Abstract
In this study, we identified an antisense transcript to ZIM2 (zinc finger imprinted gene 2) in the human, called ZIM2as. Sequence analysis of the 110 kb region spanned by this transcript revealed a cluster of tandemly repeated sequence in the human, orangutan, and chimpanzee as well as a loss of approximately 70 kb from the corresponding region in the rhesus. The homologous region in most mammals contains a cluster of olfactory receptor (OLFR) genes, but this gene cluster has been lost from the primate lineage. Expression analyses confirmed that ZIM2as is expressed in the human brain and testis. Two CpG islands near the promoter region of ZIM2as showed different methylation patterns in these three species. The CpG island distal to ZIM2as showed an allele-specific DNA methylation pattern in the human testis, while the CpG island proximal to the ZIM2as promoter showed a mosaic methylation pattern in the chimpanzee. The methylation status of several nearby zinc finger genes was unchanged among the primates tested. Overall, this study reports the presence of a previously unreported primate-specific antisense transcript in the PEG3 imprinted domain, suggesting that the formation of this transcript may coincide with the loss of the OLFR cluster.
Keywords: CpG island, imprinting, differentially methylated region, PEG3
1. Introduction
Most autosomal genes are expressed equally from two parental alleles, but up to 200 mammalian genes are only transcribed from one allele based on the parent of origin due to a process called genomic imprinting (Ideraabdullah et al., 2008). Genomic imprinting is found only in marsupials and placental mammals, organisms that utilize a unique reproductive strategy in which young offspring develop inside females’ wombs. Genomic imprinting is a critical gene dosage control mechanism for a subset of genes involved in this strategy (John and Surani, 2000). Most imprinted genes are involved in controlling fetal growth rates and nurturing behaviors (Tilghman, 1999). Proper dosage of imprinted genes is critical for the survival of mammals, and abnormalities in the dosages quite often manifest as genetic diseases in humans. Imprinting-related diseases include Beckwith-Wiedemann, Prader/Willi, Angelman, and Silver-Russell syndromes as well as autistic spectrum disorders (Ferguson-Smith et al., 2004; Ideraabdullah et al., 2008).
Although imprinted genes are found only in mammals, their imprinting status of these genes is not always conserved among all mammals. For example, Igf2r (Insulin-like growth factor type-2 receptor) is imprinted in the mouse, but is biallelically expressed in the human (Kalscheur et al., 1993). This domain is an example of one common feature of imprinted domains: the expression of antisense transcripts that regulate imprinting (Pauler et al., 2007). Another common feature of imprinted domains is the presence of CpG islands with allele-specific methylation patterns (differentially methylated regions: DMR) (Ferguson-Smith et al., 2004). Some of these DMRs inherit their methylation as a gametic signal from the previous generations, and these DMRs play critical roles for maintaining the imprinting and transcription of a given domain (Edwards and Ferguson-Smith, 2007). Abnormal methylation levels of these DMRs, either hyper or hypomethylation, are also often associated with many types of human diseases as ‘epimutations’ (Hatchwell and Greally, 2007).
An evolutionarily conserved imprinted region, the PEG3 (Paternally expressed gene 3) domain, is found on human chromosome 19q13.4/chimpanzee chromosome 19/rhesus macaque chromosome 19/proximal mouse chromosome 7. In the mouse, this region contains several imprinted genes including Peg3 and Zim2 (Zinc finger imprinted gene 2). The overall structure of the PEG3 imprinted domain is generally well conserved among mammals, but there have been several lineage specific changes in this region. For example, a cluster of olfactory receptor genes that is present in most mammals has been lost from the primate lineage (Huang and Kim, 2009). Since this deletion of olfactory receptor genes is adjacent to the PEG3 imprinted domain, we analyzed the effects of this change in terms of sequence structure of the region and methylation and expression levels of nearby genes. This study revealed the presence of an antisense transcript to ZIM2, called ZIM2as, in this region of the human, orangutan and chimpanzee genomes.
2. Methods
2.1 Sequence analysis
The sequences covering the 250 kb upstream of PEG3 were obtained from the UCSC genome browser for four species: human (hg18-chr19: 61,742,129-62,050,982), chimpanzee (panTro2-chr19: 62,386,300-62,704,467), orangutan (ponAbe2-chr19:58,643,223-58,893,596), and rhesus macaque (rheMac2-chr19: 62,502,756-62,749,741). Pairwise comparisons between human and chimpanzee, human and orangutan, and human and macaque were made using PipMaker (Schwartz et al., 2000). The genomic DNA was analyzed with a Perl script to predict the location of CpG islands. CpG islands were defined as a region of sequence at least 500 bp long with greater than 55% C+G content and an observed/expected CpG dinucleotide ratio of at least 0.65 (Takai and Jones, 2002).
2.2 COBRA (COmbined Bisulfite Restriction Analysis) and bisulfite sequencing
Human genomic DNA derived from normal tissues were obtained from a commercial firm (Biochain). Chimpanzee (Pan troglodytes) and rhesus macaque (Macaca mulatta) fibroblast genomic DNA were the generous gift of Dr. Mark Batzer. Each DNA (2 μg) was modified with the bisulfite conversion reaction according to the manufacturer protocol (EZ DNA methylation kit, Zymo Research). The converted DNA was eluted with 15 μl of TE. Each converted DNA (1 μl) was used as a template for PCR with primers designed using the MethPrimer. The PCR amplification was performed using the Maxime PCR premix kit (Intron Biotech). Information regarding the primer sequences and detailed PCR conditions for each tested region are provided as Supplementary Material 1.
The amplified PCR products were analyzed using restriction enzyme digestion (COBRA) (Xiong and Laird, 1997). Each PCR product was analyzed with two sets of restriction enzymes. First, the efficiency of the bisulfite conversion reaction was monitored with a set of enzymes that contain non-CpG cytosines in their recognition sites (DdeI, HpaII). Any digestion by these enzymes indicates that the conversion reaction was incomplete. A second set of enzymes that distinguish between unmethylated and methylated DNA were chosen to analyze the degree of methylation in each tested region. The recognition site of each of these enzymes contains a CpG site (TaqI, BstUI, and HpyCH4IV). Since methylation inhibits the conversion of cytosines into thymidines during the bisulfite conversion, digestion by these enzymes indicates methylation on a given CpG site in vivo. Each of these restriction digestion reactions was repeated at least three times.
Selected PCR products amplified from the bisulfite-treated DNA were further analyzed through cloning and sequencing. Each of the selected PCR products was purified using the MEGA-Spin agarose gel purification kit (Intron), and then individually cloned into the pGEM-tEasy vector (Promega). At least 10 different clones were randomly selected for DNA sequencing for each PCR product. Unincorporated primers and dye terminators were removed via ethanol precipitation, and an ABI 3130 XL was used to analyze the results. To determine methylation status, the resulting electropherograms were visually inspected in BioEdit (Hall, 1999).
2.3 RT PCR
Normalized cDNA from the human brain, heart, kidney, liver, and placenta was obtained from Biochain. RT-PCR was performed using the Maxime PCR premix kit (Intron Biotech) using an annealing temperature of 56 °C and 38 cycles.
2.4 cDNA Cloning
Human brain and testis RNA were obtained from a commercial firm (Biochain). Brain and testis RNA (5 μg) were converted to cDNA using the Superscript III First-Strand Synthesis System (Invitrogen). PCR products amplified using the Zim2as-a and Zim2as-b2 primers were cloned into the pGEM-T Easy vector (Promega). Seven colonies were picked, plasmid DNA was isolated using the DNA-spin kit (Intron Biotech), and sequenced using BigDye v3.1 (Applied Biosystems). Unincorporated primers and dye terminators were removed via ethanol precipitation, and the ABI 3130 XL was used to analyze the results.
3. Results
3.1 Olfactory receptor deletion in primates
The region between Zfp28 (Zinc finger protein 28) and Zim2 contains a cluster of olfactory receptor genes in the cow, dog, mouse, and several other mammals (Fig. 1). However, this location is devoid of olfactory receptors in the rhesus macaque, orangutan, chimpanzee, and human. This deletion was accompanied by a loss of approximately 90 kb between the mouse and rhesus macaque regions. Comparison of the rhesus, orangutan, chimpanzee, and human sequences revealed the presence of a cluster of tandem repeats in this region in the human, orangutan, and chimpanzee genomes only, which are responsible for an approximately 70 kb size difference between this region in the rhesus macaque and the same region in humans, orangutans and chimpanzees (Fig. 2). Analysis of ESTs suggested the possibility of a transcript spanning this region (AI829612.1, DA176232.1, BG707577, AL554662, BX365418.2, BM710959, AW850989), and cDNA cloning and sequencing confirmed the identity of the ZIM2as transcript in the human (Genbank accession No. FJ997633) (Fig. 2B). The exons of ZIM2as are also detected in other primates, chimpanzee and orangutan, with 99% and 97% sequence identity, respectively. Furthermore, one orangutan EST (CR629606) was found to be derived from the 5′-end portion of ZIM2as. These results strongly suggest that ZIM2as is evolutionarily conserved in the great apes.
Figure 1. Organization of the region between ZFP28 and PEG3 in the mouse, rhesus macaque, chimpanzee, and human genomes.
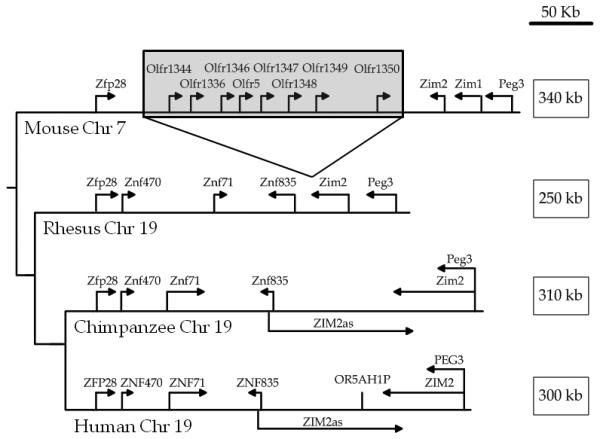
Since the size of this region is different in each organism, this figure is drawn to the scale indicated in the upper right corner. The location for the sequences shown in this figure are human (hg18)-chr19:61,742,129-62,050,982; chimpanzee (panTro2)-chr19:62,386,300-62,704,467; and rhesus (rheMac2)-chr19:62,502,756-62,749,741. Arrows show the direction of transcription. The mouse region is shown as a representative of the non-primate mammals that possess a cluster of olfactory receptor genes in this region. The tree is only representative of the relative relationship between these four animals, and branch lengths are not to scale.
Figure 2. Structure of the ZIM2as transcript and the surrounding sequence.
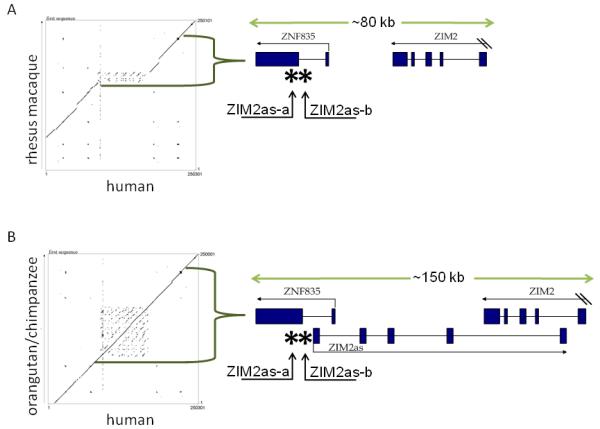
Dot plot comparison of the sequence structure of the region between ZNF71 and PEG3 in the rhesus macaque and chimpanzee against human. The locations of the sequences used for this analysis are human (hg18)-chr19:61,797,100-62,047,400; chimpanzee (panTro2)-chr19:62,451,000-62,701,000; orangutan (ponAbe2)-chr19:58,643,223-58,893,596; and rhesus macaque (rheMac2)-chr19:62,498,300-62,748,400. Each identity between the two sequences is plotted as a dot, so the diagonal line seen in each graph shows a region of sequence similarity. The human sequence is on the x axis of each graph. In the diagrams on the right, two CpG islands in the promoter region of ZIM2as are indicated by asterisks (*) and labeled with the names of the primers used to analyze methylation in this region.
A) Rhesus vs. human. The corresponding rhesus macaque sequence is on the y axis of the dot plot, and the diagram on the right shows the structure of the genes in the indicated region of the rhesus genome. Exons are indicated by boxes, introns by lines, and arrows indicate the direction of transcription.
B) Orangutan/chimpanzee vs. human. Similar results were seen using chimpanzee and orangutan sequence, and the chimpanzee sequence is shown on the y axis of the dot plot. The diagram on the right shows the structure of the genes in the studied region in the human, orangutan, and chimpanzee genomes, including ZIM2as. The ZIM2as transcript is 568 bp long, has five exons, and spans over 118 kb of genomic region. The first exon is found within the first intron of ZNF835 and the final exon is found within an intron of ZIM2. Exons are indicated by boxes, introns by lines, and arrows indicate the direction of transcription.
3.2 ZIM2as transcript formation
The ZIM2as transcript is composed of five exons and is 568 bp long, but lacks an ORF (Supplemental Table 1). It spans over 100 kb between ZNF835 (Zinc finger protein 835) and ZIM2, beginning within the first intron of ZNF835 and ending in the fourth from last intron of ZIM2 (Fig. 2). The 5′-end of ZIM2as is located in a nuclease accessible site (Boyle et al., 2008) that shows enrichment in histone 3 lysine 4 dimethylation, a mark associated with transcriptional start sites (Mikkelsen et al., 2007), suggesting that the transcription of ZIM2as likely starts at this site. We performed RT-PCR-based expression analyses using primers specific to the ZNF835, ZIM2as, ZIM2, and PEG3 transcripts (Fig. 3). Expression of each of these transcripts was assayed using a commercially available panel containing normalized cDNA from human brain, heart, kidney, liver, and placenta as well as human testis. Out of all the tissues studied, ZIM2as expression was detected in the brain and testis (Fig. 3). Expression of the ZNF835 transcript was detected in all tested tissues, most strongly in the heart. Both the ZIM2 and PEG3 transcripts were expressed in all tissues tested with relatively high levels in brain and placenta.
Figure 3. Expression Analysis of genes in the vicinity of ZIM2as.
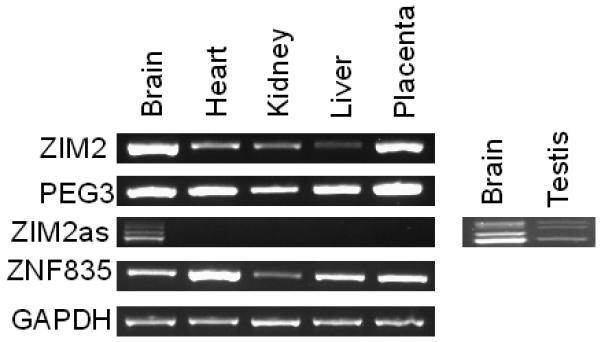
Expression patterns of ZIM2as and several nearby transcripts, ZNF835, ZIM2, and PEG3 were analyzed by RT-PCR. GAPDH was used as an internal control. The tissue origin of each cDNA is indicated on the top, and the transcript analyzed is indicated on the left. The two additional lanes shown for the ZIM2as transcript represent results from the cDNA used to clone and sequence this transcript. These two cDNA samples are not normalized with respect to the lanes containing results from the commercial cDNA.
3.3 DNA Methylation analysis
Since long antisense transcripts are often associated with the maintenance of imprinted domains, we tested the methylation status of several genes near ZIM2as. Briefly, genomic DNA was converted using the bisulfite method, and analyzed via restriction enzyme digestion (COBRA, Xiong and Laird, 1997) and/or cloning and sequencing. We analyzed the methylation levels of the DNA that were derived from human brain and testis, rhesus and chimpanzee fibroblast cell lines. We first analyzed the two CpG islands near the promoter region of ZIM2as. The one farther from ZIM2as (named ZIM2as-a) was totally digested by HpyCH4 IV in chimpanzee, rhesus macaque, and human brain DNA, indicating complete methylation (Fig. 4A). However, only 50% of the PCR product was digested in the human testis. Bisulfite sequencing and SNP analysis of this region showed allele-specific methylation in the testis (Fig. 4B, T vs. G). However, we are unable to determine the parent-of-origin of the methylated allele. The CpG island closer to ZIM2as (named ZIM2as-b) showed species-specific methylation patterns (Fig. 4A): the human samples showed no digestion (indicating a lack of methylation), the rhesus macaque sample showed complete digestion, but the chimpanzee sample showed approximately 50% digestion after incubation with HpyCH4 IV. Subsequent bisulfite sequencing of the same chimpanzee sample revealed a mosaic pattern of methylation, with no clone being entirely methylated (Fig. 4B). In sum, the two CpG islands of ZIM2as displayed species-specific variations among primates, but one of these CpG islands (ZIM2as-a) showed an allele-specific methylation pattern in human testis DNA.
Figure 4. DNA Methylation analysis of ZIM2as.
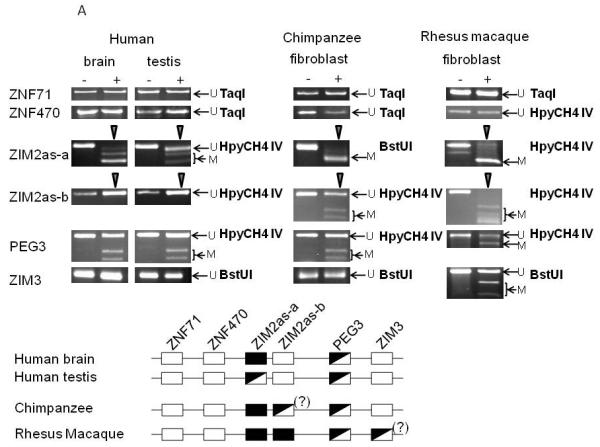
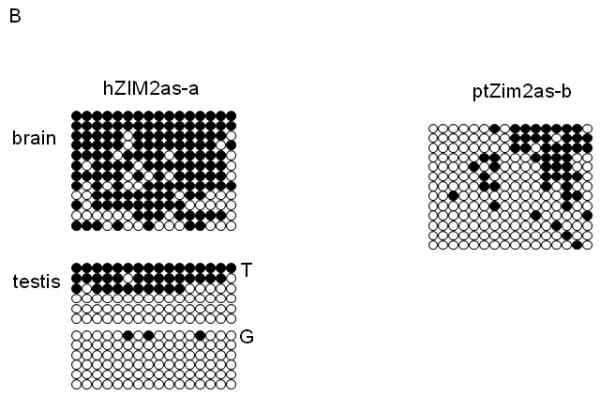
The methylation status of CpG islands associated with several genes upstream of the ZIM2as transcript was assayed by bisulfite conversion of each DNA sample followed by COBRA and/or cloning and sequencing. Each analyzed CpG island is indicated by the name of the associated gene.
A) COBRA. The type of DNA sample is indicated above each column, along with (-) for DNA that was incubated without enzyme, and (+) for DNA that was incubated with the appropriate enzyme. The enzyme used for each COBRA is indicated to the right of each gel image. Arrows labeled U and M indicate the position of the unmethylated and methylated DNA bands, respectively. Each reaction was performed at least three times, and a representative image is shown. The bottom portion of the figure contains a summary of the COBRA results in which open boxes indicate a lack of methylation, filled boxes indicate complete methylation, and half-filled boxed indicate a DMR region. The regions that show different methylation patterns among the human, chimpanzee, and rhesus macaque are indicated by diamonds above each lane.
B) Bisulfite sequencing. The associated gene is shown above and the type of DNA sample is shown on the left. The T and G on the right of the human testis bisulfite sequencing result indicate the SNP used to separate the two alleles. Each row of the bisulfite sequencing results indicates a different clone, and each column is an individual cytosine within a CpG dinucleotide. Filled circles indicate methylation, and empty circles denote a lack of methylation.
To investigate the possibility that the ZIM2as transcript formation coincided with the expansion of this imprinted region, we also tested methylation status at several upstream genes, including ZNF71 (zinc finger gene 71) and ZNF470 (zinc finger gene 470). The CpG islands associated with each of these genes were unmethylated in the human brain and testis as well as in the chimpanzee and rhesus macaque fibroblast, based on lack of digestion when incubated with the appropriate restriction enzyme (Fig. 4A). This indicates an overall similar pattern between different primates. We also analyzed two CpG islands on the downstream side of ZIM2as. The PEG3-CpG island showed a typical DMR pattern in all three species. The ZIM3 (Zinc finger gene imprinted 3)-CpG island showed a similar pattern of non-digestion (unmethylation) between human and chimpanzee. However, some fraction of the ZIM3-CpG island from rhesus was digested, indicating some levels of DNA methylation, which warrants further investigation in the near future.
In summary, the above data show that an antisense transcript has formed in the region upstream of the PEG3 imprinted domain in the human and chimpanzee. The two CpG islands close to the promoter of this primate-specific transcript gene showed specific-specific variations in terms of their DNA methylation status. This is in stark contrast to the stable and conserved DNA methylation pattern observed from the nearby zinc finger genes. One of these CpG islands, Zim2as-a, showed allele-specific methylation in the human testis, suggesting that ZIM2as is probably imprinted in the human (Fig. 4B).
4. Discussion
In this study, we report the formation of a new primate-specific antisense transcript, ZIM2as, in a region containing a cluster of olfactory receptor genes in non-primate mammals (Fig. 2). In the human, this transcript is expressed in the brain and testis (Fig. 3), and a CpG island within its promoter region shows allele-specific methylation in the testis, suggesting that this transcript may be imprinted (Fig. 4). The presence and possible imprinting of this antisense transcript gene suggest that the PEG3 imprinted domain may have expanded in the great apes.
The overall structure of the PEG3 imprinted domain is generally well conserved among mammals, but there have been several lineage specific changes in this region. Figure 5 shows these changes in the structure of the area covered by ZIM2as and highlights the probable timing of two major events. First, a cluster of olfactory receptor genes that is present in most mammals has been lost from the primate lineage (Fig. 1). Second, an antisense transcript to ZIM2, called ZIM2as, is found in the homologous regions of the human, chimpanzee and orangutan genomes. These two changes might be related to each other. Clusters of olfactory genes have been shown to be involved as insulators between differently regulated chromatin domains (Valenzuela and Kamakaka, 2006). This cluster may have originally acted as a boundary to prevent spreading of imprinting outside of the PEG3 domain, and its loss might have triggered the formation of an antisense transcript gene, ZIM2as, in the primates. So far, all the primate genomes analyzed lack this olfactory cluster. On the other hand, sequence analysis and EST searches confirmed the presence of ZIM2as only in the great apes, but not in the rhesus genome. This suggests that the loss of the OLFR cluster may predate the formation of ZIM2as, and further predicts that the formation of this antisense gene was around 15 million years ago. This antisense transcript gene is most likely imprinted in the humans based on the allele-specific DNA methylation observed from the human testis sample (Fig. 4B). We also predict that this antisense transcript gene is imprinted in other primates based on the patterns observed from similar antisense transcripts in other imprinted domains (Ideraabdullah et al., 2008). In particular, many antisense transcripts associated with imprinted domains also begin in intronic regions, e.g Kcnq1ot1 and 91H (Ideraabdullah et al., 2008; Bertaux et al., 2008). Following this pattern, the beginning of ZIM2as is located within the first intron of ZNF835. If it is the case that ZIM2as is imprinted, it will be interesting to test if the PEG3 imprinted domain has indeed expanded in the primate lineage. In particular, it would be of great interest to investigate the potential imprinting of several zinc finger genes located in the region immediately adjacent to ZIM2as, in particular ZNF835.
Figure 5. Primate-specific changes in the PEG3 region.
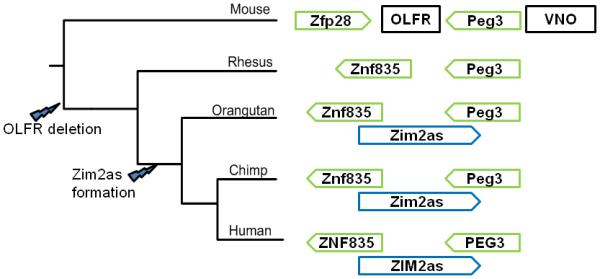
The tree is only representative of the relative relationship between these four animals, and branch lengths are not to scale. For simplicity, only selected genes are shown in this figure. The directional arrows show the direction of transcription of selected genes in this region, and the boxes show the positions of the olfactory receptor (OLFR) and vomeronasal organ receptor (VNO) gene clusters that flank the Peg3 imprinted domain in the mouse. The approximate timings of the olfactory receptor cluster deletion and Zim2as transcript formation are shown on the tree.
It is interesting to note that DNA methylation patterns are not conserved between great apes. The methylation status of two CpG islands adjacent to this end of the transcript differs between the chimpanzee and human. The chimpanzee lacks methylation at the first CpG island (Zim2as-a) and has a mosaic methylation pattern at the second (Zim2as-b), while in the human ZIM2as-a is a tissue specific DMR and ZIM2as-b is completely methylated (Fig. 3B). This result is consistent with previous reports that human and chimpanzee methylation often differs (Enard et al., 2004). Several possibilities could account for this result. First, the chimpanzee DNA was derived from a cell line, and cell lines have been reported to show altered methylation patterns (Chang et al., 2009). However, four other CpG islands (ZNF470, ZNF471, PEG3, and ZIM3) showed the same methylation pattern between chimpanzee and human. Second, Zim2as-a could represent a testis-specific DMR. Without access to chimpanzee testis tissue, we are unable to determine if this is the case. Finally, the lineage-specific differences in methylation could be due to the recent appearance of this transcript. There has probably not been enough evolutionary time for its regulation to undergo selection pressure, so the imprinting status of this transcript might not have been fixed due to the very young age of this gene.
In conclusion, both the human and chimpanzee genomes have lost a cluster of olfactory receptors from the same region in which the ZIM2as transcript has formed, and the ZIM2as promoter region shows allele-specific methylation in the human testis. Although the loss of the olfactory cluster and the formation of ZIM2as were not simultaneous, the methylation data and the fact that this transcript is antisense to an imprinted gene suggest that ZIM2as may be imprinted. Since OLFR clusters often function as boundary elements, the loss of this OLFR cluster may have allowed the expansion of the PEG3 imprinted domain in the primates.
Supplementary Material
Acknowledgments
We would like to thank Dr. Jeong Do Kim, Keunsoo Kang, and Kim Ngoc Tran for their technical assistance, and Deepa Srikanta for critical reading of this manuscript. We also thank Dr. Mark Batzer and Jerilyn Walker for the generous gift of the chimpanzee and rhesus DNA samples. This work was supported by NIH (R01-GM66225).
Abbreviations
- CpG
cytosine followed immediately by guanine
- DMR
differentially methylated region
- COBRA
combined bisulfite restriction analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bertaux N, Aptel N, Cathala G, Genton C, Coll J, Daccache A, Spruyt N, Hondermarck H, Dugimont T, Curgy J-J, Forne T, Adrianaenssens E. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–6745. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G, Liu S, Wang F, Zhang Y, Kou Z, Chen D, Gao S. Differential methylation status of imprinted genes in nuclear transfer derived ES (NT-ES) cells. Genomics. 2009;93:112–119. doi: 10.1016/j.ygeno.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Enard W, Fassbender A, Model F, Adorjan P, Paabo S, Olek A. Differences in DNA methylation patterns between humans and chimpanzees. Curr Biol. 2004;14:R148–149. [PubMed] [Google Scholar]
- Ferguson-Smith AC, Lin SP, Youngson N. Regulation of gene activity and repression: a consideration of unifying themes. Curr Top Dev Biol. 2004;60:197–213. doi: 10.1016/S0070-2153(04)60006-8. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hatchwell E, Greally JM. The potential role of epigenomic dysregulation in complex human disease. Trends Genet. 2007;23:588–595. doi: 10.1016/j.tig.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Huang JM, Kim J. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009 doi: 10.1016/j.gene.2009.04.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John RM, Surani MA. Genomic imprinting, mammalian evolution, and the mystery of egg-laying mammals. Cell. 2000;101:585–588. doi: 10.1016/s0092-8674(00)80870-3. [DOI] [PubMed] [Google Scholar]
- Kalscheur VM, Mariman EC, Schepens MT, Rehder H, Ropers H-H. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat Genet. 1993;5:74–78. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Isaac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander E, Bernstein BE. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–92. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—A Web Server for Aligning Two Genomic DNA Sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


