Abstract
A subset of essential cellular proteins requires the assistance of chaperonins (in E. coli, GroEL and GroES), double-ring complexes in which the two rings act alternately to bind, encapsulate and fold nascent or stress-denatured proteins1,2,3,4,5. This process starts by the trapping of a substrate protein on hydrophobic surfaces in the central cavity of a GroEL ring6,7,8,9,10. Then, binding of ATP and co-chaperonin GroES to that ring ejects the non-native protein from its binding sites, through forced unfolding or other major conformational changes, and encloses it in a hydrophilic chamber for folding11,12,13,14. ATP hydrolysis and subsequent ATP binding to the opposite ring trigger dissociation of the chamber and release of the substrate protein15,2. The bacteriophage T4 requires its own version of GroES, gp31, that forms a taller folding chamber, to fold the major viral capsid protein gp2316,17,18,19,20. Polypeptides are known to fold inside the chaperonin complex, but the conformation of an encapsulated protein has not previously been visualized. Here we present structures of gp23-chaperonin complexes, showing both the initial captured state and the final, close-to-native state with gp23 encapsulated in the folding chamber. Although the chamber is expanded, it is still barely large enough to contain the elongated gp23 monomer, explaining why the GroEL-GroES complex is not able to fold gp23 and showing how the chaperonin structure distorts to enclose a large, physiological substrate protein.
Chaperonin-substrate (binary) complexes were formed by rapidly mixing urea-denatured gp23 with GroEL in 2.5-fold molar excess over GroEL oligomer. Ternary complexes were generated by adding gp31 (bacteriophage T4 GroES homologue) and the ATP transition state analogue ADP•AlF3. Cryo electron microscopy (EM) data sets of 30-35,000 particles were collected of both preparations and initial 3D maps were obtained by treating each data set as a single structure with 7-fold symmetry. The resulting maps showed GroEL and GroEL-gp31 complexes with some additional densities in the binding cavities (Supplementary figure 1). As in our earlier study on malate dehydrogenase (MDH) folding, we expected the non-native substrate to form heterogeneous and asymmetric complexes with the chaperonins. Therefore we used a combination of multivariate statistical analysis (MSA) and competitive projection matching to sort the images into more homogeneous classes and determine their structures, without imposing any symmetry10,21.
The binary complexes were resolved into 5 classes (Supplementary Figure 2 and Supplementary Table 1). In agreement with our mass spectrometry results22, we observed empty GroEL (20% of the images) and GroEL with extra density in either one (40%) or both rings (40%). The classes displaying the largest fraction of the substrate density are shown in Figure 1, with the GroEL subunit domains fitted into the maps as separate rigid bodies. The GroEL rings deviate little from 7-fold symmetry (Figure 1c, d, g, h and rotational correlation analysis, not shown). This is unlike GroEL-MDH complexes in which the GroEL apical domains are bunched together on the side of the ring where MDH binds10. The large gp23 densities contact at least 5 of the 7 GroEL apical domains in the ring, and the density is located deep inside the cavity, mainly around Helix I and the underlying hydrophobic segment7 (Figure 1b, c, f, g and h). Whether substrate is bound or not, the open rings are very similar to one another and to apo GroEL.
Figure 1. Asymmetric reconstructions of GroEL with non-native gp23 in one or both rings.
a-d: Binary complex with gp23 in one ring (red density) with the crystal structures of GroEL domains fitted into the maps, shown from the side (a), as a central section (b), from the top (c) and from the bottom (d). e-h: The same views of the binary complex with gp23 in both rings (red density). The EM density maps were sharpened between 20 and 10 Å. Automated docking of the atomic coordinates of the 42 GroEL domains as rigid bodies into each complex gave excellent fits to the maps, with hinge residues of neighbouring domains in proximity to each other, except for a few regions such as the intermediate domains of some subunits. Helices H and I are shown as cyan cylinders. The C-termini of some subunits are visible and either contact the substrate (b, upper ring) or bend away from it (f, lower ring). Interaction of the flexible GroEL C-termini with substrates is consistent with earlier reports23,24. The resolution of the maps is around 11 Å at 0.5 Fourier shell correlation.
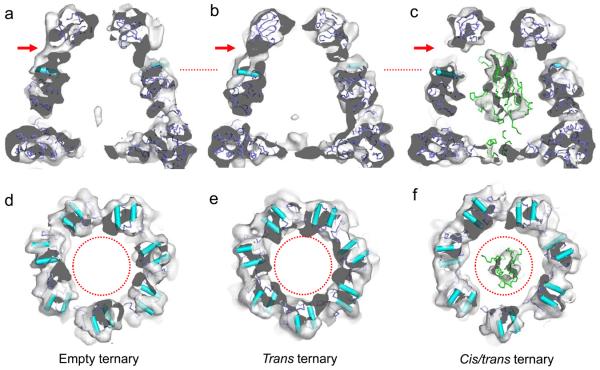
Most of the ternary complexes fell into three well-defined structural classes (Figure 2 and Supplementary Table 1): ‘empty’ (no apparent substrate density, 33% of the images), substrate bound in the trans (open) ring (32%), and substrate bound in both cis and trans rings (35%). Consistent with the presence of the ATP analogue, the trans ring of all three complexes displayed the same intra-ring β-sheet contact between neighbouring equatorial domains as in the GroEL-GroES-ATP complex25,20, although there is some variation in orientation of the apical domains. Less substrate density is resolved in the trans ring than in the binary complexes but it is found in a very similar location, i.e, deep in the cavity and interacting with most of the apical domains (Figure 2e, f, h and i). In the cis chamber, the substrate density fills most of the space, without strong contacts to either GroEL or gp31 (Figure 2h). At lower density thresholds, the cis substrate density extends to contact several regions of the gp31 lid and the GroEL apical domains (not shown). The atomic structure of gp23 is not known, but sequence alignment and genetic analysis have shown that it is closely related to the T4 vertex protein, gp24, for which the structure has been determined26. The encapsulated gp23 density is remarkably similar in shape to the major domain of gp24 (Figure 3c). Unexpectedly, we found that the cis chamber of the trans-occupied complex is slightly compressed, estimated as a 7% decrease in volume, and is significantly expanded in the cis/trans occupied complex, with a 12 % increase in volume, relative to the cis chamber in the empty complex. This can be seen by the differences in density of the mobile loops of gp31, which interact with the apical domains of GroEL (arrows, Figure 3a-c), and in the cross-sections through the cis apical domains which are almost losing contact at some positions (Figure 3d-f). The expansion suggests that the folding substrate is exerting pressure on the folding chamber. No density is visible for the small, mobile insertion domain of gp23, but there is space at the base of chamber to accommodate it, in a disordered form close to the major domain (Figure 3c). The compression of the folding chamber in the trans-bound complex reveals an allosteric effect of the trans substrate, and suggests a mechanism for newly bound substrates to prime dissociation of the folding chamber. The nature of this allosteric effect was surprising since the connections between GroEL and gp31 are stronger, rather than weaker as expected in preparation for the release of gp31.
Figure 2. Asymmetric reconstructions of GroEL-gp31 without visible substrate, with gp23 in the open ring and with gp23 in both rings.
a-c: Empty ternary complex with the crystal structures of GroEL and gp31 fitted as 49 individual domains shown from the side (a), as a central section (b) and from the bottom (c). d-f: Same views of the trans-bound ternary complex. g-i: Same views of the cis/trans bound ternary complex. Gp23 density in the trans rings is shown in red and in the cis ring in green. The resolution of the maps is around 10 Å at 0.5 Fourier shell correlation and they were therefore sharpened between 20 and 10 Å. The GroEL and gp31 domain coordinates fit very well into the density maps, with only a few minor mismatches and no significant perturbation from 7-fold symmetry.
Figure 3. Folding chambers of the GroEL-gp23-gp31 complexes.
a-c, side view sections and d-f, cross sections through the apical domains of the folding chambers (at the position of the red dotted lines in a-c). Empty complex (a and d), the trans-occupied complex (b and e) and the cis/trans occupied complex (c and f). The red arrows in a-c show the loss of density at the contact between gp31 and GroEL. Correspondingly, the dotted red circles in d-f are all the same size (45 Å in diameter) and highlight the expansion of the apical domain ring in the cis/trans complex and the contraction of the trans-occupied apical domain ring. The major domain of the gp24 structure fits very well into the cis substrate density (c, f), but not the mobile insertion domain of gp24 in the extended conformation seen in the crystal structure, where it makes an inter-subunit contact. In this position it clashes with the GroEL C-termini, but there is clearly space available for it closer to the major domain.
Extracting the gp23 densities from the maps revealed remarkably well-defined structures for both the initially captured, non-native form and the almost fully folded state in the folding chamber of the GroEL-gp31 complex (Figure 4). The cryo EM density for gp23 bound to the top rings of the different binary complexes represents over half of the native molecular volume (Figure 4a and b), showing that the non-native protein is constrained in position and shape when bound to the chaperonin complex (disordered regions are not seen in these averaged structures). The substrate densities in the second ring of doubly occupied complexes (bottom rings) are less well defined, possibly because one substrate dominates the alignment and classification, and the second substrate is not necessarily in a fixed position relative to the first. In the trans occupied ternary complex, the observation of less substrate density suggests that it is less well ordered in this class (Figure 4c). In the cis complex, the encapsulated density is clearly recognisable as a low resolution version of the major domain in the gp24 crystal structure (Figure 4d and e).
Figure 4. Substrate densities isolated from the binary and ternary complexes.
Top and side views of substrate densities isolated from the GroEL-gp23 binary complex (a), the GroEL-gp232 binary complex (b), the trans-only GroEL-gp23-gp31 ternary complex (c) and the cis ternary complex (d) compared to the low resolution filtered density of the gp24 crystal structure (e). The isolated substrate densities were low-pass filtered at 15 Å and their approximate molecular masses were determined at a density threshold of 1σ of the complete complex. A larger observed mass in the class average reflects a more homogeneous class and therefore a more consistent structure for that sub-population. The green line in (e) indicates the major domain of gp24.
A recent FRET study shows that another large, stringent GroEL substrate, Rubisco, occupies a more compact conformation in the trans ring of a ternary complex than in the open ring of a binary complex14. Our findings do not show a more compact conformation for gp23 in the trans ring of the ternary complex. However, the resolution of our study is not sufficient to discriminate subtle differences between the non-native gp23 conformations. On the other hand, our mass spectrometry data shows that Rubisco binds to GroEL with strong negative cooperativity between the rings, but that gp23 binds to both rings27,28. Therefore, these two large substrates may have distinct modes of interaction with GroEL. A diversity of GroEL-substrate interaction modes can be anticipated, depending on the folding pathways and intermediates of different substrate proteins.
It is estimated that all of the GroEL in E. coli would be required to fold the large number of gp23 subunits produced during T4 infection29. It has been proposed that gp23 monopolizes the cell's GroEL through specific N-terminal regions of gp23 that pause its translation and target the nascent chain to GroEL. Our observation of well-defined electron density for the large domain of gp23 bound to GroEL, in a relatively small number of structural classes, is consistent with the notion that there is a specific binding region on gp23 that targets it to GroEL.
In our previous study of GroEL-MDH complexes, most of the MDH density was observed in a similar region of the GroEL surface to what we report here for gp23, namely the lower part of helix I and the underlying segment10. However MDH (33 kDa) only interacts with 3 of the 7 apical domains whereas gp23 (56 kDa) contacts at least 5 of the 7 sites and occupies more of the binding cavity than MDH. The more asymmetric interaction of MDH with the cavity appears to perturb the GroEL symmetry in both rings10. The internal location of the substrate around helix I and the underlying segment leaves the helix H/I groove accessible to the mobile loop of the co-chaperonin even with a large substrate protein bound in the open cavity. However, it was reported that glutamine synthetase (51 kDa) occupies a more external binding site30. It is possible that the external binding site preferentially binds more folded states, similar to that suggested for one class of MDH complexes10.
Once gp23 is bound to GroEL, gp31 (but not GroES) can encapsulate and fold the capsid protein19. The inability of GroES to encapsulate gp23 can be inferred from the elongated shape adopted by newly folded gp23 inside the GroEL-gp31 chamber, and implies that the folding intermediate formed upon ATP and co-chaperonin binding is too bulky or too extended for GroES to bind. In this respect, it should be noted that gp31 has longer mobile loops and a larger internal space, which we show here to be stretched to the limit in order to encapsulate gp23.
This paper presents the first visualisation of a newly folded substrate protein trapped in a largely native conformation inside the folding chamber of GroEL. The well-defined gp23 density in the GroEL-gp31 cis chamber shows that this physiological substrate protein is trapped in a unique position and orientation in the chaperonin chamber. The small insertion domain, which makes inter-subunit contacts in the viral capsid26 is likely to be mobile and disordered until assembly of the capsid hexamers, which form as soon as the folded gp23 is released from GroEL-gp3122. The encapsulated substrate causes a significant expansion of the folding chamber in the region of the apical domains, and is exerting pressure on the connections between GroEL and gp31 and the inter-subunit contacts between apical domains of the cis ring. Thus, even though the folding chamber formed by GroEL-gp31 is larger than that formed by GroEL-GroES, it is still under pressure to contain the gp23 monomer. Previous studies of GroEL-GroES and GroEL-gp31 complexes in various nucleotide states did not reveal any significant changes in the conformation of the folding chamber25,20 (and Supplementary results and discussion). The expansion and compression observed here show that folding substrates directly and indirectly affect the conformation of the chamber. In conclusion, this study reveals a remarkable view of the chaperonin folding chamber strained to encapsulate a physiological substrate protein.
Methods summary
The GroEL, gp23 and gp31 were expressed and purified as previously described19. The binary GroEL-gp23 complexes were prepared as previously described28, but with an additional buffer exchange step included in order to reduce the residual urea concentration to 100 mM with 1 μM GroEL oligomer and 2.5 μM gp23 monomer. GroEL-gp23-gp31 ternary complexes were formed by adding a 2-fold molar excess of gp31 heptamer over GroEL tetradecamer to preformed binary complexes, along with ADP to give a final concentration of 2.5 mM. After incubation at 24°, KF and KAl(SO4)2 were added, giving final concentrations of 20 mM KF and 2 mM KAl(SO4)2, to form the ATP transition state analogue ADP•AlF3 to generate folding-active complexes31.
Images were recorded on a 200 kV FEG microscope on photographic film and processed at 2.8 Å/pixel, with final data sets of 30,000 and 35,000 side views of the binary and ternary complexes respectively. A starting model for the binary complex was obtained by angular reconstitution in IMAGIC32, and our previously determined GroEL-ADP-gp31 structure20 was used as a starting model for the ternary complexes. The data sets were sorted into classes showing different substrate features by a combination of MSA and competitive projection matching10, and the atomic structures of the GroEL subunit domains, gp31 and gp24 subunits were docked into the final, asymmetric maps as separate rigid bodies using URO33.
Supplementary Material
Acknowledgements
We thank Richard Westlake, Dave Houldershaw and Luchun Wang for computing and EM support. This work was carried out at the School of Crystallography, Birkbeck College, and was supported by a Wellcome Trust programme grant, EU 3D EM Network of Excellence and 3D Repertoire grants.
Footnotes
Full methods are presented in the Supplementary Material.
References
- 1.Sigler PB, et al. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 1998;67:581. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 2.Rye HS, et al. GroEL-GroES cycling: ATP and nonnative polypeptide direct alternation of folding-active rings. Cell. 1999;97:325. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 3.Houry WA, et al. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 4.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 6.Braig K, et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 7.Fenton WA, Kashi Y, Furtak K, Horwich AL. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 1994;371:614. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 8.Lin Z, Schwartz FP, Eisenstein E. The hydrophobic nature of GroEL-substrate binding. J. Biol. Chem. 1995;270:1011. doi: 10.1074/jbc.270.3.1011. [DOI] [PubMed] [Google Scholar]
- 9.Farr GW, et al. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell. 2000;100:561. doi: 10.1016/s0092-8674(00)80692-3. [DOI] [PubMed] [Google Scholar]
- 10.Elad N, et al. Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell. 2007;26:415. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 12.Lin Z, Rye HS. Expansion and compression of a protein folding intermediate by GroEL. Mol. Cell. 2004;16:23. doi: 10.1016/j.molcel.2004.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motojima F, et al. Substrate polypeptide presents a load on the apical domains of the chaperonin GroEL. Proc. Natl. Acad. Sci. USA. 2004;101:15005. doi: 10.1073/pnas.0406132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Madan D, Rye HS. GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol. 2008;15:303. doi: 10.1038/nsmb.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayhew M, et al. Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature. 1996;379:420. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK, Beguin F, Gujer-Kellenberger G. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J Mol Biol. 1970;47:69. doi: 10.1016/0022-2836(70)90402-x. [DOI] [PubMed] [Google Scholar]
- 17.van der Vies SM, Gatenby AA, Georgopoulos C. Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature. 1994;368:654. doi: 10.1038/368654a0. [DOI] [PubMed] [Google Scholar]
- 18.Hunt JF, van der Vies SM, Henry L, Deisenhofer J. Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage. Cell. 1997;90:361. doi: 10.1016/s0092-8674(00)80343-8. [DOI] [PubMed] [Google Scholar]
- 19.Bakkes PJ, Faber BW, van Heerikhuizen H, van der Vies SM. The T4-encoded cochaperonin, gp31, has unique properties that explain its requirement for the folding of the T4 major capsid protein. Proc. Natl. Acad. Sci. USA. 2005;102:8144. doi: 10.1073/pnas.0500048102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clare DK, et al. An Expanded Protein Folding Cage in the GroEL-gp31 Complex. J. Mol. Biol. 2006;358:905. doi: 10.1016/j.jmb.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 21.Elad N, Clare DK, Saibil HR, Orlova EV. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J Struct Biol. 2008;162:108. doi: 10.1016/j.jsb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 22.van Duijn E, et al. Monitoring macromolecular complexes involved in the chaperonin-assisted protein folding cycle by mass spectrometry. Nat. Methods. 2005;2:371. doi: 10.1038/nmeth753. [DOI] [PubMed] [Google Scholar]
- 23.Tang YC, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Machida K, et al. Hydrophilic Residues 526KNDAAD531 in the Flexible C-terminal Region of the Chaperonin GroEL Are Critical for Substrate Protein Folding within the Central Cavity. J Biol Chem. 2008;283:6886. doi: 10.1074/jbc.M708002200. [DOI] [PubMed] [Google Scholar]
- 25.Ranson NA, et al. Allosteric signaling of ATP hydrolysis in GroEL-GroES complexes. Nat. Struct. Biol. 2006;13:147. doi: 10.1038/nsmb1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fokine A, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Duijn E, et al. Tandem mass spectrometry of intact GroEL-substrate complexes reveals substrate-specific conformational changes in the trans ring. J. Am. Chem. Soc. 2006;128:4694. doi: 10.1021/ja056756l. [DOI] [PubMed] [Google Scholar]
- 28.van Duijn E, Heck AJ, van der Vies SM. Inter-ring communication allows the GroEL chaperonin complex to distinguish between different substrates. Protein Sci. 2007;16:956. doi: 10.1110/ps.062713607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder L, Tarkowski HJ. The N terminus of the head protein of T4 bacteriophage directs proteins to the GroEL chaperonin. J Mol Biol. 2005;345:375. doi: 10.1016/j.jmb.2004.10.052. [DOI] [PubMed] [Google Scholar]
- 30.Falke S, et al. The 13 angstroms structure of a chaperonin GroEL-protein substrate complex by cryo-electron microscopy. J. Mol. Biol. 2005;348:219. doi: 10.1016/j.jmb.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 31.Chaudhry C, et al. Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics. EMBO J. 2003;22:4877. doi: 10.1093/emboj/cdg477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Heel M, et al. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 33.Navaza J, et al. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1820. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.