouldin C, Sims J, Hung H, Rehr JJ, Ankudinov AL. Rapid calculation of x-ray absorption near edge structure using parallel computation. X-Ray Spectr. 2001;30:431–434. [Google Scholar]
- Boussac A, Un S, Horner O, Rutherford AW. High-spin states (S >= 5/2) of the photosystem II manganese complex. Biochemistry. 1998;37:4001–4007. doi: 10.1021/bi9728710. [DOI] [PubMed] [Google Scholar]
- Boussac A, Zimmermann JL, Rutherford AW. Epr Signals From Modified Charge Accumulation States of the Oxygen Evolving Enzyme in Ca2+-Deficient Photosystem-II. Biochemistry. 1989;28:8984–8989. doi: 10.1021/bi00449a005. [DOI] [PubMed] [Google Scholar]
- Britt RD, Campbell KA, Peloquin JM, Gilchrist ML, Aznar CP, Dicus MM, Robblee J, Messinger J. Recent pulsed EPR studies of the photosystem II oxygen-evolving complex: implications as to water oxidation mechanisms. Biochim. Biophys. Acta. 2004;1655:158–171. doi: 10.1016/j.bbabio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Britt RD, Peloquin JM, Campbell KA. Pulsed and parallel-polarization EPR characterization of the photosystem II oxygen-evolving complex. Ann. Rev. Biophys. Biomol. Struct. 2000;29:463–495. doi: 10.1146/annurev.biophys.29.1.463. [DOI] [PubMed] [Google Scholar]
- Britt RD, Tang XS, Gilchrist ML, Lorigan GA, Larsen BS, Diner BA. Histidine at the Catalytic Site of the Photosynthetic Oxygen-Evolving Complex. Biochem. Soc. Trans. 1994;22:343–347. doi: 10.1042/bst0220343. [DOI] [PubMed] [Google Scholar]
- Brudvig GW, Crabtree RH. Mechanism For Photosynthetic O2 Evolution. Proc. Natl. Acad. Sci. U.S.A. 1986;83:4586–4588. doi: 10.1073/pnas.83.13.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady CW, Incarvito C, Brudvig GW, Crabtree RH. Secondary bonding in a six-coordinate Mn(II) complex as a model of associative substitution. Inorg. Chim. Acta. 2006;359:2509–2512. [Google Scholar]
- Chu H, Hillier W, Debus RJ. Evidence that the C-terminus of the D1 polypeptide of photosystem II is ligated to the manganese ion that undergoes oxidation during the S1 to S2 transition: an isotope-edited FTIR study. Biochemistry. 2004;43:3152–3166. doi: 10.1021/bi035915f. [DOI] [PubMed] [Google Scholar]
- Chu HA, Debus RJ, Babcock GT. D1-Asp170 is structurally coupled to the oxygen evolving complex in photosystem II as revealed by light-induced Fourier transform infrared difference spectroscopy. Biochemistry. 2001;40:2312–2316. doi: 10.1021/bi0022994. [DOI] [PubMed] [Google Scholar]
- Chu HA, Nguyen AP, Debus RJ. Site-Directed Photosystem-II Mutants with Perturbed Oxygen-Evolving Properties .2. Increased Binding or Photooxidation of Manganese in the Absence of the Extrinsic 33-Kda Polypeptide in-Vivo. Biochemistry. 1994;33:6150–6157. doi: 10.1021/bi00186a014. [DOI] [PubMed] [Google Scholar]
- Chu HA, Nguyen AP, Debus RJ. Amino-Acid-Residues That Influence the Binding of Manganese or Calcium to Photosystem-II .2. The Carboxy-Terminal Domain of the D1 Polypeptide. Biochemistry. 1995;34:5859–5882. doi: 10.1021/bi00017a017. [DOI] [PubMed] [Google Scholar]
- Clausen J, Debus RJ, Junge W. Time-resolved oxygen production by PSII: chasing chemical intermediates. Biochim. Biophys. Acta. 2004;1655:184–194. doi: 10.1016/j.bbabio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules (vol 117, pg 5179, 1995) J. Am. Chem. Soc. 1996;118:2309–2309. [Google Scholar]
- Dapprich S, Komaromi I, Byun KS, Morokuma K, Frisch MJ. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct.- Theochem. 1999;462:1–21. [Google Scholar]
- Dasgupta J, van Willigen RT, Dismukes GC. Consequences of structural and biophysical studies for the molecular mechanism of photosynthetic oxygen evolution: functional roles for calcium and bicarbonate. Phys. Chem. Chem. Phys. 2004;6:4793–4802. [Google Scholar]
- Dau H, Iuzzolino L, Dittmer J. The tetra-manganese complex of photosystem II during its redox cycle - X-ray absorption results and mechanistic implications. Biochim. Biophys. Acta. 2001;1503:24–39. doi: 10.1016/s0005-2728(00)00230-9. [DOI] [PubMed] [Google Scholar]
- Dau H, Liebisch P, Haumann M. X-ray absorption spectroscopy to analyze nuclear geometry and electronic structure of biological metal centers - potential and questions examined with special focus on the tetra-nuclear manganese complex of oxygenic photosynthesis. Anal. Bioanal. Chem. 2003;376:562–583. doi: 10.1007/s00216-003-1982-2. [DOI] [PubMed] [Google Scholar]
- Dau H, Liebisch P, Haumann M. The structure of the manganese complex of photosystem II in its dark-stable S1 state - EXAFS results in relation to recent crystallographic data. Phys. Chem. Chem. Phys. 2004;6:4781–4792. [Google Scholar]
- Debus RJ. The Manganese and Calcium-Ions of Photosynthetic Oxygen Evolution. Biochim. Biophys. Acta. 1992;1102:269–352. doi: 10.1016/0005-2728(92)90133-m. [DOI] [PubMed] [Google Scholar]
- Debus RJ. Amino acid residues that modulate the properties of tyrosine Y-Z and the manganese cluster in the water oxidizing complex of photosystem II. Biochim. Biophys. Acta. 2001;1503:164–186. doi: 10.1016/s0005-2728(00)00221-8. [DOI] [PubMed] [Google Scholar]
- Debus RJ, Aznar C, Campbell KA, Gregor W, Diner BA, Britt RD. Does aspartate 170 of the D1 polypeptide ligate the manganese cluster in photosystem II? An EPR and ESEEM study. Biochemistry. 2003;42:10600–10608. doi: 10.1021/bi034859f. [DOI] [PubMed] [Google Scholar]
- Debus RJ, Barry BA, Babcock GT, McIntosh L. Site-directed mutagenesis identifies a tyrosine radical involved in the photosynthetic oxygen-evolving system. Proc. Natl. Acad. Sci. U.S.A. 1988a;85:427–430. doi: 10.1073/pnas.85.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debus RJ, Barry BA, Sithole I, Babcock GT, McIntosh L. Directed mutagenesis indicates that the donor to P680+ in photosystem II is tyrosine-161 of the D1 polypeptide. Biochemistry. 1988b;27:9071–9074. doi: 10.1021/bi00426a001. [DOI] [PubMed] [Google Scholar]
- Debus RJ, Campbell KA, Gregor W, Li ZL, Burnap RL, Britt RD. Does histidine 332 of the D1 polypeptide ligate the manganese cluster in photosystem II? An electron spin echo envelope modulation study. Biochemistry. 2001;40:3690–3699. doi: 10.1021/bi002394c. [DOI] [PubMed] [Google Scholar]
- Debus RJ, Campbell KA, Peloquin JM, Pham DP, Britt RD. Histidine 332 of the D1 polypeptide modulates the magnetic and redox properties of the manganese cluster and tyrosine Y-Z in photosystem II. Biochemistry. 2000;39:470–478. doi: 10.1021/bi9917737. [DOI] [PubMed] [Google Scholar]
- Debus RJ, Strickler MA, Walker LM, Hillier W. No evidence from FTIR difference spectroscopy that aspartate-170 of the D1 polypeptide ligates a manganese ion that undergoes oxidation during the S0 to S1, S1 to S2, or S2 to S3 transitions in photosystem II. Biochemistry. 2005;44:1367–1374. doi: 10.1021/bi047558u. [DOI] [PubMed] [Google Scholar]
- Deeth RJ, Elding LI. Theoretical modeling of water exchange on Pd(H2O)(4) (2+), Pt(H2O)(4) (2+), and trans-PtCl2(H2O)(2) Inorg. Chem. 1996;35:5019–5026. doi: 10.1021/ic950335v. [DOI] [PubMed] [Google Scholar]
- Derose VJ, Mukerji I, Latimer MJ, Yachandra VK, Sauer K, Klein MP. Comparison of the Manganese Oxygen-Evolving Complex in Photosystem-Ii of Spinach and Synechococcus Sp with Multinuclear Manganese Model Compounds by X-Ray-Absorption Spectroscopy. J. Am. Chem. Soc. 1994;116:5239–5249. [Google Scholar]
- Diner BA. Amino acid residues involved in the coordination and assembly of the manganese cluster of photosystem II. Proton-coupled electron transport of the redox-active tyrosines and its relationship to water oxidation. Biochim. Biophys. Acta. 2001;1503:147–163. doi: 10.1016/s0005-2728(00)00220-6. [DOI] [PubMed] [Google Scholar]
- Diner BA, Babcock GT. Structure, dynamics, and energy conversion efficiency in photosystem II. In: Ort DR, Yocum CF, editors. Oxygenic photosynthesis: the light reactions. vol. 4. Kluwer Academic Publishers; Dordrecht: 1996. pp. 213–247. [Google Scholar]
- Diner BA, Bautista JA, Nixon PJ, Berthomieu C, Hienerwadel R, Britt RD, Vermaas WFJ, Chisholm DA. Coordination of proton and electron transfer from the redox-active tyrosine, Y-Z, of Photosystem II and examination of the electrostatic influence of oxidized tyrosine, Y-D(center dot)(H+) Phys. Chem. Chem. Phys. 2004;6:4844–4850. [Google Scholar]
- Diner BA, Britt RD. The Redox-Active Tyrosines YZ and YD. In: Wydrzynski TJ, Satoh K, editors. Photosystem II: The Light-Driven Water:Plastoquinone Oxidoreductase. vol. 22. Springer; Dordrecht: 2005. pp. 207–233. [Google Scholar]
- Diner BA, Force DA, Randall DW, Britt RD. Hydrogen bonding, solvent exchange, and coupled proton and electron transfer in the oxidation and reduction of redox-active tyrosine Y-z in Mn-depleted core complexes of Photosystem II. Biochemistry. 1998;37:17931–17943. doi: 10.1021/bi981894r. [DOI] [PubMed] [Google Scholar]
- Diner BA, Nixon PJ. Evidence for D1-His190 as the proton acceptor implicated in the oxidation of redox-active tyrosine YZ of PSII. In: Garab G, editor. Photosynthesis: Mechanisms and Effects. vol. 2. Kluwer Academic Publishers; Dordrecht: 1998. pp. 1177–1180. [Google Scholar]
- Diner BA, Rappaport F. Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Ann. Rev. Plant Biol. 2002;53:551–580. doi: 10.1146/annurev.arplant.53.100301.135238. [DOI] [PubMed] [Google Scholar]
- Faller P, Rutherford AW, Debus RJ. Tyrosine D oxidation at cryogenic temperature in photosystem II. Biochemistry. 2002;41:12914–12920. doi: 10.1021/bi026588z. [DOI] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- Force DA, Randall DW, Britt RD. Proximity of acetate, manganese, and exchangeable deuterons to tyrosine YZ· in acetate-inhibited photosystem II membranes: Implications for the direct involvement of YZ· in water-splitting. Biochemistry. 1997;36:12062–12070. doi: 10.1021/bi9712504. [DOI] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Jr., J. AM, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. 2003.
- Gascon JA, Leung SSF, Batista ER, Batista VS. A self-consistent space-domain decomposition method for QM/MM computations of protein electrostatic potentials. J. Chem. Theor. Comput. 2006;2:175–186. doi: 10.1021/ct050218h. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Persson BJ, Taylor PR. Ab initio multiconfiguration reference perturbation theory calculations on the energetics of low-energy spin states of iron(III) porphyrins. J. Biolog. Inorg. Chem. 2003;8:507–511. doi: 10.1007/s00775-002-0435-2. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Steene E. High-valent transition metal centers and noninnocent ligands in metalloporphyrins and related molecules: a broad overview based on quantum chemical calculations. J. Biolog. Inorg. Chem. 2001;6:739–752. doi: 10.1007/s007750100275. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Taylor PR. High-level ab initio calculations on the energetics of low-lying spin states of biologically relevant transition metal complexes: first progress report. Curr. Op. Chem. Biol. 2003;7:113–124. doi: 10.1016/s1367-5931(02)00023-6. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Taylor PR. Iron(IV) porphyrin difluoride does not exist: Implications for DFT calculations on heme protein reaction pathways. J. Chem. Theor. Comput. 2005;1:597–600. doi: 10.1021/ct050086s. [DOI] [PubMed] [Google Scholar]
- Grabolle M, Dau H. Energetics of primary and secondary electron transfer in Photosystem II membrane particles of spinach revisited on basis of recombination-fluorescence measurements. Biochim. Biophys. Acta. 2005;1708:209–218. doi: 10.1016/j.bbabio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Grabolle M, Haumann M, Muller C, Liebisch P, Dau H. Rapid loss of structural motifs in the manganese complex of oxygenic photosynthesis by X-ray irradiation at 10 - 300 K. J. Biolog. Chem. 2006;281:4580–4588. doi: 10.1074/jbc.M509724200. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Clark T, van Eldik R. Water exchange reactions and hydrolysis of hydrated titanium(III) ions. A density functional theory study. J. Phys. Chem. A. 1999;103:9899–9905. [Google Scholar]
- Hartmann M, Clark T, vanEldik R. Hydration and water exchange of zinc(II) ions. Application of density functional theory. J. Am. Chem. Soc. 1997;119:7843–7850. [Google Scholar]
- Harvey JN. Computational Organometallic Chemistry. Marcel Dekker; New York: 2001. [Google Scholar]
- Harvey JN, Poli R, Smith KM. Understanding the reactivity of transition metal complexes involving multiple spin states. Coord. Chem. Rev. 2003;238:347–361. [Google Scholar]
- Hasegawa K, Ono T, Inoue T, Kusunoki M. How to evaluate the structure of a tetranuclear Mn cluster from magnetic and EXAFS data: Case of the S2-state Mn-cluster in photosystem II. Bull. Chem. Soc. Japan. 1999;72:1013–1023. [Google Scholar]
- Haumann M, Bogershausen O, Cherepanov D, Ahlbrink R, Junge W. Photosynthetic oxygen evolution: H/D isotope effects and the coupling between electron and proton transfer during the redox reactions at the oxidizing side of Photosystem II. Photosynth. Res. 1997;51:193–208. [Google Scholar]
- Haumann M, Junge W. Protons and Charge Indicators in Oxygen Evolution. In: Ort DR, Yocum CF, editors. Oxygenic Photosynthesis: The Light Reactions. vol. 4. Kluwer Academic Publishers; Dordrecht: 1996. pp. 165–192. [Google Scholar]
- Haumann M, Junge W. Evidence for impaired hydrogen-bonding of tyrosine Y-Z in calcium-depleted Photosystem II. Biochim. Biophys. Acta. 1999;1411:121–133. doi: 10.1016/s0005-2728(99)00045-6. [DOI] [PubMed] [Google Scholar]
- Haumann M, Liebisch P, Muller C, Barra M, Grabolle M, Dau H. Photosynthetic O2 formation tracked by time-resolved X-ray experiments. Science. 2005a;310:1019–1021. doi: 10.1126/science.1117551. [DOI] [PubMed] [Google Scholar]
- Haumann M, Muller C, Liebisch P, Iuzzolino L, Dittmer J, Grabolle M, Neisius T, Meyer-Klaucke W, Dau H. Structural and oxidation state changes of the photosystem II manganese complex in four transitions of the water oxidation cycle (S0 > S1, S1 > S2, S2 > S3, and S3, S4 > S0) characterized by X-ray absorption spectroscopy at 20 K and room temperature. Biochemistry. 2005b;44:1894–1908. doi: 10.1021/bi048697e. [DOI] [PubMed] [Google Scholar]
- Hays AMA, Vassiliev IR, Golbeck JH, Debus RJ. Role of D1-His190 in proton-coupled electron transfer reactions in photosystem II: a chemical complementation study. Biochemistry. 1998;37:11352–11365. doi: 10.1021/bi980510u. [DOI] [PubMed] [Google Scholar]
- Hays AMA, Vassiliev IR, Golbeck JH, Debus RJ. Role of D1-His190 in the proton-coupled oxidation of tyrosine YZ in manganese-depleted photosystem II. Biochemistry. 1999;38:11851–11865. doi: 10.1021/bi990716a. [DOI] [PubMed] [Google Scholar]
- Helm L, Merbach AE. Water exchange on metal ions: experiments and simulations. Coord. Chem. Rev. 1999;187:151–181. [Google Scholar]
- Hendry G, Wydrzynski T. The two substrate-water molecules are already bound to the oxygen-evolving complex in the S2 state of photosystem II. Biochemistry. 2002;41:13328–13334. doi: 10.1021/bi026246t. [DOI] [PubMed] [Google Scholar]
- Hendry G, Wydrzynski T. 18O isotope exchange measurements reveal that calcium is involved in the binding of one substrate-water molecule to the oxygen-evolving complex in photosystem II. Biochemistry. 2003;42:6209–6217. doi: 10.1021/bi034279i. [DOI] [PubMed] [Google Scholar]
- Hillier W, Messinger J. Mechanism of Photosynthetic Oxygen Evolution. In: Wydrzynski TJ, Satoh K, editors. Photosystem II: The Light-Driven Water:Plastoquinone Oxidoreductase. vol. 22. Springer; Dordrecht: 2005. pp. 567–608. [Google Scholar]
- Hillier W, Messinger J, Wydrzynski T. Kinetic determination of the fast exchanging substrate water molecule in the S3 state of photosystem II. Biochemistry. 1998;37:16908–16914. doi: 10.1021/bi980756z. [DOI] [PubMed] [Google Scholar]
- Hillier W, Wydrzynski T. The affinities for the two substrate water binding sites in the O2 evolving complex of photosystem II vary independently during S-state turnover. Biochemistry. 2000;39:4399–4405. doi: 10.1021/bi992318d. [DOI] [PubMed] [Google Scholar]
- Hillier W, Wydrzynski T. Oxygen ligand exchange at metal sites - implications for the O-2 evolving mechanism of photosystem II. Biochim. Biophys. Acta. 2001;1503:197–209. doi: 10.1016/s0005-2728(00)00225-5. [DOI] [PubMed] [Google Scholar]
- Hillier W, Wydrzynski T. Substrate water interactions within the Photosystem II oxygen evolving complex. Phys. Chem. Chem. Phys. 2004;6:4882–4889. [Google Scholar]
- Hoganson CW, Babcock GT. A metalloradical mechanism for the generation of oxygen from water in photosynthesis. Science. 1997;277:1953–1956. doi: 10.1126/science.277.5334.1953. [DOI] [PubMed] [Google Scholar]
- Hoganson CW, Lydakis-Simantiris N, Tang XS, Tommos C, Warncke K, Babcock GT, Diner BA, McCracken J, Styring S. A hydrogen-atom abstraction model for the function of YZ in photosynthetic oxygen evolution. Photosyn. Res. 1995;46:177–184. doi: 10.1007/BF00020428. [DOI] [PubMed] [Google Scholar]
- Holthausen MC. Benchmarking approximate density functional theory. I. s/d excitation energies in 3d transition metal cations. J. Comput. Chem. 2005;26:1505–1518. doi: 10.1002/jcc.20279. [DOI] [PubMed] [Google Scholar]
- Houston JR, Richens DT, Casey WH. Distinct water-exchange mechanisms for trinuclear transition-metal clusters. Inorg. Chem. 2006;45:7962–7967. doi: 10.1021/ic0609608. [DOI] [PubMed] [Google Scholar]
- Ishikita H, Saenger W, Loll B, Biesiadka J, Knapp EW. Energetics of a possible proton exit pathway for water oxidation in photosystem II. Biochemistry. 2006;45:2063–2071. doi: 10.1021/bi051615h. [DOI] [PubMed] [Google Scholar]
- Iuzzolino L, Dittmer J, Dorner W, Meyer-Klaucke W, Dau H. X-ray absorption spectroscopy on layered photosystem II membrane particles suggests manganese-centered oxidation of the oxygen-evolving complex for the S0 > S1, S1 > S2, and S2 > S3 transitions of the water oxidation cycle. Biochemistry. 1998;37:17112–17119. doi: 10.1021/bi9817360. [DOI] [PubMed] [Google Scholar]
- Jaguar 5.5. Schroedinger . Portland, OR: 19912003. L. L. C. [Google Scholar]
- Joliot P, Barbieri G, Chabaud R. A New Model of Photochemical Centers in System-2. Photochem. Photobiol. 1969;10:309. [Google Scholar]
- Junge W, Haumann M, Ahlbrink R, Mulkidjanian A, Clausen J. Electrostatics and proton transfer in photosynthetic water oxidation. Philos.Trans. Royal Soc. London Ser. B-Biol. Sci. 2002;357:1407–1417. doi: 10.1098/rstb.2002.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Shen JR. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-angstrom resolution. Proc. Natl. Acad. Sci. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Mizusawa N, Ishii A, Nakazawa S, Ono T. Changes in structural and functional properties of oxygen-evolving complex induced by replacement of D1-glutamate 189 with glutamine in photosystem II. J. Biol. Chem. 2005a;280:37895–37900. doi: 10.1074/jbc.M505887200. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Mizusawa N, Yamanari T, Ishii A, Ono T. Structural changes of D1 C-terminal alpha-carboxylate during S-state cycling in photosynthetic oxygen evolution. J. Biol. Chem. 2005b;280:2078–2083. doi: 10.1074/jbc.M410627200. [DOI] [PubMed] [Google Scholar]
- Knoepfle N, Bricker TM, Putnam-Evans C. Site-directed mutagenesis of basic arginine residues 305 and 342 in the CP 43 protein of photosystem II affects oxygen-evolving activity in Synechocystis 6803. Biochemistry. 1999;38:1582–1588. doi: 10.1021/bi9821418. [DOI] [PubMed] [Google Scholar]
- Koch W, Holthausen MCA. A Chemist’s Guide to Density Functional Theory. Wiley-VCH; Weinheim: 2001. [Google Scholar]
- Kok B, Forbush B, McGloin M. Cooperation of charges in photosynthetic O2 evolution. 1. A linear four step mechanism. Photochem. Photobiol. 1970;11:457–475. doi: 10.1111/j.1751-1097.1970.tb06017.x. [DOI] [PubMed] [Google Scholar]
- Kramer DM, Roffey RA, Govindjee, Sayre RT. The a(T) Thermoluminescence band from Chlamydomonas reinhardtii and the effects of mutagenesis of histidine residues on the donor side of the photosystem II D1 polypeptide. Biochim. Biophys. Acta. 1994;1185:228–237. doi: 10.1016/0005-2728(94)90240-2. [DOI] [PubMed] [Google Scholar]
- Krishtalik LI. Energetics of multielectron reactions - photosynthetic oxygen evolution. Biochim. Biophys. Acta. 1986;849:162–171. [Google Scholar]
- Kronig RD. The quantum theory of dispersion in metallic conductors - II. Proceedings of the Royal Society of London Series a-Containing Papers of a Mathematical and Physical Character. 1931;133:255–265. [Google Scholar]
- Kronig RD, Penney WG. Quantum mechanics of electrons on crystal lattices. Proceedings of the Royal Society of London Series a-Containing Papers of a Mathematical and Physical Character. 1931;130:499–513. [Google Scholar]
- Kühne H, Szalai VA, Brudvig GW. Competitive binding of acetate and chloride in photosystem II. Biochemistry. 1999;38:6604–6613. doi: 10.1021/bi990341t. [DOI] [PubMed] [Google Scholar]
- Kulik L, Epel B, Messinger J, Lubitz W. Pulse EPR, Mn-55-ENDOR and ELDOR-detected NMR of the S-2-state of the oxygen evolving complex in Photosystem II. Photosyn. Res. 2005a;84:347–353. doi: 10.1007/s11120-005-2438-7. [DOI] [PubMed] [Google Scholar]
- Kulik LV, Epel B, Lubitz W, Messinger J. Mn-55 pulse ENDOR at 34 GHz of the S0 and S2 states of the oxygen-evolving complex in photosystem II. J. Am. Chem. Soc. 2005b;127:2392–2393. doi: 10.1021/ja043012j. [DOI] [PubMed] [Google Scholar]
- Kulik LV, Lubitz W, Messinger J. Electron Spin-Lattice Relaxation of the S0 State of the Oxygen-Evolving Complex in Photosystem II and of Dinuclear Manganese Model Complexes. Biochemistry. 2005c;44:9368–9374. doi: 10.1021/bi050411y. [DOI] [PubMed] [Google Scholar]
- Kuzek D, Pace RJ. Probing the Mn oxidation states in the OEC. Insights from spectroscopic, computational and kinetic data. Biochim. Biophys. Acta. 2001;1503:123–137. doi: 10.1016/s0005-2728(00)00218-8. [DOI] [PubMed] [Google Scholar]
- Lavergne J, Junge W. Proton Release During the Redox Cycle of the Water Oxidase. Photosyn. Res. 1993;38:279–296. doi: 10.1007/BF00046752. [DOI] [PubMed] [Google Scholar]
- Lee PA, Pendry JB. Theory of Extended X-Ray Absorption Fine-Structure. Phys. Rev. B. 1975;11:2795–2811. [Google Scholar]
- Lewis NS, Nocera DG. Powering the planet: Chemical challenges in solar energy utilization. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15729–15735. doi: 10.1073/pnas.0603395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WC, Roelofs TA, Cinco RM, Rompel A, Latimer MJ, Yu WO, Sauer K, Klein MP, Yachandra VK. Structural change of the Mn cluster during the S-2 -> S-3 state transition of the oxygen-evolving complex of photosystem II. Does it reflect the onset of water/substrate oxidation? Determination by Mn X-ray absorption spectroscopy. J. Am. Chem. Soc. 2000;122:3399–3412. doi: 10.1021/ja992501u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- Lundberg M, Blomberg MRA, Siegbahn PEM. Modeling water exchange on monomeric and dimeric Mn centers. Theor. Chem. Acc. 2003;110:130–143. [Google Scholar]
- Lundberg M, Siegbahn PEM. Theoretical investigations of structure and mechanism of the oxygen-evolving complex in PSII. Phys. Chem. Chem. Phys. 2004;6:4772–4780. [Google Scholar]
- Lundberg M, Siegbahn PEM. Agreement between experiment and hybrid DFT calculations for O-H bond dissociation enthalpies in manganese complexes. J. Comput. Chem. 2005a;26:661–667. doi: 10.1002/jcc.20206. [DOI] [PubMed] [Google Scholar]
- Lundberg M, Siegbahn PEM. Quantifying the effects of the self-interaction error in DFT: When do the delocalized states appear? J. Chem. Phys. 2005b;122 doi: 10.1063/1.1926277. [DOI] [PubMed] [Google Scholar]
- Matsukawa T, Mino H, Yoneda D, Kawamori A. Dual-mode EPR study of new signals from the S-3-state of oxygen-evolving complex in photosystem II. Biochemistry. 1999;38:4072–4077. doi: 10.1021/bi9818570. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Brudvig GW. Structure-based mechanism of photosynthetic water oxidation. Phys. Chem. Chem. Phys. 2004;6:4754–4763. [Google Scholar]
- McEvoy JP, Brudvig GW. Water-Splitting Chemistry of Photosystem II. Chem. Rev. 2006;106:4455–4483. doi: 10.1021/cr0204294. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Gascon JA, Batista VS, Brudvig GW. The mechanism of photosynthetic water splitting. Photochem. Photobiol. Sci. 2005a;4:940–949. doi: 10.1039/b506755c. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Gascon JA, Sproviero EM, Batista VS, Brudvig GW. Computational structural model of the oxygen evolving complex in photosystem II: Complete ligation by protein, water and chloride. In: Bruce D, van der Est A, editors. Photosynthesis: Fundamental Aspects to Global Perspectives. vol. 1. Allen Press; Lawrence, Kansas: 2005b. pp. 278–280. [Google Scholar]
- Messinger J. Evaluation of different mechanistic proposals for water oxidation in photosynthesis on the basis of Mn4OxCa structures for the catalytic site and spectroscopic data. Phys. Chem. Chem. Phys. 2004;6:4764–4771. [Google Scholar]
- Messinger J, Badger M, Wydrzynski T. Detection of One Slowly Exchanging Substrate Water Molecule in the S-3 State of Photosystem II. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3209–3213. doi: 10.1073/pnas.92.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinger J, Nugent JHA, Evans MCW. Detection of an EPR multiline signal for the S0 state in photosystem II. Biochemistry. 1997a;36:11055–11060. doi: 10.1021/bi9711285. [DOI] [PubMed] [Google Scholar]
- Messinger J, Robblee JH, Yu WO, Sauer K, Yachandra VK, Klein MP. The S0 state of the oxygen-evolving complex in photosystem II is paramagnetic: detection of EPR multiline signal. J. Am. Chem. Soc. 1997b;119:11349–11350. doi: 10.1021/ja972696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz JG, Nixon PJ, Rogner M, Brudvig GW, Diner BA. Directed alteration of the D1 polypeptide of photosystem II: Evidence that tyrosine-161 is the redox component, Z, connecting the oxygen-evolving complex to the primary electron donor, P680. Biochemistry. 1989;28:6960–6969. doi: 10.1021/bi00443a028. [DOI] [PubMed] [Google Scholar]
- Miller AF, Brudvig GW. A Guide to Electron-Paramagnetic Resonance Spectroscopy of Photosystem II Membranes. Biochim. Biophys. Acta. 1991;1056:1–18. doi: 10.1016/s0005-2728(05)80067-2. [DOI] [PubMed] [Google Scholar]
- Mino H, Kawamori A. EPR studies of the water oxidizing complex in the S-1 and the higher S states: the manganese cluster and Y-Z radical. Biochim. Biophys. Acta. 2001;1503:112–122. doi: 10.1016/s0005-2728(00)00229-2. [DOI] [PubMed] [Google Scholar]
- Mishra A, Yano J, Pushkar Y, Yachandra VK, Abboud KA, Christou G. Heteronuclear Mn-Ca/Sr complexes, and Ca/Sr EXAFS spectral comparisons with the oxygen-evolving complex of photosystem II. Chem. Comm. 2007:1538–1540. doi: 10.1039/b701355h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Diner BA. Aspartate-170 of the Photosystem-II Reaction Center Polypeptide-D1 Is Involved in the Assembly of the Oxygen-Evolving Manganese Cluster. Biochemistry. 1992;31:942–948. doi: 10.1021/bi00118a041. [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Trost JT, Diner BA. Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis Sp Pcc 6803 - assembly requires a free carboxyl group at C-terminal position 344. Biochemistry. 1992;31:10859–10871. doi: 10.1021/bi00159a029. [DOI] [PubMed] [Google Scholar]
- Noodleman L. Valence bond description of anti-ferromagnetic coupling in transition-metal dimers. J. Chem. Phys. 1981;74:5737–5743. [Google Scholar]
- Noodleman L, Case DA. Density functional theory of spin polarization and spin coupling in iron-sulfur clusters. Adv. Inorg. Chem. 1992;38:423–470. [Google Scholar]
- Noodleman L, Davidson ER. Ligand spin polarization and antiferromagnetic coupling in transition-metal dimers. Chem. Phys. 1986;109:131–143. [Google Scholar]
- Noodleman L, Peng CY, Case DA, Mouesca JM. Orbital interactions, electron delocalization and spin coupling in iron-sulfur clusters. Coord. Chem. Rev. 1995;144:199–244. [Google Scholar]
- Nugent JHA, Rich AM, Evans MCW. Photosynthetic water oxidation: towards a mechanism. Biochim. Biophys. Acta. 2001;1503:138–146. doi: 10.1016/s0005-2728(00)00223-1. [DOI] [PubMed] [Google Scholar]
- Nugent JHA, Turconi S, Evans MCW. EPR investigation of water oxidizing photosystem II: Detection of new EPR signals at cryogenic temperatures. Biochemistry. 1997;36:7086–7096. doi: 10.1021/bi962179p. [DOI] [PubMed] [Google Scholar]
- Pecoraro VL, Baldwin MJ, Caudle MT, Hsieh WY, Law NA. A proposal for water oxidation in photosystem II. Pure Appl. Chem. 1998;70:925–929. [Google Scholar]
- Pecoraro VL, Baldwin MJ, Gelasco A. Interaction of Manganese with Dioxygen and Its Reduced Derivatives. Chem. Rev. 1994;94:807–826. [Google Scholar]
- Peloquin JM, Britt RD. EPR/ENDOR characterization of the physical and electronic structure of the OEC Mn cluster. Biochim. Biophys. Acta. 2001;1503:96–111. doi: 10.1016/s0005-2728(00)00219-x. [DOI] [PubMed] [Google Scholar]
- Peloquin JM, Campbell KA, Britt RD. Mn-55 pulsed ENDOR demonstrates that the Photosystem II “split” EPR signal arises from a magnetically-coupled mangano-tyrosyl complex. J. Am. Chem. Soc. 1998;120:6840–6841. [Google Scholar]
- Peloquin JM, Campbell KA, Randall DW, Evanchik MA, Pecoraro VL, Armstrong WH, Britt RD. Mn-55 ENDOR of the S-2-state multiline EPR signal of photosystem II: Implications on the structure of the tetranuclear Mn cluster. J. Am. Chem. Soc. 2000;122:10926–10942. [Google Scholar]
- Petrie S, Stranger R. DFT and metal-metal bonding: A dys-functional treatment for multiply charged complexes? Inorg. Chem. 2004;43:2597–2610. doi: 10.1021/ic034525e. [DOI] [PubMed] [Google Scholar]
- Poli R, Harvey JN. Spin forbidden chemical reactions of transition metal compounds. New ideas and new computational challenges. Chem. Soc. Rev. 2003;32:1–8. doi: 10.1039/b200675h. [DOI] [PubMed] [Google Scholar]
- Poluektov OG, Paschenko SV, Utschig LM, Lakshmi KV, Thurnauer MC. Bidirectional electron transfer in photosystem I: Direct evidence from high-frequency time-resolved EPR spectroscopy. J. Am. Chem. Soc. 2005;127:11910–11911. doi: 10.1021/ja053315t. [DOI] [PubMed] [Google Scholar]
- Puustinen A, Wikstrom M. Proton exit from the heme-copper oxidase of Escherichia coli. Proc. Natl. Acad. Sci. 1999;96:35–37. doi: 10.1073/pnas.96.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Mills DA, Geren L, Wang K, Hoganson CW, Schmidt B, Hiser C, Babcock GT, Durham B, Millett F, Ferguson-Miller S. Role of the conserved arginine pair in proton and electron transfer in cytochrome c oxidase. Biochemistry. 2004;43:5748–5756. doi: 10.1021/bi036279o. [DOI] [PubMed] [Google Scholar]
- Rappaport F, Guergova-Kuras M, Nixon PJ, Diner BA, Lavergne J. Kinetics and pathways of charge recombination in photosystem II. Biochemistry. 2002;41:8518–8527. doi: 10.1021/bi025725p. [DOI] [PubMed] [Google Scholar]
- Razeghifard MR, Pace RJ. EPR kinetic studies of oxygen release in thylakoids and PSII membranes: A kinetic intermediate in the S-3 to S-0 transition. Biochemistry. 1999;38:1252–1257. doi: 10.1021/bi9811765. [DOI] [PubMed] [Google Scholar]
- Reiher M, Salomon O, Hess BA. Reparameterization of hybrid functionals based on energy differences of states of different multiplicity. Theor. Chem. Acc. 2001a;107:48–55. [Google Scholar]
- Reiher M, Salomon O, Sellmann D, Hess BA. Dinuclear diazene iron and ruthenium complexes as models for studying nitrogenase activity. Chem.-a Eur. J. 2001b;7:5195–5202. doi: 10.1002/1521-3765(20011203)7:23<5195::aid-chem5195>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Renger G. Photosynthetic water oxidation to molecular oxygen: apparatus and mechanism. Biochim. Biophys. Acta. 2001;1503:210–228. doi: 10.1016/s0005-2728(00)00227-9. [DOI] [PubMed] [Google Scholar]
- Riggs-Gelasco PJ, Mei R, Yocum CF, PennerHahn JE. Reduced derivatives of the Mn cluster in the oxygen-evolving complex of photosystem II: An EXAFS study. J. Am. Chem. Soc. 1996;118:2387–2399. [Google Scholar]
- Robblee JH, Cinco RM, Yachandra VK. X-ray spectroscopy-based structure of the Mn cluster and mechanism of photosynthetic oxygen evolution. Biochim. Biophys. Acta. 2001;1503:7–23. doi: 10.1016/s0005-2728(00)00217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robblee JH, Messinger J, Cinco RM, McFarlane KL, Fernandez C, Pizarro SA, Sauer K, Yachandra VK. The Mn cluster in the S0 state of the oxygen-evolving complex of photosystem II studied by EXAFS spectroscopy: Are there three di-mu-oxo-bridged Mn2 moieties in the tetranuclear Mn complex? J. Am. Chem. Soc. 2002;124:7459–7471. doi: 10.1021/ja011621a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs TA, Liang WC, Latimer MJ, Cinco RM, Rompel A, Andrews JC, Sauer K, Yachandra VK, Klein MP. Oxidation states of the manganese cluster during the flash-induced S-state cycle of the photosynthetic oxygen-evolving complex. Proc. Natl. Acad. Sci. U.S.A. 1996;93:3335–3340. doi: 10.1073/pnas.93.8.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffey RA, Kramer DM, Govindjee, Sayre RT. Lumenal side histidine mutations in the D1 protein of photosystem II affect donor side electron-transfer in Chlamydomonas reinhardtii. Biochim. Biophys. Acta. 1994;1185:257–270. doi: 10.1016/0005-2728(94)90240-2. [DOI] [PubMed] [Google Scholar]
- Rotzinger FP. Mechanism of water exchange for the di-and trivalent metal hexaaqua ions of the first transition series. J. Am. Chem. Soc. 1997;119:5230–5238. [Google Scholar]
- Rotzinger FP. Performance of molecular orbital methods and density functional theory in the computation of geometries and energies of metal aqua ions. J. Phys. Chem. B. 2005;109:1510–1527. doi: 10.1021/jp045407v. [DOI] [PubMed] [Google Scholar]
- Sauer K, Yachandra VK. The water-oxidation complex in photosynthesis. Biochim. Biophys. Acta. 2004;1655:140–148. doi: 10.1016/j.bbabio.2003.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Yano J, Yachandra VK. X-ray spectroscopy of the Mn4Ca cluster in the water-oxidation complex of Photosystem II. Photosyn. Res. 2005;85:73–86. doi: 10.1007/s11120-005-0638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers DE, Stern EA, Lytle FW. New Technique for Investigating Noncrystalline Structures - Fourier Analysis of Extended X-Ray - Absorption Fine Structure. Phys. Rev. Lett. 1971;27:1204-&. [Google Scholar]
- Schlodder E, Witt HT. Stoichiometry of proton release from the catalytic center in photosynthetic water oxidation - Reexamination by a glass electrode study at pH 5.5-7.2. J. Biol. Chem. 1999;274:30387–30392. doi: 10.1074/jbc.274.43.30387. [DOI] [PubMed] [Google Scholar]
- Schroder D, Shaik S, Schwarz H. Two-state reactivity as a new concept in organometallic chemistry. Acc. Chem. Res. 2000;33:139–145. doi: 10.1021/ar990028j. [DOI] [PubMed] [Google Scholar]
- Shaik S, de Visser SP, Ogliaro F, Schwarz H, Schroder D. Two-state reactivity mechanisms of hydroxylation and epoxidation by cytochrome P-450 revealed by theory. Curr. Op. Chem. Biol. 2002;6:556–567. doi: 10.1016/s1367-5931(02)00363-0. [DOI] [PubMed] [Google Scholar]
- Siegbahn PEM. Quantum chemical studies of manganese centers in biology. Curr. Op. Chem. Biol. 2002;6:227–235. doi: 10.1016/s1367-5931(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Siegbahn PEM. The performance of hybrid DFT for mechanisms involving transition metal complexes in enzymes. J. Biol. Inorg. Chem. 2006;11:695–701. doi: 10.1007/s00775-006-0137-2. [DOI] [PubMed] [Google Scholar]
- Siegbahn PEM, Crabtree RH. Manganese oxyl radical intermediates and O-O bond formation in photosynthetic oxygen evolution and a proposed role for the calcium cofactor in photosystem II. J. Am. Chem. Soc. 1999;121:117–127. [Google Scholar]
- Siegbahn PEM, Lundberg M. The mechanism for dioxygen formation in PSII studied by quantum chemical methods. Photochem. Photobiol. Sci. 2005;4:1035–1043. doi: 10.1039/b506746b. [DOI] [PubMed] [Google Scholar]
- Siegbahn PEM, Lundberg M. Hydroxide instead of bicarbonate in the structure of the oxygen evolving complex. J. Inorg. Biochem. 2006;100:1035–1040. doi: 10.1016/j.jinorgbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Sinclair J. The Influence of Anions on Oxygen Evolution by Isolated Spinach-Chloroplasts. Biochim. Biophys. Acta. 1984;764:247–252. [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Characterization of synthetic oxomanganese complexes and the inorganic core of the O2-evolving complex in photosystem II: Evaluation of the DFT/B3LYP level of theory. J. Inorg. Biochem. 2006a;100:786–800. doi: 10.1016/j.jinorgbio.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. QM/MM models of the O2-evolving complex of photosystem II. J. Chem. Theor. Comput. 2006b;2:1119–1134. doi: 10.1021/ct060018l. [DOI] [PubMed] [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Effect of oxidation state transitions on the vibrational properties of the proteinaceous ligands of the oxygen evolving catalytic center in photosystem II. Biochemistry. 2007a in prep. [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Ligation of the C-terminus of the D1-polypeptide of photosystem II to the Oxygen Evolving Complex of Photosystem II. Proceedings 14th Intl. Conference Photosynth. 2007b in press. [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Structural Models of the Oxygen-Evolving Complex of Photosystem II. Curr. Op. Struct. Biol. 2007c;17:173–180. doi: 10.1016/j.sbi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. The structure of the oxygen evolving catalytic center in photosystem II. J. Am. Chem. Soc. 2007d doi: 10.1021/ja801979n. in prep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Study of the catalytic cycle for water splitting in photosystem II. J. Am. Chem. Soc. 2007e doi: 10.1021/ja076130q. submitted. [DOI] [PubMed] [Google Scholar]
- Sproviero EM, Shinopoulos K, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. QM/MM computational studies of substrate water binding to the oxygen evolving complex of Photosystem II. Phil. Trans. Royal Soc. London Series B - Biol. Sci. 2007f doi: 10.1098/rstb.2007.2210. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler TL, Sossong TM, Goldstein JI, Ash DE, Elgren TE, Krutz DM, PennerHahn JE. EXAFS comparison of the dimanganese core structures of manganese catalase, arginase, and manganese-substituted ribonucleotide reductase and hemerythrin. Biochemistry. 1997;36:9847–9858. doi: 10.1021/bi9702795. [DOI] [PubMed] [Google Scholar]
- Stern EA. Theory of Extended X-Ray-Absorption Fine-Structure. Phys. Rev. B. 1974;10:3027–3037. [Google Scholar]
- Strickler MA, Hillier W, Debus RJ. No evidence from FTIR difference spectroscopy that glutamate-189 of the D1 polypeptide ligates a Mn ion that undergoes oxidation during the S0 to S1, S1 to S2, or S2 to S3 transitions in photosystem II. Biochemistry. 2006:8801–8811. doi: 10.1021/bi060583a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler MA, Walker LM, Hillier W, Britt RD, Debus RJ. No evidence from FTIR spectroscopy that aspartate-342 of the D1 polypeptide ligates a Mn ion that undergoes oxidation during the S0 to S1, S1 to S2, or S2 to S3 transitions in photosystem II. Biochemistry. 2007;46:3151–3160. doi: 10.1021/bi062195e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler MA, Walker LM, Hillier W, Debus RJ. Evidence from Biosynthetically Incorporated Strontium and FTIR Difference Spectroscopy that the C-Terminus of the D1 Polypeptide of Photosystem II Does Not Ligate Calcium. Biochemistry. 2005;44:8571–8577. doi: 10.1021/bi050653y. [DOI] [PubMed] [Google Scholar]
- Szalai VA, Brudvig GW. Reversible binding of nitric oxide to tyrosyl radicals in photosystem II. Nitric oxide quenches formation of the S3 EPR signal species in acetate-inhibited photosystem II. Biochemistry. 1996;35:15080–15087. doi: 10.1021/bi961117w. [DOI] [PubMed] [Google Scholar]
- Tagore R, Chen H, Crabtree RH, Brudvig GW. Determination of u-oxo exchange rates in di-u-oxo dimanganese complexes by electrospray ionization mass spectrometry. J. Am. Chem. Soc. 2006;128:9457–9465. doi: 10.1021/ja061348i. [DOI] [PubMed] [Google Scholar]
- Tagore R, Crabtree RH, Brudvig GW. Distinct mechanisms of bridging-oxo exchange in Di-mu-O dimanganese complexes with and without water-binding sites: Implications for water binding in the O-2-evolving complex of photosystem II. Inorg. Chem. 2007;46:2193–2203. doi: 10.1021/ic061968k. [DOI] [PubMed] [Google Scholar]
- Tang XS, Diner BA, Larsen BS, Gilchrist ML, Lorigan GA, Britt RD. Identification of Histidine at the Catalytic Site of the Photosynthetic Oxygen-Evolving Complex. Proc. Natl. Acad. Sci. U.S.A. 1994;91:704–708. doi: 10.1073/pnas.91.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XS, Zheng M, Chisholm DA, Dismukes GC, Diner BA. Investigation of the differences in the local protein environments surrounding tyrosine radicals Y-Z(center dot) and Y-D(center dot) in photosystem II using wild-type and the D2-Tyr160Phe mutant of Synechocystis 6803. Biochemistry. 1996;35:1475–1484. doi: 10.1021/bi951489p. [DOI] [PubMed] [Google Scholar]
- Tommos C, Tang XS, Warncke K, Hoganson CW, Styring S, McCracken J, Diner BA, Babcock GT. Spin-Density Distribution, Conformation, and Hydrogen-Bonding of the Redox-Active Tyrosine Y-Z in Photosystem-Ii from Multiple Electron Magnetic-Resonance Spectroscopies - Implications for Photosynthetic Oxygen Evolution. J. Am. Chem. Soc. 1995;117:10325–10335. [Google Scholar]
- Tsutsui Y, Wasada H, Funahashi S. Reaction mechanism of water exchange on di-and trivalent cations of the first transition series and structural stability of seven-coordinate species. J. Mol. Struct.-Theochem. 1999;462:379–390. [Google Scholar]
- Vallet V, Wahlgren U, Schimmelpfennig B, Szabo Z, Grenthe I. The mechanism for water exchange in UO2(H2O)(5) (2+) and UO2(oxalate)(2)(H2O)(2-), as studied by quantum chemical methods. J. Am. Chem. Soc. 2001;123:11999–12008. doi: 10.1021/ja015935+. [DOI] [PubMed] [Google Scholar]
- Vrettos JS, Limburg J, Brudvig GW. Mechanism of photosynthetic water oxidation: combining biophysical studies of photosystem II with inorganic model chemistry. Biochim. Biophys. Acta. 2001a;1503:229–245. doi: 10.1016/s0005-2728(00)00214-0. [DOI] [PubMed] [Google Scholar]
- Vrettos JS, Stone DA, Brudvig GW. Quantifying the ion selectivity of the Ca2+ site in photosystem II: Evidence for direct involvement of Ca2+ in O2 formation. Biochemistry. 2001b;40:7937–7945. doi: 10.1021/bi010679z. [DOI] [PubMed] [Google Scholar]
- Vreven T, Morokuma K. The ONIOM (our own N-layered integrated molecular orbital plus molecular mechanics) method for the first singlet excited (S-1) state photoisomerization path of a retinal protonated Schiff base. J. Chem. Phys. 2000;113:2969–2975. [Google Scholar]
- Whitesides GM, Crabtree GW. Don’t forget long-term fundamental research in energy. Science. 2007;315:796–798. doi: 10.1126/science.1140362. [DOI] [PubMed] [Google Scholar]
- Wincencjusz H, van Gorkom HJ, Yocum CF. The photosynthetic oxygen evolving complex requires chloride for its redox state S-2->S-3 and S-3->S-0 transitions but not for S-0->S-1 or S-1->S-2 transitions. Biochemistry. 1997;36:3663–3670. doi: 10.1021/bi9626719. [DOI] [PubMed] [Google Scholar]
- Witt HT. Primary reactions of oxygenic photosynthesis. Berichte Der Bunsen-Gesellschaft-Physical Chemistry Chemical Physics. 1996;100:1923–1942. [Google Scholar]
- Yachandra VK. Structure of the manganese complex in photosystem II: insights from X-ray spectroscopy. Phil. Trans. Royal Soc. London Ser. B-Biol. Sci. 2002;357:1347–1357. doi: 10.1098/rstb.2002.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachandra VK. The Catalytic Manganese Cluster: Organization of the Metal Ions. In: Wydrzynski TJ, Satoh K, editors. Photosystem II: The Light-Driven Water:Plastoquinone Oxidoreductase. vol. 22. Springer; Dordrecht: 2005. pp. 235–260. [Google Scholar]
- Yachandra VK, Derose VJ, Latimer MJ, Mukerji I, Sauer K, Klein MP. Where Plants Make Oxygen - a Structural Model for the Photosynthetic Oxygen-Evolving Manganese Cluster. Science. 1993;260:675–679. doi: 10.1126/science.8480177. [DOI] [PubMed] [Google Scholar]
- Yachandra VK, Guiles RD, McDermott A, Britt RD, Dexheimer SL, Sauer K, Klein MP. The State of Manganese in the Photosynthetic Apparatus .4. Structure of the Manganese Complex in Photosystem-II Studied Using Exafs Spectroscopy - the S1 State of the O2-Evolving Photosystem-II Complex from Spinach. Biochimica Et Biophysica Acta. 1986;850:324–332. [Google Scholar]
- Yachandra VK, Guiles RD, McDermott AE, Cole JL, Britt RD, Dexheimer SL, Sauer K, Klein MP. Comparison of the Structure of the Manganese Complex in the S-1 and S-2 States of the Photosynthetic O-2-Evolving Complex - an X-Ray Absorption-Spectroscopy Study. Biochemistry. 1987;26:5974–5981. doi: 10.1021/bi00393a005. [DOI] [PubMed] [Google Scholar]
- Yachandra VK, Sauer K, Klein MP. Manganese cluster in photosynthesis: Where plants oxidize water to dioxygen. Chem. Rev. 1996;96:2927–2950. doi: 10.1021/cr950052k. [DOI] [PubMed] [Google Scholar]
- Yano J, Kern J, Irrgang KD, Latimer MJ, Bergmann U, Glatzel P, Pushkar Y, Biesiadka J, Loll B, Sauer K, Messinger J, Zouni A, Yachandra VK. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. U.S.A. 2005a;102:12047–12052. doi: 10.1073/pnas.0505207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J, Kern J, Sauer K, Latimer MJ, Pushkar Y, Biesiadka J, Loll B, Saenger W, Messinger J, Zouni A, Yachandra VK. Where Water is Oxidized to Dioxygen: Structure of the Photosynthetic Mn4Ca Cluster. Science. 2006;314:821–825. doi: 10.1126/science.1128186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J, Pushkar Y, Glatzel P, Lewis A, Sauer K, Messinger J, Bergmann U, Yachandra VK. High-resolution Mn EXAFS of the oxygen-evolving complex in photosystem II: structural implications for the Mn4Ca cluster. J. Am. Chem. Soc. 2005b;127:14974–14975. doi: 10.1021/ja054873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Dismukes GC. Orbital configuration of the valence electrons, ligand field symmetry, and manganese oxidation states of the photosynthetic water oxidizing complex: Analysis of the S-2 state multiline EPR signals. Inorg. Chem. 1996;35:3307–3319. doi: 10.1021/ic9512340. [DOI] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 angstrom resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]